Submitted:
01 August 2023
Posted:
03 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. SNN and the STAM-SNN concept from [7]
2.1. Spiking neural networks (SNN)
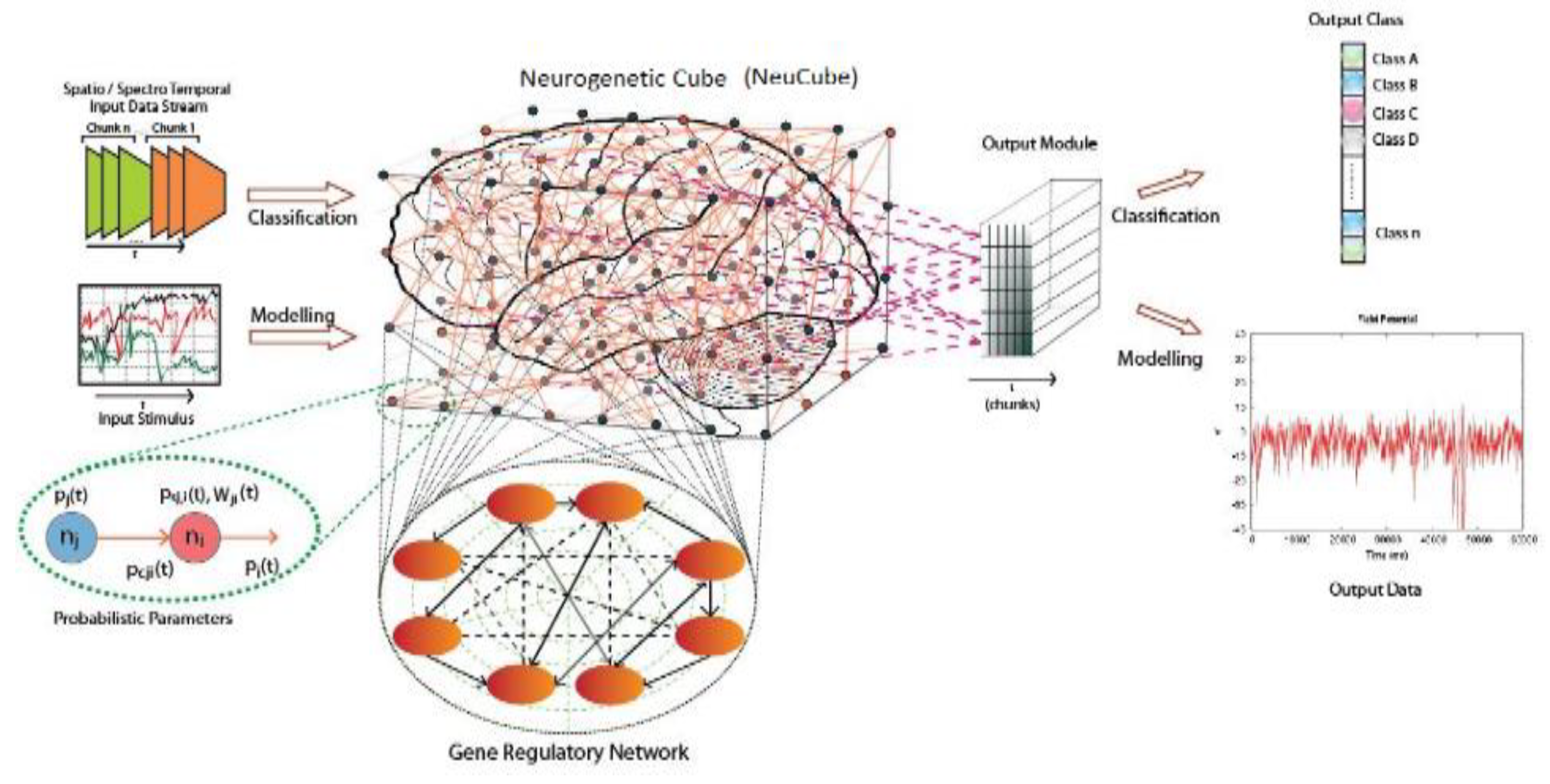
2.2. STAM on the NeuCube Framework [7]
- Input data encoding module.
- 3D SNN reservoir module (SNNcube).
- Output function (classification) module, such as deSNN [11].
- Gene regulatory network (GRN) module (optional).
- Parameter optimization module (optional).
- Temporal association accuracy: validating the full model on partial temporal data of the same variables and same data.
- Spatial association accuracy: validation of the full model on full temporal data and on a subset of variables, using the same data set.
- Temporal generalization accuracy: validation of the full model on partial temporal data of the same variables or a subset of them, but on a new data set.
- Spatial generalization accuracy: validation of the full model on full or partial temporal data and on a subset of variables, using a new data set.
3. STAM-EEG for classification
3.1. The proposed STAM-EEG classification method
- Defining the spatial and the temporal components of the EEG data for the classification task, e.g., EEG channels and EEG time series data.
- Designing a SNNcube that is structured according to a brain template suitable for the EEG data (e.g., Talairach, or MNI, etc.).
