Submitted:
01 August 2023
Posted:
04 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Protease homology search
2.2. Properties of protein sequence homologs
2.3. Protein structure modeling
| Algorithm | Application | Server address | Reference |
| AlphaFold 2 andColabFold | Three-dimensional protein structure modeling | https://github.com/sokrypton/ColabFold | [22,64] |
| HHpred | Protein sequence homology detection | https://toolkit.tuebingen.mpg.de/tools/hhpred | [59] |
| MAFFT | Multiple protein sequence alignment | https://toolkit.tuebingen.mpg.de/tools/mafft | [60] |
| NetSurfP-3.0 | Prediction of secondary protein structures and protein intrinsic disorder regions | https://services.healthtech.dtu.dk/ service.php?NetSurfP-3.0 | [65] |
| SignalP6 | Secretory signal peptide and cleavage site prediction | https://services.healthtech.dtu.dk/service.php?SignalP | [58] |
| T-REKS | Identification of tandem repeat patterns in protein sequences | https://bioinfo.crbm.cnrs.fr/index.php?route=tools&tool=3 | [61] |
| XSTREAM | Identification of tandem repeat patterns in protein sequences | https://amnewmanlab.stanford.edu/xstream/ | [62] |
3. Results
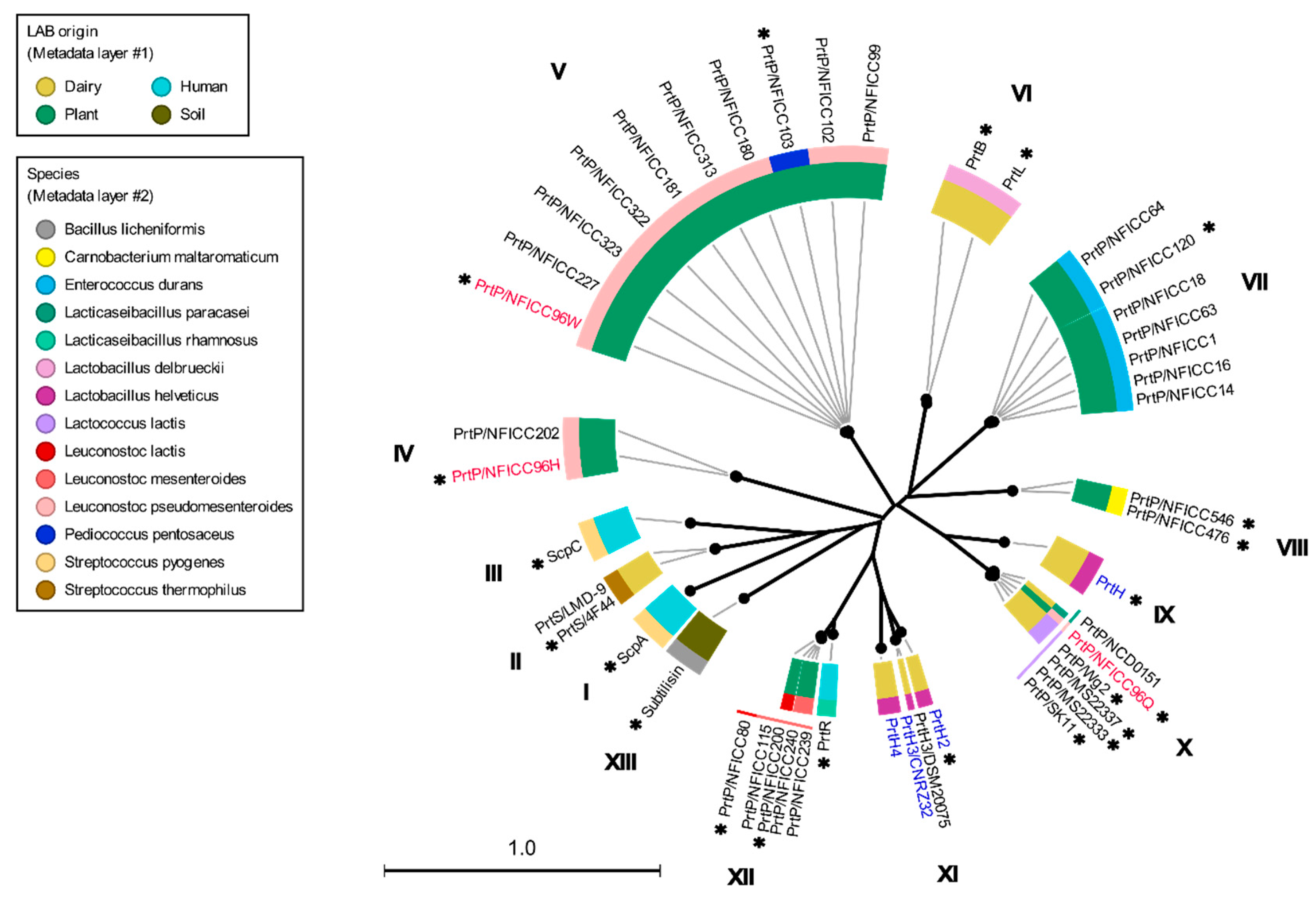
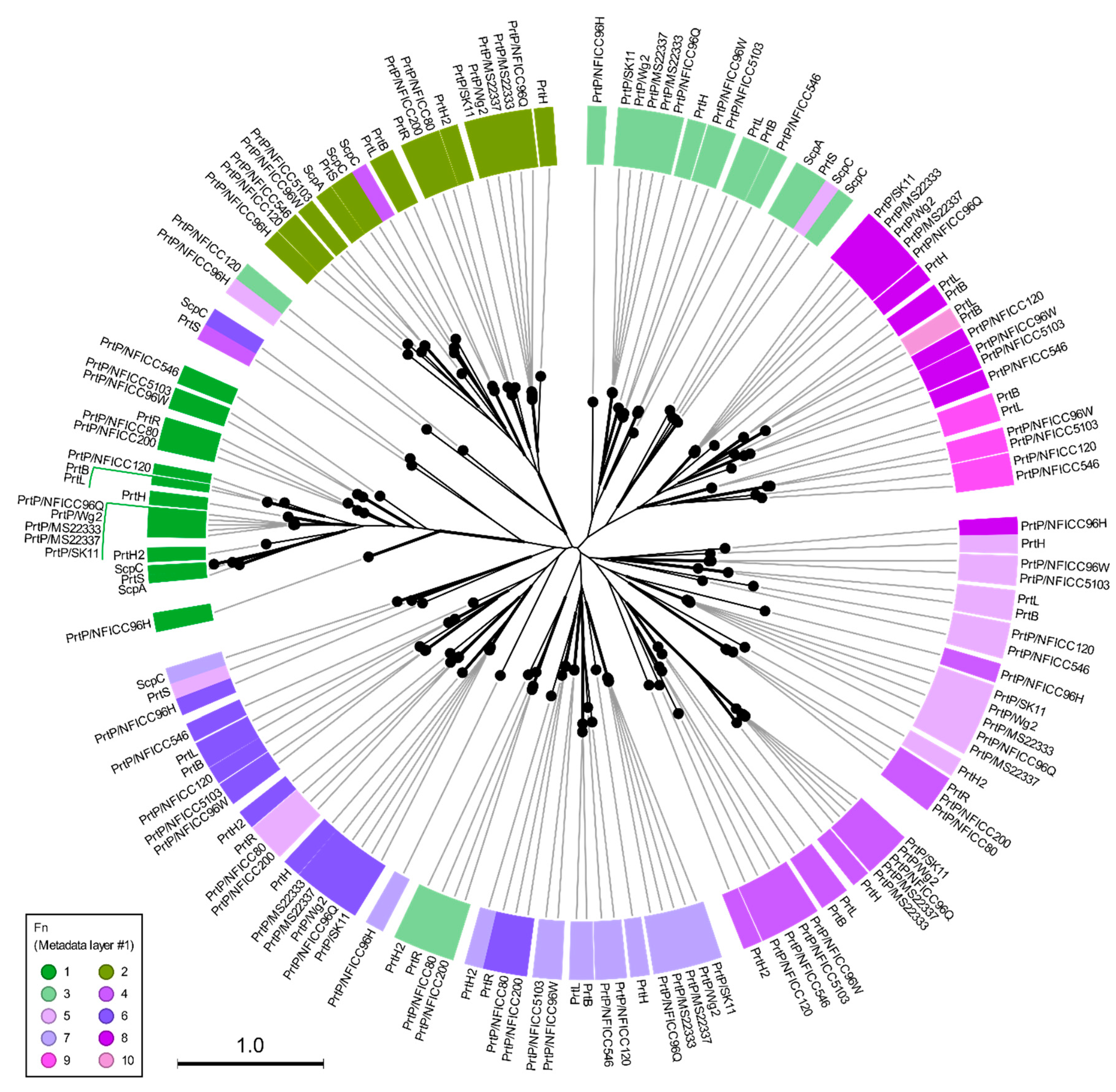
3.1. Phylogenetic clustering of diverse PrtP homologs
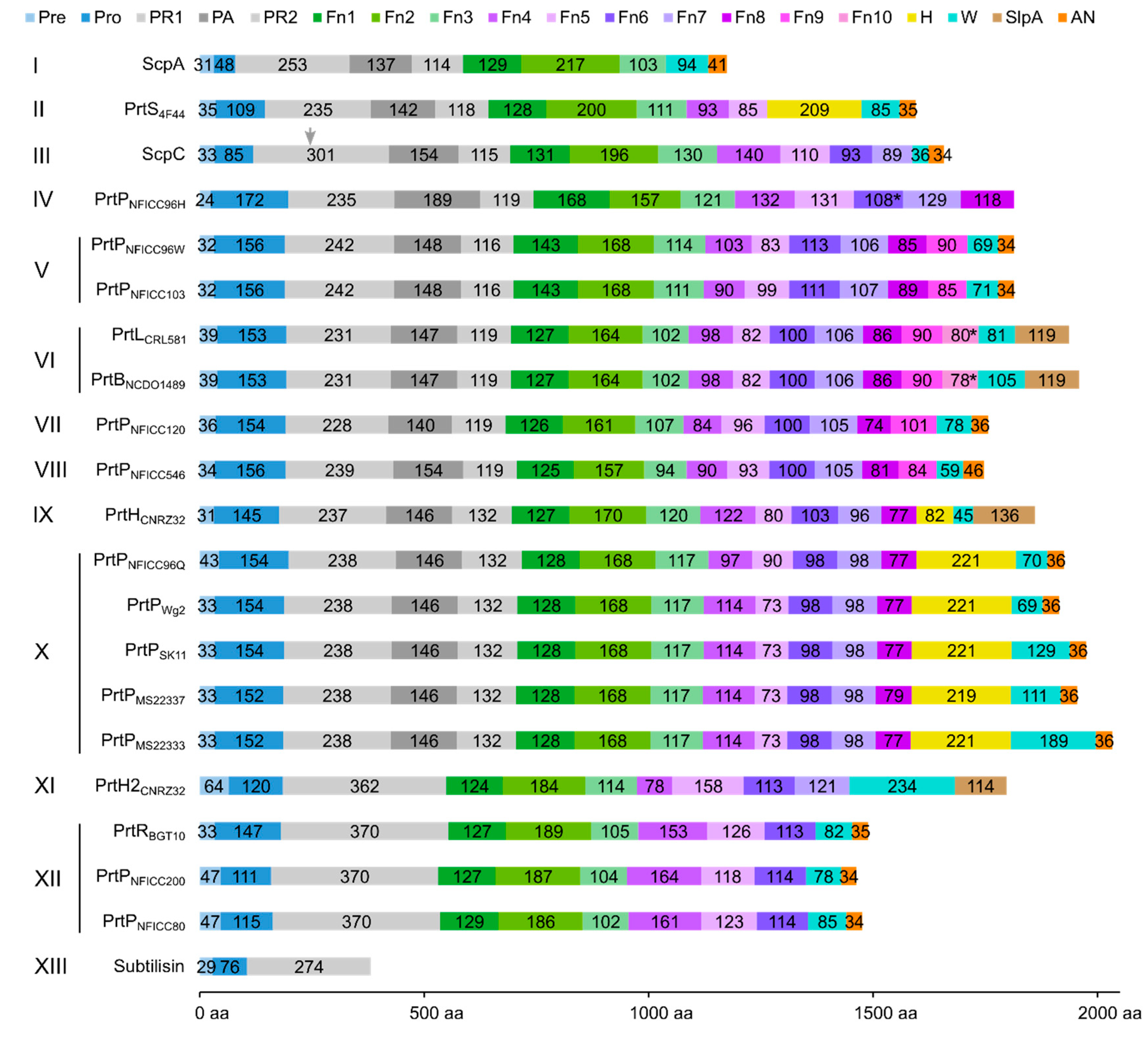
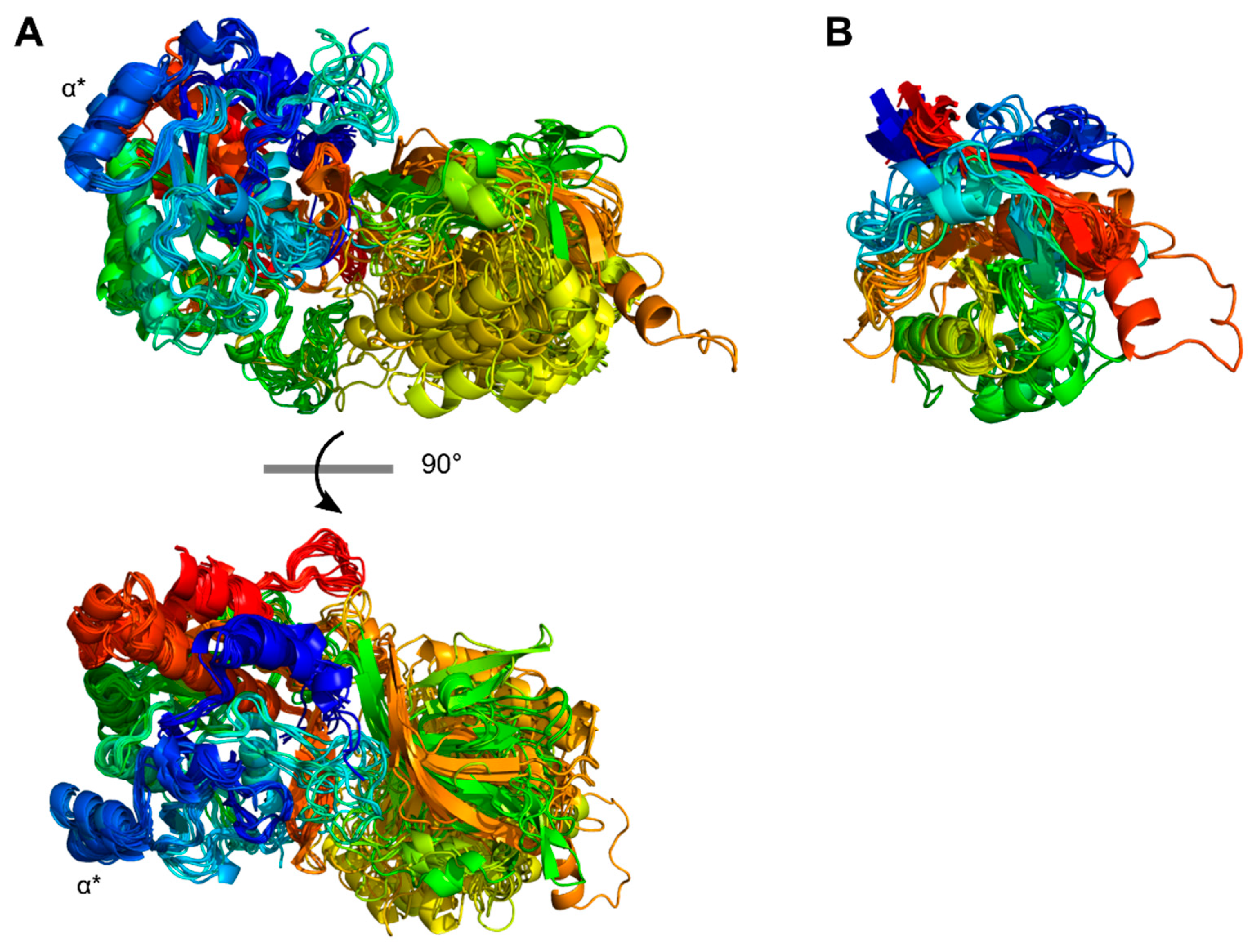
3.2. Multi-domain structures of PrtP homologous proteins
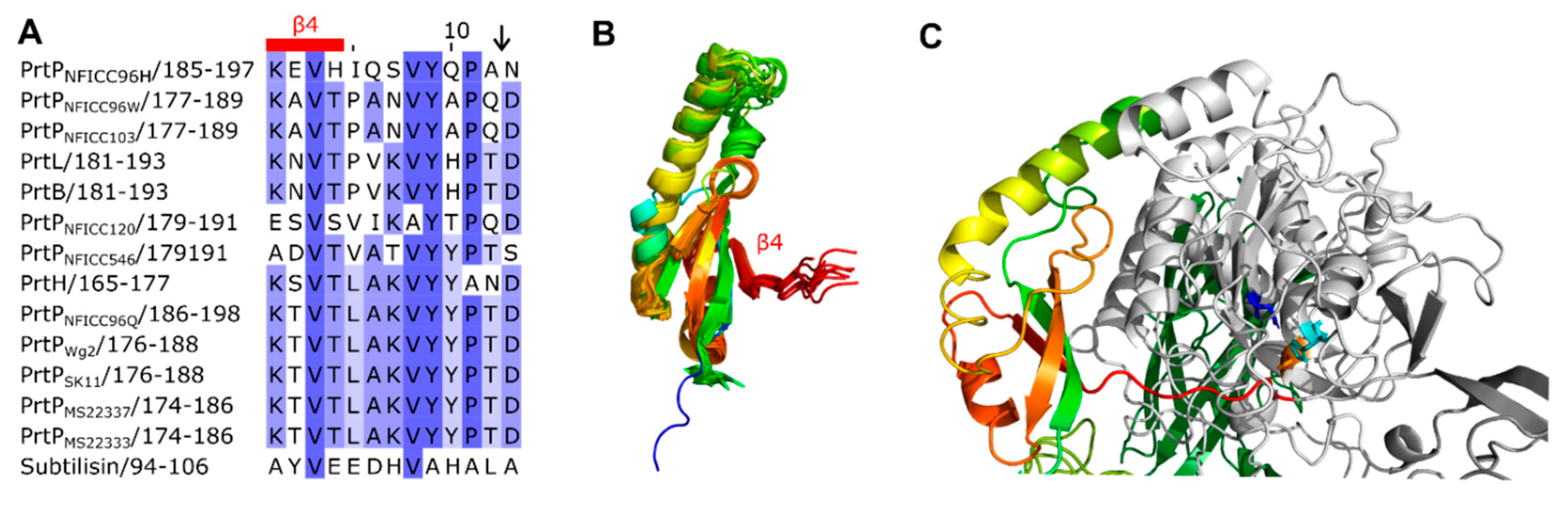
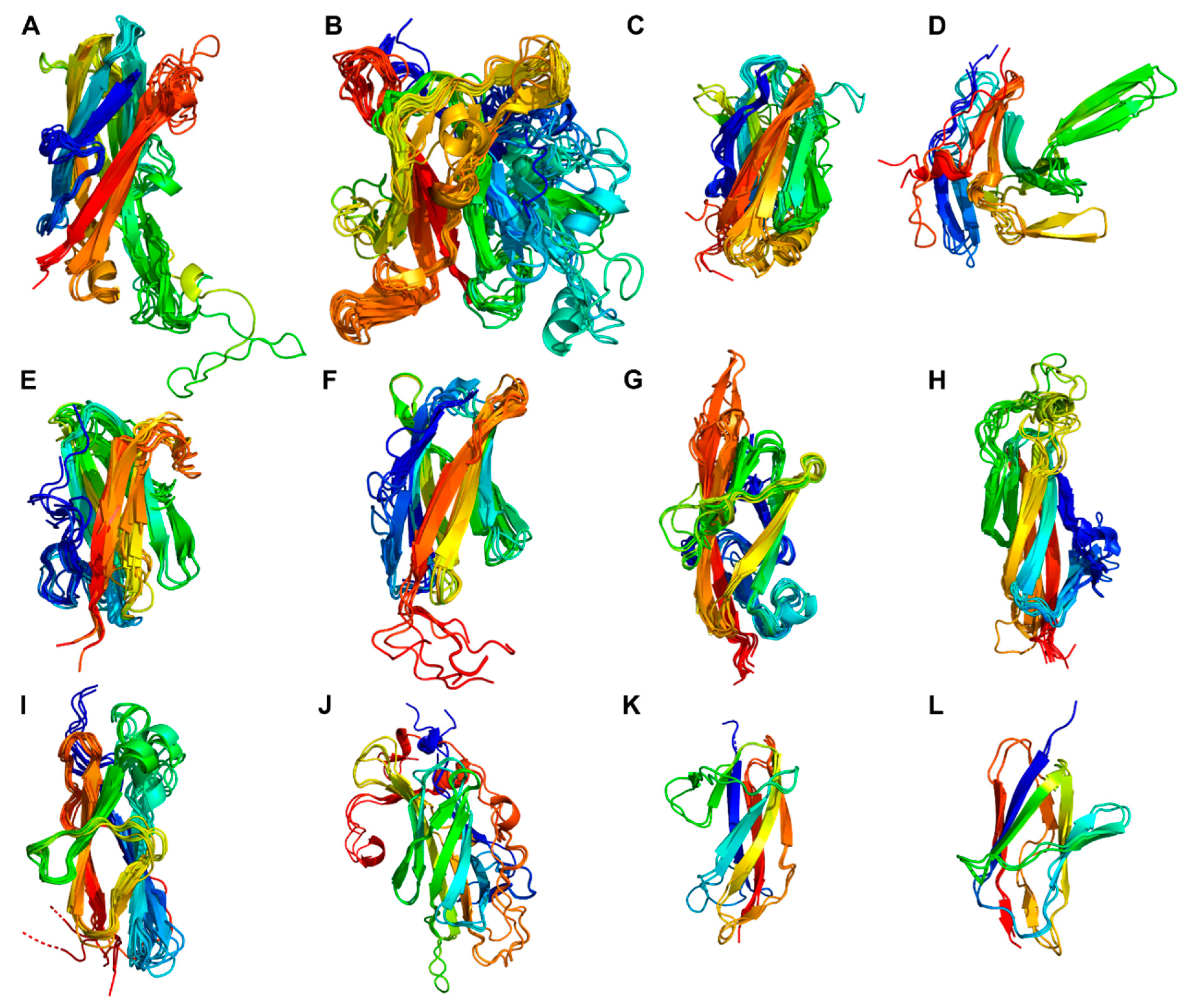