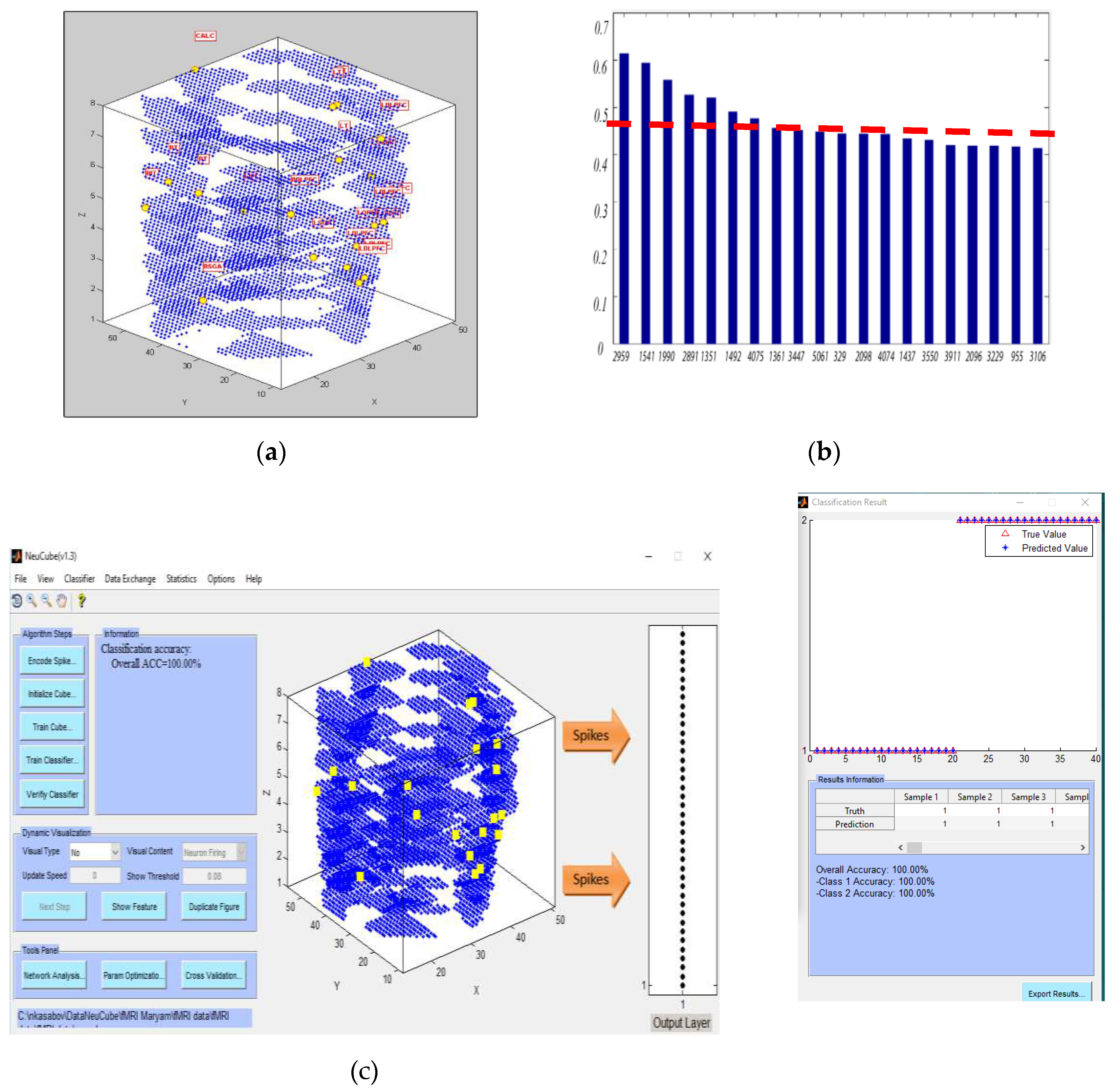
- Defining the mapping in the input EEG channels into the SNNcube 3D structure (see Figure 3a as an example of mapping 14 EEG channels in a Talairach structured SNNcube).
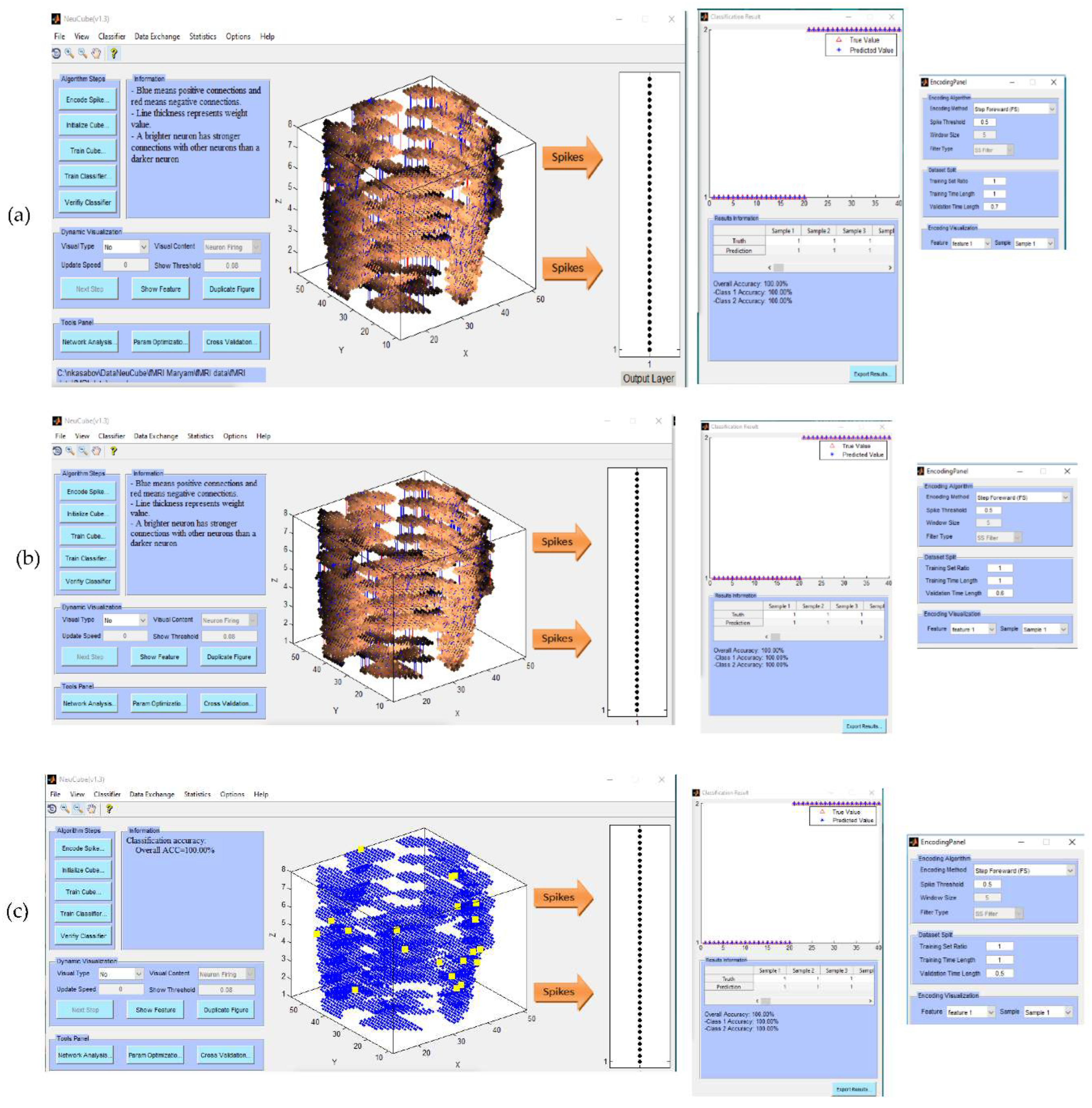
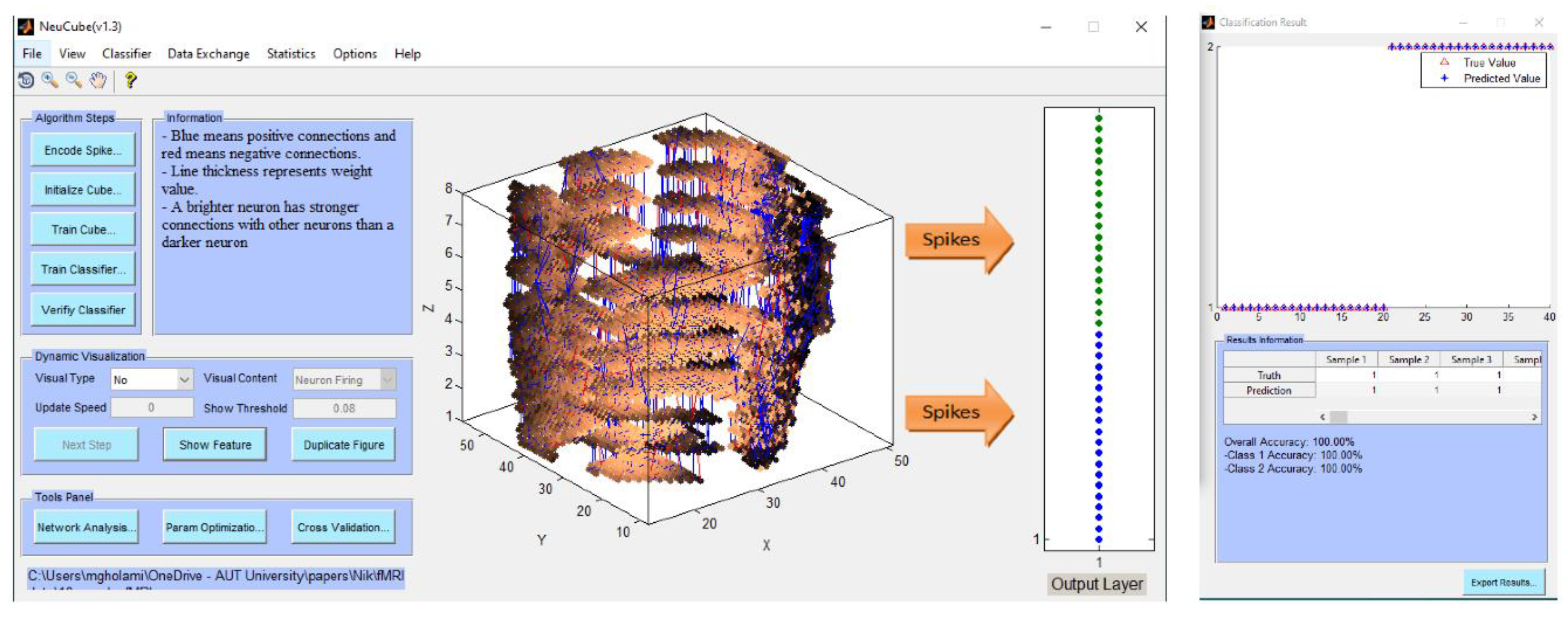
- Encode data and train a NeuCube model to classify a complete spatio-temporal EEG data, having K EEG channels measured over time T.