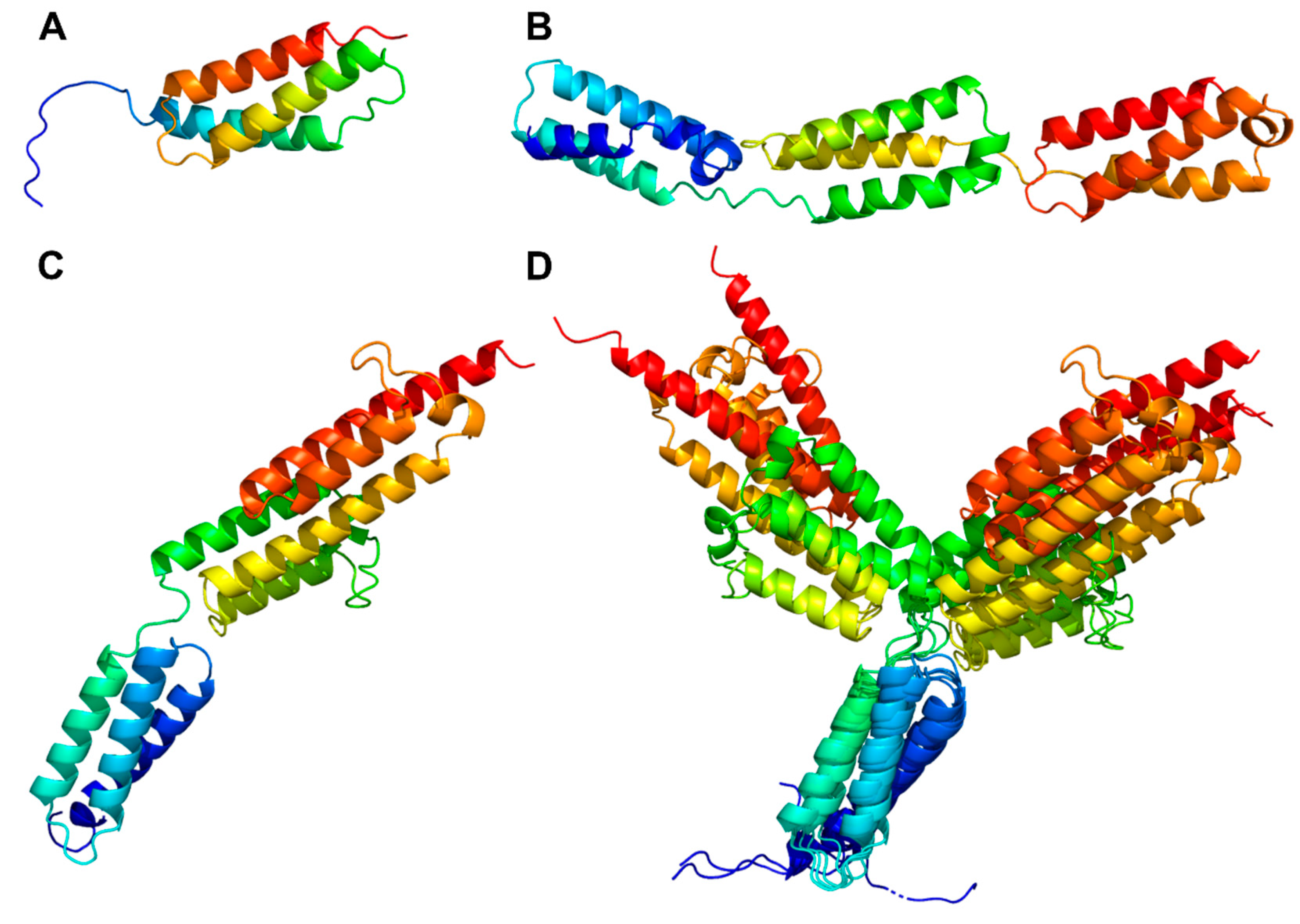
3.3. Structural characteristics of the distinctive domains
3.3.1. Pre-propeptide domain
3.3.2. Catalytic region
3.3.3. Tail of fibronectin-like domains
3.3.4. Helix domain
3.3.5. Cell wall spacing domain
3.3.6. Cell wall attachment domains
4. Discussion
4.1. Conserved domains in novel domain architectures
4.2. Conformational maturation and stability
4.3. Substrate selectivity and specificity
4.4. Adhesion potential
4.5. Distribution and environmental constraints
5. Conclusion
Supplementary Materials
Funding
References
- Douillard, F.P.; de Vos, W.M. Functional genomics of lactic acid bacteria: from food to health. Microb. Cell Factories 2014, 13, S8. [Google Scholar] [CrossRef]
- Ji, D.; Ma, J.; Xu, M.; Agyei, D. Cell-envelope proteinases from lactic acid bacteria: Biochemical features and biotechnological applications. Compr. Rev. Food Sci. Food Saf. 2020, 20, 369–400. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef] [PubMed]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie van Leeuwenhoek 1999, 76, 139–155. [Google Scholar] [CrossRef]
- Sun, Z.; Harris, H.M.B.; McCann, A.; Guo, C.; Argimón, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef]
- Boulay, M.; Metton, C.; Mézange, C.; Correia, L.O.; Meylheuc, T.; Monnet, V.; Gardan, R.; Juillard, V. Three Distinct Proteases Are Responsible for Overall Cell Surface Proteolysis in Streptococcus thermophilus. Appl. Environ. Microbiol. 2021, 87, e01292-21. [Google Scholar] [CrossRef]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: a genomic comparison. BMC Genom. 2010, 11, 36–36. [Google Scholar] [CrossRef]
- A Exterkate, F.; Alting, A.C.; Bruinenberg, P.G. Diversity of cell envelope proteinase specificity among strains of Lactococcus lactis and its relationship to charge characteristics of the substrate-binding region. Appl. Environ. Microbiol. 1993, 59, 3640–3647. [Google Scholar] [CrossRef]
- Børsting, M.; Qvist, K.; Brockmann, E.; Vindeløv, J.; Pedersen, T.; Vogensen, F.; Ardö, Y. Classification of Lactococcus lactis cell envelope proteinase based on gene sequencing, peptides formed after hydrolysis of milk, and computer modeling. J. Dairy Sci. 2015, 98, 68–77. [Google Scholar] [CrossRef]
- Holck, A.; NAeS, H. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO 151. J. Gen. Microbiol. 1992, 138, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Villegas, J.M.; Brown, L.; de Giori, G.S.; Hebert, E.M. Characterization of the mature cell surface proteinase of Lactobacillus delbrueckii subsp. lactis CRL 581. Appl. Microbiol. Biotechnol. 2014, 99, 4277–4286. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Atlan, D.; Blanc, B.; Portailer, R.; E Germond, J.; Lapierre, L.; Mollet, B. A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbruekii subsp. bulgaricus. J. Bacteriol. 1996, 178, 3059–3065. [Google Scholar] [CrossRef] [PubMed]
- Pastar, I.; Tonic, I.; Golic, N.; Kojic, M.; van Kranenburg, R.; Kleerebezem, M.; Topisirovic, L.; Jovanovic, G. Identification and Genetic Characterization of a Novel Proteinase, PrtR, from the Human Isolate Lactobacillus rhamnosus BGT10. Appl. Environ. Microbiol. 2003, 69, 5802–5811. [Google Scholar] [CrossRef]