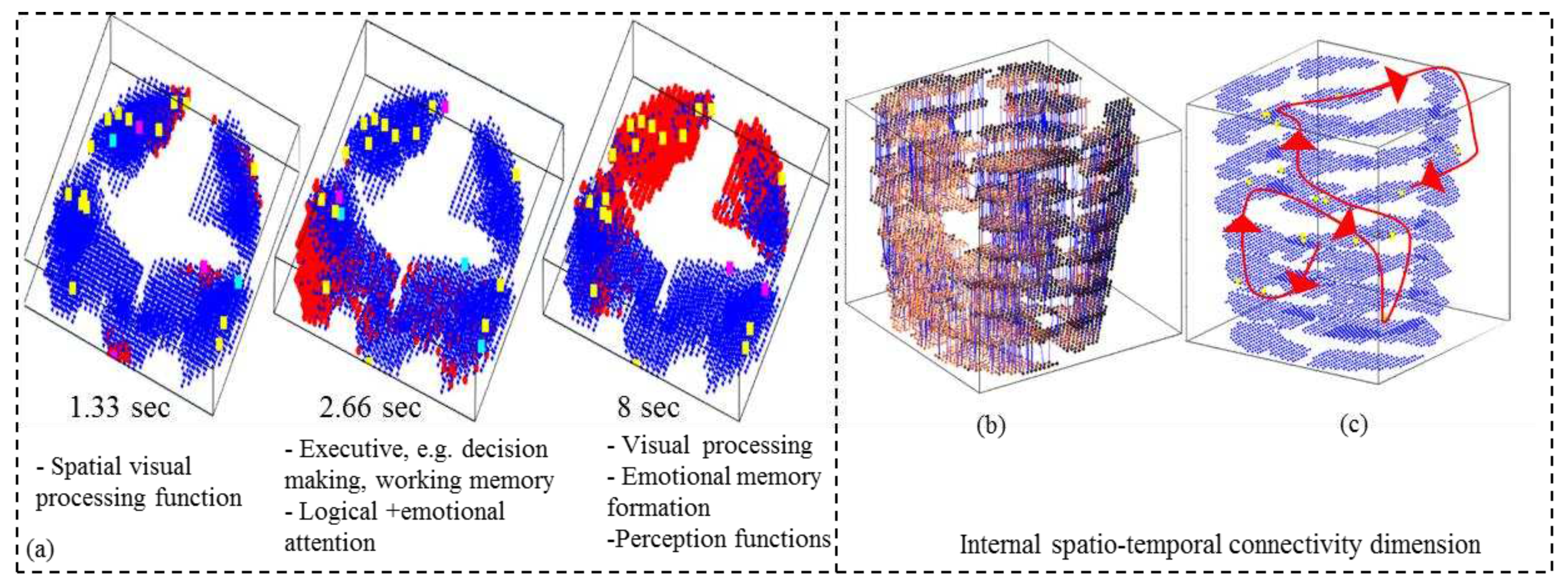
- Analyse the model through cluster analysis, spiking activity and the EEG channel spiking proportional diagram (Figs. 3b, c,d,e).
- Recall the STAM-EEG model on the same data and same variables but measured over time T1 < T to calculate the classification temporal association accuracy.
- Recall the STAM-EEG model on K1<K EEG channels to evaluate the classification spatial association accuracy.
- Recall the model on the same variables, measured over time T or T1 < T on a new data to calculate the classification temporal generalization accuracy.
- Recall the NeuCube model on K1<K EEG channels to evaluate the classification spatial generalization accuracy using a new EEG dataset.
- Evaluate the K1 EEG channels as potential classification brain biomarkers according to the problem at hand.
3.2. Experimental results
3.3. Why STAM-EEG are needed?
4. STAM-fMRI for classification
4.1. The proposed STAM-fMRI classification method
- Defining the spatial and the temporal components of the fMRI data for the classification task, e.g., fMRI voxels and the time series measurement.
- Encode data and train a NeuCube model to classify a complete spatio-temporal fMRI data, having K voxel inputs measured over time T.
- Analyse the model through connectivity and spiking activity analysis around the input voxels (Table 3).
- Recall the STAM-fMRI model on the same data and same variables but measured over time T1 < T to calculate the classification temporal association accuracy.
- Recall the STAM-fMRI on K1<K EEG channels to evaluate the classification spatial association accuracy.
- Recall the model on the same variables, measured over time T or T1 < T on a new data to calculate the classification temporal generalization accuracy.
- Recall the NeuCube model on K1<K voxel variables to evaluate the classification spatial generalization accuracy using a new fMRI dataset.
- Evaluate the K1 fMRI voxel variables as potential classification brain biomarkers (section 4.4).
4.2. STAM-fMRI for classification on experimental fMRI data
- Experiment 1 (Section 4.2): training and testing the SNN model using the whole space and time information of fMRI dat. The results are shown in Figure 4a-c.