- Awussi, A.A.; Roux, E.; Humeau, C.; Hafeez, Z.; Maigret, B.; Chang, O.K.; Lecomte, X.; Humbert, G.; Miclo, L.; Genay, M.; et al. Role of the Sortase A in the Release of Cell-Wall Proteinase PrtS in the Growth Medium of Streptococcus thermophilus 4F44. Microorganisms 2021, 9, 2380. [Google Scholar] [CrossRef]
- Kagawa, T.F.; O’Connell, M.R.; Mouat, P.; Paoli, M.; O’Toole, P.W.; Cooney, J.C. Model for Substrate Interactions in C5a Peptidase from Streptococcus pyogenes: A 1.9 Å Crystal Structure of the Active Form of ScpA. J. Mol. Biol. 2009, 386, 754–772. [Google Scholar] [CrossRef]
- Jobichen, C.; Tan, Y.C.; Prabhakar, M.T.; Nayak, D.; Biswas, D.; Pannu, N.S.; Hanski, E.; Sivaraman, J. Structure of ScpC, a virulence protease from Streptococcus pyogenes, reveals the functional domains and maturation mechanism. Biochem. J. 2018, 475, 2847–2860. [Google Scholar] [CrossRef]
- Teçza, M.; Kagawa, T.F.; Jain, M.; Cooney, J.C. Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease. Comput. Struct. Biotechnol. J. 2021, 19, 2356–2365. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- McKenna, S.; Malito, E.; Rouse, S.L.; Abate, F.; Bensi, G.; Chiarot, E.; Micoli, F.; Mancini, F.; Moriel, D.G.; Grandi, G.; et al. Structure, dynamics and immunogenicity of a catalytically inactive CXC chemokine-degrading protease SpyCEP from Streptococcus pyogenes. Comput. Struct. Biotechnol. J. 2020, 18, 650–660. [Google Scholar] [CrossRef]
- Hansen, E.B.; Marcatili, P. Modeled Structure of the Cell Envelope Proteinase of Lactococcus lactis. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.F.; García-Béjar, B.; Bang-Berthelsen, C.H.; Hansen, E.B. Extracellular microbial proteases with specificity for plant proteins in food fermentation. Int. J. Food Microbiol. 2022, 381, 109889. [Google Scholar] [CrossRef] [PubMed]
- Martoglio, B.; Dobberstein, B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998, 8, 410–415. [Google Scholar] [CrossRef]
- Desvaux, M.; Hébraud, M.; Talon, R.; Henderson, I.R. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009, 17, 139–145. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Anderson, E.T.; Wetherell, M.G.; Winter, L.A.; Olmsted, S.B.; Cleary, P.P.; Matsuka, Y.V. Processing, stability, and kinetic parameters of C5a peptidase fromStreptococcus pyogenes. JBIC J. Biol. Inorg. Chem. 2002, 269, 4839–4851. [Google Scholar] [CrossRef]
- Takahashi, S.; Leiss, M.; Moser, M.; Ohashi, T.; Kitao, T.; Heckmann, D.; Pfeifer, A.; Kessler, H.; Takagi, J.; Erickson, H.P.; et al. The RGD motif in fibronectin is essential for development but dispensable for fibril assembly. J. Cell Biol. 2007, 178, 167–178. [Google Scholar] [CrossRef]
- Gajic, O. Relationships between MDR Proteins, Bacteriocin Production and Proteolysis in Lactococcus Lactis. 2003. Available online: https://research.rug.nl/en/publications/relationships-between-mdr-proteins-bacteriocin-production-and-pro.