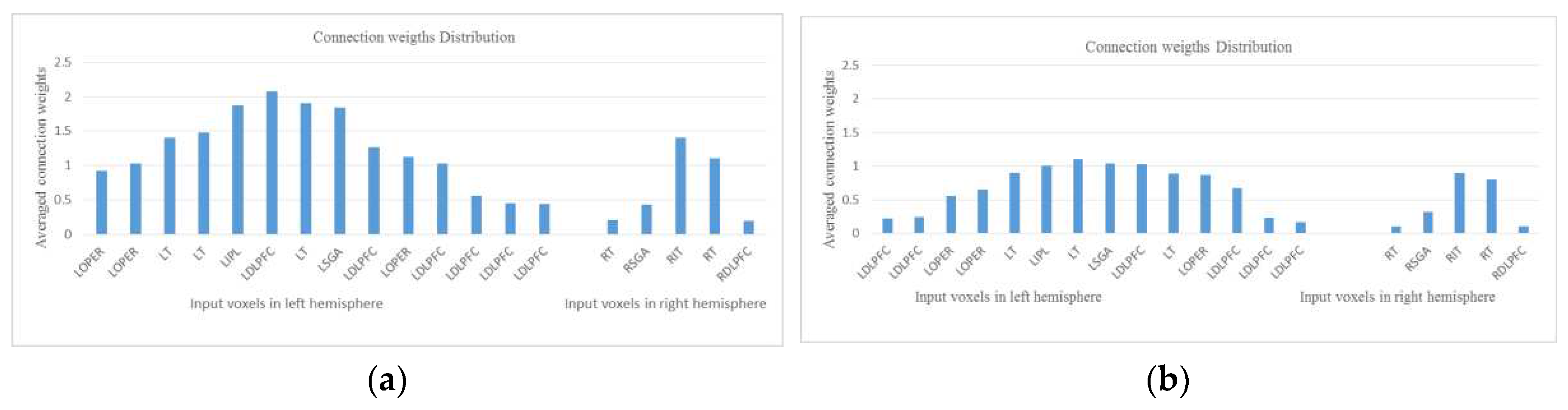
- Experiment 2 (Section 4.3): training the SNN model using the whole space (voxels) and time information of fMRI data but testing the model using a smaller temporal length of fMRI data. The results are shown in Figure 5.
- Experiment 3 (Section 4.4): training the SNN model using the whole space (voxels) and time information of fMRI data but testing the model using a smaller portion of the spatial information (a smaller number of fMRI variables/voxels). The results are shown in Figure 6.
| area | LT | LOPER | LIPL | LOPER | LDLPFC | LOPER | LT | LDLPFC | RT | CALC | |
| Neg | 1.4 | 0.92 | 1.87 | 1.03 | 2.08 | 1.12 | 1.48 | 0.44 | 0.2 | 0.89 | |
| Aff | 0.9 | 0.56 | 1.01 | 0.87 | 1.03 | 0.65 | 0.89 | 0.23 | 0.1 | 0.43 | |
| area | LSGA | LDLPFC | LT | LDLPFC | RT | LDLPFC | LDLPFC | RDLPFC | RSGA | RIT | Avg |
| Neg | 1.84 | 1.03 | 1.9 | 0.45 | 1.1 | 1.26 | 0.56 | 0.19 | 0.43 | 1.4 | 1.7 |
| Aff | 1.04 | 0.68 | 1.1 | 0.17 | 0.8 | 0.24 | 0.22 | 0.11 | 0.32 | 0.9 | 0.6 |
4.3. STAM-fMRI recalled on partial temporal fMRI data
4.4. STAM-fMRI, recalled on partial spatial fMRI data
4.5. Potential marker discovery from the STAM-fMRI
4.6. STAM for longitudinal MRI neuroimaging
5. Discussions, conclusions, and directions for further research
Acknowledgments
References
- Squire, L. R. Memory and Brain Systems: 1969 –2009. The Journal of Neuroscience 2009, 29, 12711–12716. [Google Scholar] [CrossRef] [PubMed]
- Squire, L.R. Memory systems of the brain: A brief history and current perspective. Neurobiol. Learn. Mem. 2004, 82, 171–177. [Google Scholar] [CrossRef]
- Hopfield, J.J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci. 1982, 79, 2554–2558. [Google Scholar] [CrossRef] [PubMed]
- Kosko, B. Bidirectional associative memories. IEEE Trans. Syst. Man, Cybern. 1988, 18, 49–60. [Google Scholar] [CrossRef]
- Haga, T.; Fukai, T. Extended Temporal Association Memory by Modulations of Inhibitory Circuits. Phys. Rev. Lett. 2019, 123, 078101. [Google Scholar] [CrossRef] [PubMed]
- Kasabov, N.K. , "NeuCube: A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data," Neural Networks, vol. 52, pp. 62-76, 2014.
- Kasabov, Nikola (2023). Spatio-Temporal Associative Memories in Brain-inspired Spiking Neural Networks: Concepts and Perspectives. TechRxiv. Preprint. [CrossRef]
- Song, S.; Miller, K.D.; Abbott, L.F. Competitive Hebbian learning through spike-timing-dependent synaptic plasticity. Nat. Neurosci. 2000, 3, 919–926. [Google Scholar] [CrossRef]
- Kasabov, N.K. Time-Space, Spiking Neural Networks and Brain-Inspired Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar] [CrossRef]
- Kasabov, N. , NeuroCube EvoSpike Architecture for Spatio-Temporal Modelling and Pattern Recognition of Brain Signals, in: Mana, Schwenker and Trentin (Eds) ANNPR, Springer LNAI 7477, 2012, 225-243.
- Kasabov, N.; Dhoble, K.; Nuntalid, N.; Indiveri, G. Dynamic evolving spiking neural networks for on-line spatio- and spectro-temporal pattern recognition. Neural Networks 2013, 41, 188–201. [Google Scholar] [CrossRef]
- Talairach, J. and P. Tournoux, "Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System - an Approach to Cerebral Imaging", Thieme Medical Publishers, New York, NY, 1988.
- Zilles, K. and K. Amunts, Centenary of Brodmann’s map — conception and fate, Nature Reviews Neuroscience, vol.11, 10, 139-145. 20 February.
- Mazziotta, J., A. W. Toga, A. Evans, P. Fox and J. Lancaster, "A Probablistic Atlas of the Human Brain: Theory and Rationale for Its Development", NeuroImage 2:89-101, 1995.
- Abeles,M.(1991). Corticonics, Cambridge University Press, NY.
- Izhikevich, E.M. Polychronization: Computation with Spikes. Neural Comput. 2006, 18, 245–282. [Google Scholar] [CrossRef]
- Neftci, E. and Chicca, E. and Indiveri, G. and Douglas, R. A systematic method for configuring vlsi networks of spiking neurons, Neural computation, 23:(10) 2457-2497, 2011.
- Szatmáry, B.; Izhikevich, E.M. Spike-Timing Theory of Working Memory. PLOS Comput. Biol. 2010, 6, e1000879. [Google Scholar] [CrossRef]
- Humble J, Denham S and Wennekers T (2012) Spatio-temporal pattern recognizers using spiking neurons and spike-timing-dependent plasticity. Front. Comput. Neurosci., 6, 84.
- Kasabov, N.K.; Doborjeh, M.G.; Doborjeh, Z.G. Mapping, Learning, Visualization, Classification, and Understanding of fMRI Data in the NeuCube Evolving Spatiotemporal Data Machine of Spiking Neural Networks. IEEE Trans. Neural Networks Learn. Syst. 2017, 28, 887–899. [Google Scholar] [CrossRef]
- Mitchel, T. et al, Learning to Decode Cognitive States from Brain Images, Machine Learning, 2004, 57, 145–175. [Google Scholar]
- Doborjeh, A.Merkin, H.Bahrami, A.Sumich, R.Krishnamurthi, O. Medvedev, M.Crook-Rumsey, C. Morgan, I.Kirk, P.Sachdev, H. Brodaty, K. Kang, W.Wen, V. Feigin, N. Kasabov, Personalised Predictive Modelling with Spiking Neural Networks of Longitudinal MRI Neuroimaging Cohort and the Case Study ofr Dementia, Neural Networks, vol.144, Dec.2021, 522-539. [CrossRef]
- Chong, B.; Wang, A.; Borges, V.; Byblow, W.D.; Barber, P.A.; Stinear, C. Investigating the structure-function relationship of the corticomotor system early after stroke using machine learning. NeuroImage: Clin. 2022, 33, 102935. [Google Scholar] [CrossRef] [PubMed]
- Karim, M., Chakraborty, S., & Samadiani, N. Stroke Lesion Segmentation using Deep Learning Models: A Survey. IEEE Access 2021, 9, 44155–44177.
- Li, H., Shen, D., & Wang, L. A Hybrid Deep Learning Framework for Alzheimer's Disease Classification Based on Multimodal Brain Imaging Data. Frontiers in Neuroscience 2021, 15, 625534.
- Niazi, F., Bourouis, S., & Prasad, P. W. C. Deep Learning for Diagnosis of Mild Cognitive Impairment: A Systematic Review and Meta-Analysis. Frontiers in Aging Neuroscience 2020, 12, 244.
- Sona, C., Siddiqui, S. A., & Mehmood, R. Classification of Depression Patients and Healthy Controls Using Machine Learning Techniques. IEEE Access 2021, 9, 26804–26816.
- Fanaei, M., Davari, A., & Shamsollahi, M. B. Autism Spectrum Disorder Diagnosis Based on fMRI Data Using Deep Learning and 3D Convolutional Neural Networks. Sensors 2020, 20, 4600.
- Zhang, X., Liu, T., & Qian, Z. A Comprehensive Review on Parkinson's Disease Using Deep Learning Techniques. Frontiers in Aging Neuroscience 2021, 13, 702474.
- Hjelm, R. D., Calhoun-Sauls, A., & Shiffrin, R. M. Deep Learning and the Audio-Visual World: Challenges and Frontiers. Frontiers in Neuroscience 2020, 14, 219.
- Poline, J.-B.; Poldrack, R.A. Frontiers in Brain Imaging Methods Grand Challenge. Front. Neurosci. 2012, 6, 96. [Google Scholar] [CrossRef]
- Pascual-Leone, A.; Hamilton, R. Chapter 27 The metamodal organization of the brain. 2001, 134, 427–445. [CrossRef]
- Honey, C.J.; Kötter, R.; Breakspear, M.; Sporns, O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl. Acad. Sci. 2007, 104, 10240–10245. [Google Scholar] [CrossRef] [PubMed]
- Nicolelis, M. Mind in Motion, Scientific American, vol. 307, Number 2012, 3, 44–49. [Google Scholar]
- Paulun, V. C., Beer, A. L., & Thompson-Schill, S. L. Distinct contributions of functional and deep neural network features to representational similarity of scenes in human brain and behavior. eLife 2019, 8, e42848.