- Hartford, O.; Francois, P.; Vaudaux, P.; Foster, T.J. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 1997, 25, 1065–1076. [Google Scholar] [CrossRef]
- Michon, C.; Langella, P.; Eijsink, V.G.H.; Mathiesen, G.; Chatel, J.M. Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb. Cell Factories 2016, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Schneewind, O.; Model, P.; Fischetti, V.A. Sorting of protein a to the staphylococcal cell wall. Cell 1992, 70, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Boekhorst, J.; de Been, M.W.H.J.; Kleerebezem, M.; Siezen, R.J. Genome-Wide Detection and Analysis of Cell Wall-Bound Proteins with LPxTG-Like Sorting Motifs. J. Bacteriol. 2005, 187, 4928–4934. [Google Scholar] [CrossRef]
- Marraffini, L.A.; DeDent, A.C.; Schneewind, O. Sortases and the Art of Anchoring Proteins to the Envelopes of Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 192–221. [Google Scholar] [CrossRef] [PubMed]
- Kruger, R.G.; Otvos, B.; Frankel, B.A.; Bentley, M.; Dostal, P.; McCafferty, D.G. Analysis of the Substrate Specificity of the Staphylococcus aureus Sortase Transpeptidase SrtA. Biochemistry 2004, 43, 1541–1551. [Google Scholar] [CrossRef]
- Boot, H.J.; Kolen, C.P.; Pouwels, P.H. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J. Bacteriol. 1995, 177, 7222–7230. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Exterkate, F.A.; Alting, A.C. Role of Calcium in Activity and Stability of the Lactococcus lactis Cell Envelope Proteinase. Appl. Environ. Microbiol. 1999, 65, 1390–1396. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef]
- Gallagher, T.; Gilliland, G.; Wang, L.; Bryan, P. The prosegment–subtilisin BPN′ complex: crystal structure of a specific ‘foldase’. Structure 1995, 3, 907–914. [Google Scholar] [CrossRef]
- Vos, P.; van Asseldonk, M.; van Jeveren, F.; Siezen, R.; Simons, G.; de Vos, W.M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J. Bacteriol. 1989, 171, 2795–2802. [Google Scholar] [CrossRef] [PubMed]
- Vance, T.D.; Ye, Q.; Conroy, B.; Davies, P.L. Essential role of calcium in extending RTX adhesins to their target. J. Struct. Biol. X 2020, 4, 100036. [Google Scholar] [CrossRef] [PubMed]
- Bruinenberg, P.G.; De Vos, W.M.; Siezen, R.J. Deletion of Various Carboxy-Terminal Domains of Lactococcus lactis SK11 Proteinase: Effects on Activity, Specificity, and Stability of the Truncated Enzyme. Appl. Environ. Microbiol. 2000, 66, 2859–2865. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; Bruinenberg, P.G.; Vos, P.; Van Alen-Boerrigter, I.; Nijhuis, M.; Alting, A.C.; Exterkate, F.A.; Vos, W.M. Engineering of the substrate-binding region of the sublilisin-like, cell-envelop proteinase of Lactococcus lactis. Protein Eng. Des. Sel. 1993, 6, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Bruinenberg, P.G.; Doesburg, P.; Alting, A.C.; Exterkate, F.A.; Vos, W.M.; Siezen, R.J. Evidence for a large dispensable segment in the subtilisin-like catalytic domain of the Lactococcus lactis cell-envelope proteinas. Protein Eng. Des. Sel. 1994, 7, 991–996. [Google Scholar] [CrossRef]
- Ottmann, C.; Rose, R.; Huttenlocher, F.; Cedzich, A.; Hauske, P.; Kaiser, M.; Huber, R.; Schaller, A. Structural basis for Ca 2+ -independence and activation by homodimerization of tomato subtilase 3. Proc. Natl. Acad. Sci. 2009, 106, 17223–17228. [Google Scholar] [CrossRef]
- Vos, P.; Boerrigter, I.J.; Buist, G.; Haandrikman, A.J.; Nijhuis, M.; de Reuver, M.B.; Siezen, R.J.; Venema, G.; de Vos, W.M.; Kok, J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. Des. Sel. 1991, 4, 479–484. [Google Scholar] [CrossRef]
- Fernandez-Espla, M.D.; Garault, P.; Monnet, V.; Rul, F. Streptococcus thermophilus Cell Wall-Anchored Proteinase: Release, Purification, and Biochemical and Genetic Characterization. Appl. Environ. Microbiol. 2000, 66, 4772–4778. [Google Scholar] [CrossRef]
- Habimana, O.; Le Goff, C.; Juillard, V.; Bellon-Fontaine, M.-N.; Buist, G.; Kulakauskas, S.; Briandet, R. Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol. 2007, 7, 36–36. [Google Scholar] [CrossRef]
- Radziwill-Bienkowska, J.M.; Robert, V.; Drabot, K.; Chain, F.; Cherbuy, C.; Langella, P.; Thomas, M.; Bardowski, J.K.; Mercier-Bonin, M.; Kowalczyk, M. Contribution of plasmid-encoded peptidase S8 (PrtP) to adhesion and transit in the gut of Lactococcus lactis IBB477 strain. Appl. Microbiol. Biotechnol. 2017, 101, 5709–5721. [Google Scholar] [CrossRef]
- Brown, C.K.; Gu, Z.-Y.; Matsuka, Y.V.; Purushothaman, S.S.; Winter, L.A.; Cleary, P.P.; Olmsted, S.B.; Ohlendorf, D.H.; Earhart, C.A. Structure of the streptococcal cell wall C5a peptidase. Proc. Natl. Acad. Sci. 2005, 102, 18391–18396. [Google Scholar] [CrossRef] [PubMed]
- Tarazanova, M.; Huppertz, T.; Beerthuyzen, M.; van Schalkwijk, S.; Janssen, P.; Wels, M.; Kok, J.; Bachmann, H. Cell Surface Properties of Lactococcus lactis Reveal Milk Protein Binding Specifically Evolved in Dairy Isolates. Front. Microbiol. 2017, 8, 1691. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, H.; Starrenburg, M.J.; Molenaar, D.; Kleerebezem, M.; Vlieg, J.E.v.H. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 2011, 22, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, J.; Cai, H.; Larsen, R.; Hughes, J.; Welker, D.; De Carvalho, V.; Tompkins, T.; Ardö, Y.; Vogensen, F.; De Lorentiis, A.; et al. Genetic diversity in proteolytic enzymes and amino acid metabolism among Lactobacillus helveticus strains. J. Dairy Sci. 2011, 94, 4313–4328. [Google Scholar] [CrossRef]
- Boulay, M.; Al Haddad, M.; Rul, F. Streptococcus thermophilus growth in soya milk: Sucrose consumption, nitrogen metabolism, soya protein hydrolysis and role of the cell-wall protease PrtS. Int. J. Food Microbiol. 2020, 335, 108903. [Google Scholar] [CrossRef]
- QIAGEN. QIAGEN CLC Main Workbench Version 21.0. 2022. Available online: https://digitalinsights.qiagen.com/.
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Gabler, F.; Nam, S.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Jorda, J.; Kajava, A.V. T-REKS: identification of Tandem REpeats in sequences with a K-meanS based algorithm. Bioinformatics 2009, 25, 2632–2638. [Google Scholar] [CrossRef]
- Newman, A.M.; Cooper, J.B. XSTREAM: A practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinform. 2007, 8, 382–382. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.-C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. In 2-D Proteome Analysis Protocols. Methods in Molecular Biology; Link, A.J., Ed.; Humana Press: New Jersey, 1999; pp. 531–552. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Høie, M.H.; Kiehl, E.N.; Petersen, B.; Nielsen, M.; Winther, O.; Nielsen, H.; Hallgren, J.; Marcatili, P. NetSurfP-3.0: accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef] [PubMed]
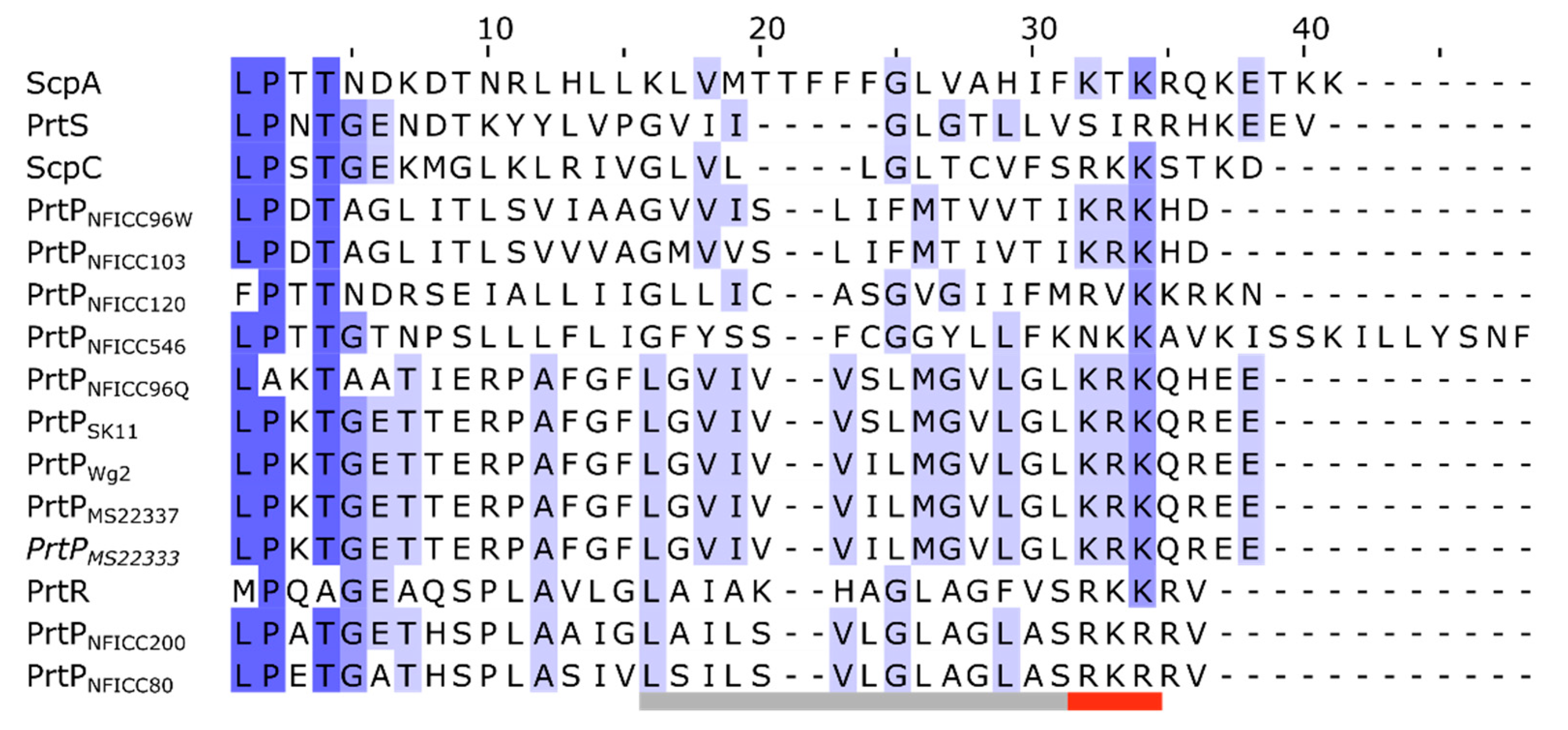
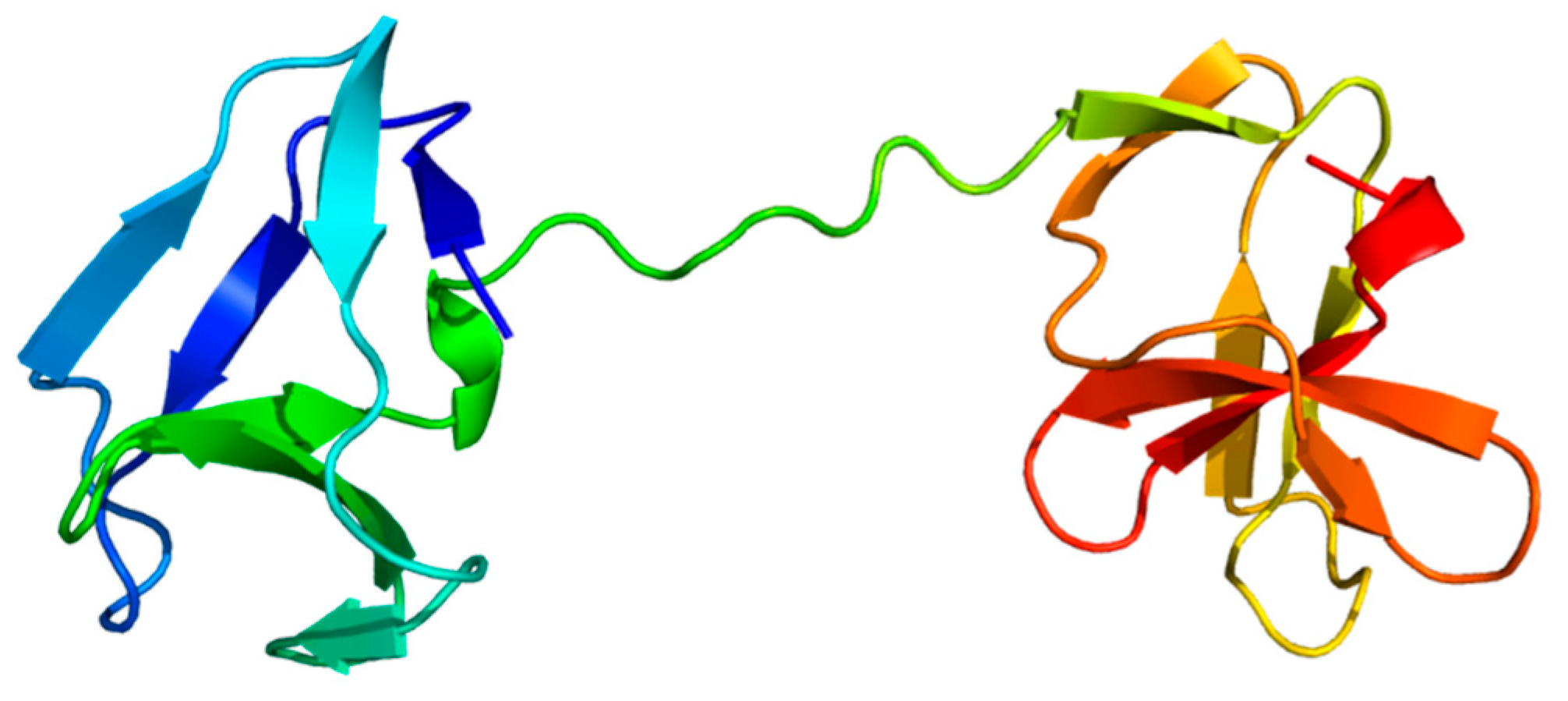
| Protease name | Sequence accession no. | Structure model | Species | Bacterial strain |
| PrtB | Q48545† | ma-uvrke§ | Lactobacillus delbrueckii subsp. bulgaricus | NCDO1489 |
| PrtH | Q9S4K2† | ma-8ai4c§ | Lactobacillus helveticus | CNRZ32 |
| PrtH2 | A4UAD8† | ma-pes1m§ | Lactobacillus helveticus | CNRZ32 |
| PrtH3CNRZ32 | G8DA68† | - | Lactobacillus helveticus | CNRZ32 |
| PrtH3DSM20075 | EEW67121.1‡ | - | Lactobacillus helveticus | DSM 20075 |
| PrtH4 | G8DA69† | - | Lactobacillus helveticus | CNRZ32 |
| PrtL | EPB98635.1‡ | ma-sthfc§ | Lactobacillus delbrueckii subsp. lactis | CRL581 |
| PrtS4F44 | D3KCP7† | ma-8wf3g§ | Streptococcus thermophilus | 4F44 |
| PrtSLMD-9 | WP_011681052.1‡ | - | Streptococcus thermophilus | LMD-9 |
| PrtPMS22333 | WWDI00000000‡ | ma-iisw7§ | Lactococcus lactis | MS22333 |
| PrtPMS22337 | WWDK00000000‡ | ma-1x50w§ | Lactococcus lactis | MS22337 |
| PrtPNCDO151 | Q02470† | - | Lacticaseibacillus paracasei subsp. paracasei | NCDO151 |
| PrtPSK11 | DQ149245.1‡ | ma-bevya§ | Lactococcus lactis subsp. cremoris | SK11 |
| PrtPWg2 | P16271† | ma-r3cva§ | Lactococcus lactis subsp. cremoris | Wg2 |
| PrtR | Q8GC13† | ma-i7gkn§ | Lacticaseibacillus rhamnosus | BGT10 |
| ScpA | P15926† | 3EIF¶ | Streptococcus pyogenes | B220 |