- Tu, E. N. Kasabov, J. Yang, Mapping Temporal Variables into the NeuCube Spiking Neural Network Architecture for Improved Pattern Recognition and Predictive Modelling, IEEE Trans. on Neural Networks and Learning Systems. 2017, 282, 1305–1317. [Google Scholar] [CrossRef]
- Laña, I.; Lobo, J.L.; Capecci, E.; Del Ser, J.; Kasabov, N. Adaptive long-term traffic state estimation with evolving spiking neural networks. Transp. Res. Part C: Emerg. Technol. 2019, 101, 126–144. [Google Scholar] [CrossRef]
- Doborjeh, M.; Doborjeh, Z.; Merkin, A.; Krishnamurthi, R.; Enayatollahi, R.; Feigin, V.; Kasabov, N. Personalized Spiking Neural Network Models of Clinical and Environmental Factors to Predict Stroke. Cogn. Comput. 2022, 14, 2187–2202. [Google Scholar] [CrossRef]
- Doborjeh, Z.; Doborjeh, M.; Sumich, A.; Singh, B.; Merkin, A.; Budhraja, S.; Goh, W.; Lai, E.M.-K.; Williams, M.; Tan, S.; et al. Investigation of social and cognitive predictors in non-transition ultra-high-risk’ individuals for psychosis using spiking neural networks. npj Schizophr. 2023, 9, 1–10. [Google Scholar] [CrossRef]
- Maciąg, P.S.; Kasabov, N.; Kryszkiewicz, M.; Bembenik, R. Air pollution prediction with clustering-based ensemble of evolving spiking neural networks and a case study for London area. Environ. Model. Softw. 2019, 118, 262–280. [Google Scholar] [CrossRef]
- Liu, H.; Lu, G.; Wang, Y.; Kasabov, N. Evolving Spiking Neural Network Model for PM2.5 Hourly Concentration Prediction Based on Seasonal Differences: A Case Study on Data from Beijing and Shanghai. Aerosol Air Qual. Res. 2021, 21, 200247. [Google Scholar] [CrossRef]
- Furber, S. , To Build a Brain, IEEE Spectrum, vol.49, Number 8, 39-41, 2012.
- Indiveri, G; Stefanini, F; Chicca, E (2010). Spike-based learning with a generalized integrate and fire silicon neuron. In: 2010 IEEE Int. Symp. Circuits and Syst. (ISCAS 2010), Paris, - 02 June 2010, 1951-1954. 30 May.