| ScpC | Q3HV58† | 5XYR¶ | Streptococcus pyogenes | - |
| Subtilisin | P00780† | AF-P00780†† | Bacillus licheniformis | - |
| Protease name | Sequence accession no.† | Structure model‡ | Species | Bacterial strain§ |
| PrtPNFICC1 | OQ263285 | - | Enterococcus durans | NFICC1 |
| PrtPNFICC14 | OQ263286 | - | Enterococcus durans | NFICC14 |
| PrtPNFICC16 | OQ263287 | - | Enterococcus durans | NFICC16 |
| PrtPNFICC18 | OQ263288 | - | Enterococcus durans | NFICC18 |
| PrtPNFICC63 | OQ263289 | - | Enterococcus durans | NFICC63 |
| PrtPNFICC64 | OQ263290 | - | Enterococcus durans | NFICC64 |
| PrtPNFICC80 | OQ263291 | ma-8yfr1 | Leuconostoc lactis | NFICC80 |
| PrtPNFICC96H | OQ263292 | ma-b49jj | Leuconostoc pseudomesenteroides | NFICC96 |
| PrtPNFICC96Q | OQ263293 | ma-eoqml | Leuconostoc pseudomesenteroides | NFICC96 |
| PrtPNFICC96W | OQ263294 | ma-5kpc0 | Leuconostoc pseudomesenteroides | NFICC96 |
| PrtPNFICC99 | OQ263295 | - | Leuconostoc pseudomesenteroides | NFICC99 |
| PrtPNFICC102 | OQ263296 | - | Leuconostoc pseudomesenteroides | NFICC102 |
| PrtPNFICC103 | OQ263297 | ma-u453p | Pediococcus pentosaceus | NFICC103 |
| PrtPNFICC115 | OQ263298 | - | Leuconostoc mesenteroides | NFICC115 |
| PrtPNFICC120 | OQ263299 | ma-gayjn | Enterococcus durans | NFICC120 |
| PrtPNFICC180 | OQ263300 | - | Leuconostoc pseudomesenteroides | NFICC180 |
| PrtPNFICC181 | OQ263301 | - | Leuconostoc pseudomesenteroides | NFICC181 |
| PrtPNFICC200 | OQ263302 | ma-ioefi | Leuconostoc mesenteroides | NFICC200 |
| PrtPNFICC202 | OQ263303 | - | Leuconostoc pseudomesenteroides | NFICC202 |
| PrtPNFICC227 | OQ263304 | - | Leuconostoc pseudomesenteroides | NFICC227 |
| PrtPNFICC239 | OQ263305 | - | Leuconostoc mesenteroides | NFICC239 |
| PrtPNFICC240 | OQ263306 | - | Leuconostoc mesenteroides | NFICC240 |
| PrtPNFICC313 | OQ263307 | - | Leuconostoc pseudomesenteroides | NFICC313 |
| PrtPNFICC322 | OQ263308 | - | Leuconostoc pseudomesenteroides | NFICC322 |
| PrtPNFICC323 | OQ263309 | - | Leuconostoc pseudomesenteroides | NFICC323 |
| PrtPNFICC476 | OQ263310 | - | Carnobacterium maltaromaticum | NFICC476 |
| PrtPNFICC546 | OQ263311 | ma-ytnir | Carnobacterium maltaromaticum | NFICC546 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