- Indiveri, G; Chicca, E; Douglas, R J Artificial cognitive systems: From VLSI networks of spiking neurons to neuromorphic cognition. Cognitive Computation 2009, 1, 119–127. [CrossRef]
- Delbruck, T. jAER open-source project, 2007. Available online: https://jaer.wiki.sourceforge.net.
- Benuskova, L, and N.Kasabov, Computational neuro-genetic modelling, Springer, New York, 2007.
- BrainwaveR Toolbox. Available online: http://www.nitrc.org/projects/brainwaver/.
- Buonomano, D. Maass, State-dependent computations: Spatio-temporal processing in cortical networks, Nature Reviews, Neuroscience. 2009, 10, 113–125. [Google Scholar]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef]
- Kasabov, N. , Tan, Y., Doborjeh, M., Tu, E., Yang, J., Goh, W., Lee, J., Transfer Learning of Fuzzy Spatio-Temporal Rules in the NeuCube Brain-Inspired Spiking Neural Network: A Case Study on EEG Spatio-temporal Data. IEEE Transactions on Fuzzy Systems 2023. [Google Scholar] [CrossRef]
- Kumarasinghe, K.; Kasabov, N.; Taylor, D. Brain-inspired spiking neural networks for decoding and understanding muscle activity and kinematics from electroencephalography signals during hand movements. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
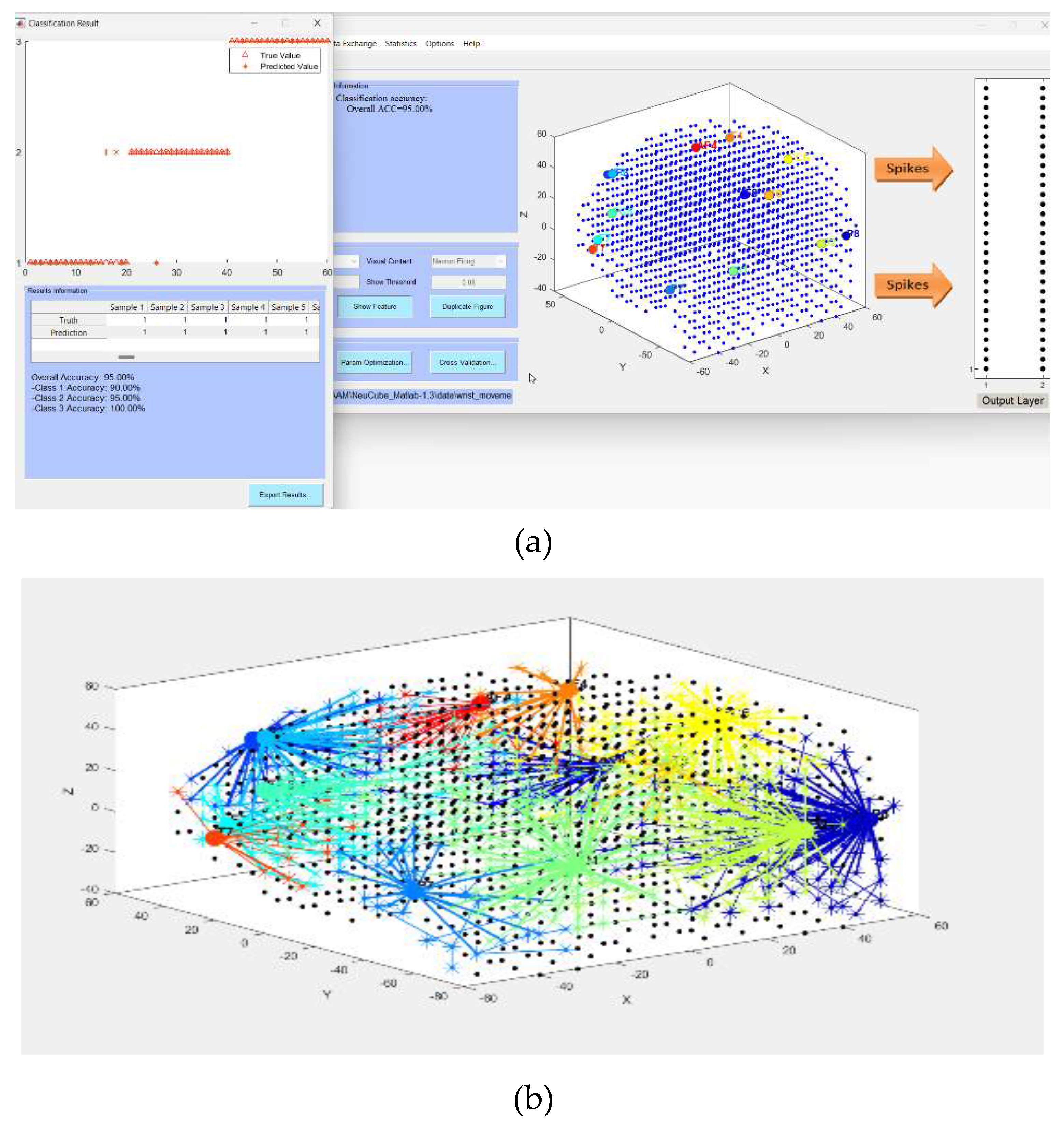
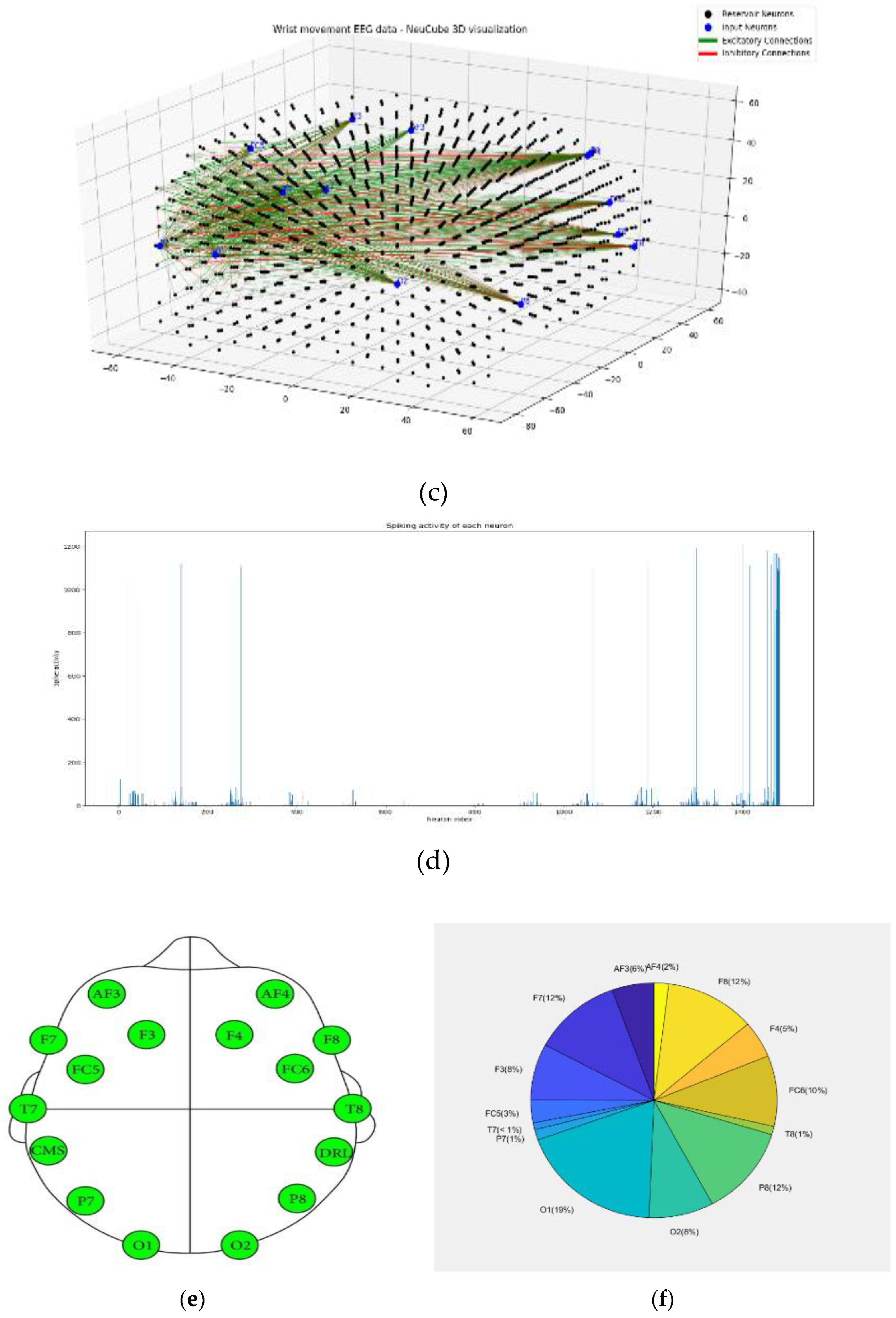
| Dataset information | Encoding method and parameters | NeuCube model | STDP parameters |
deSNNs Classifier parameters |
|---|---|---|---|---|
| sample number: 60, feature number: 14 channels, time length: 128, class number: 3. |
encoding method: Thresholding Representation (TR), spike threshold: 0.5, window size: 5, filter type: SS. |
number of neurons: 1471, brain template: Talairach, neuron model: LIF. |
potential leak rate: 0.002, STDP rate: 0.01, firing threshold: 0.5, training iteration: 1, refractory time: 6, LDC probability: 0. | mod: 0.8, drift: 0.005, K: 3, sigma: 1. |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





