Submitted:
28 July 2023
Posted:
31 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant materials
2.2. Transcriptome analysis
2.3. Metabolomics analysis
2.4. Data analyses
3. Results
3.1. Transcriptome
3.1.1. Differentially expressed genes
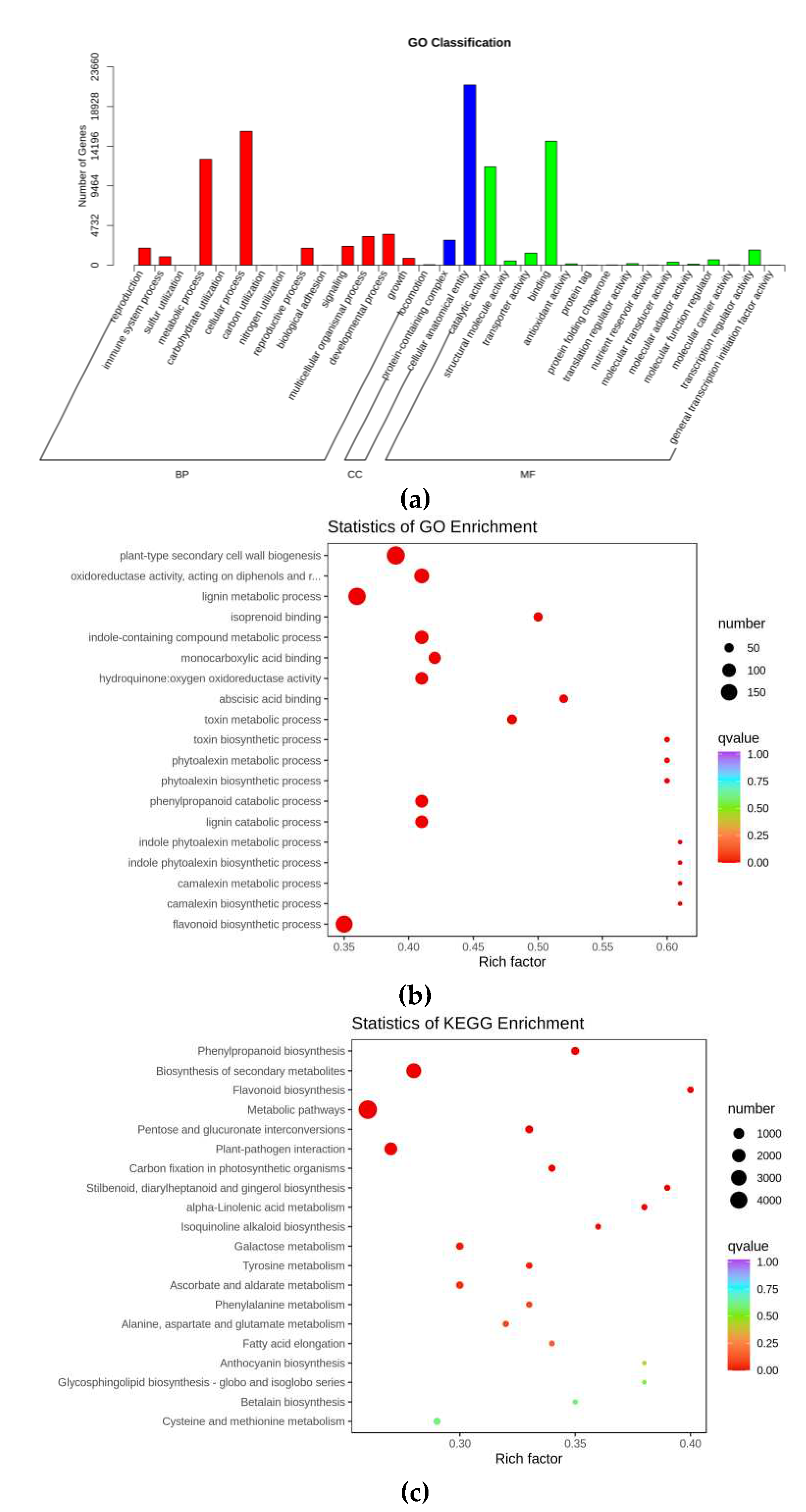
3.1.2. Annotation and enrichment analysis of DEGs
3.2. Metabolome
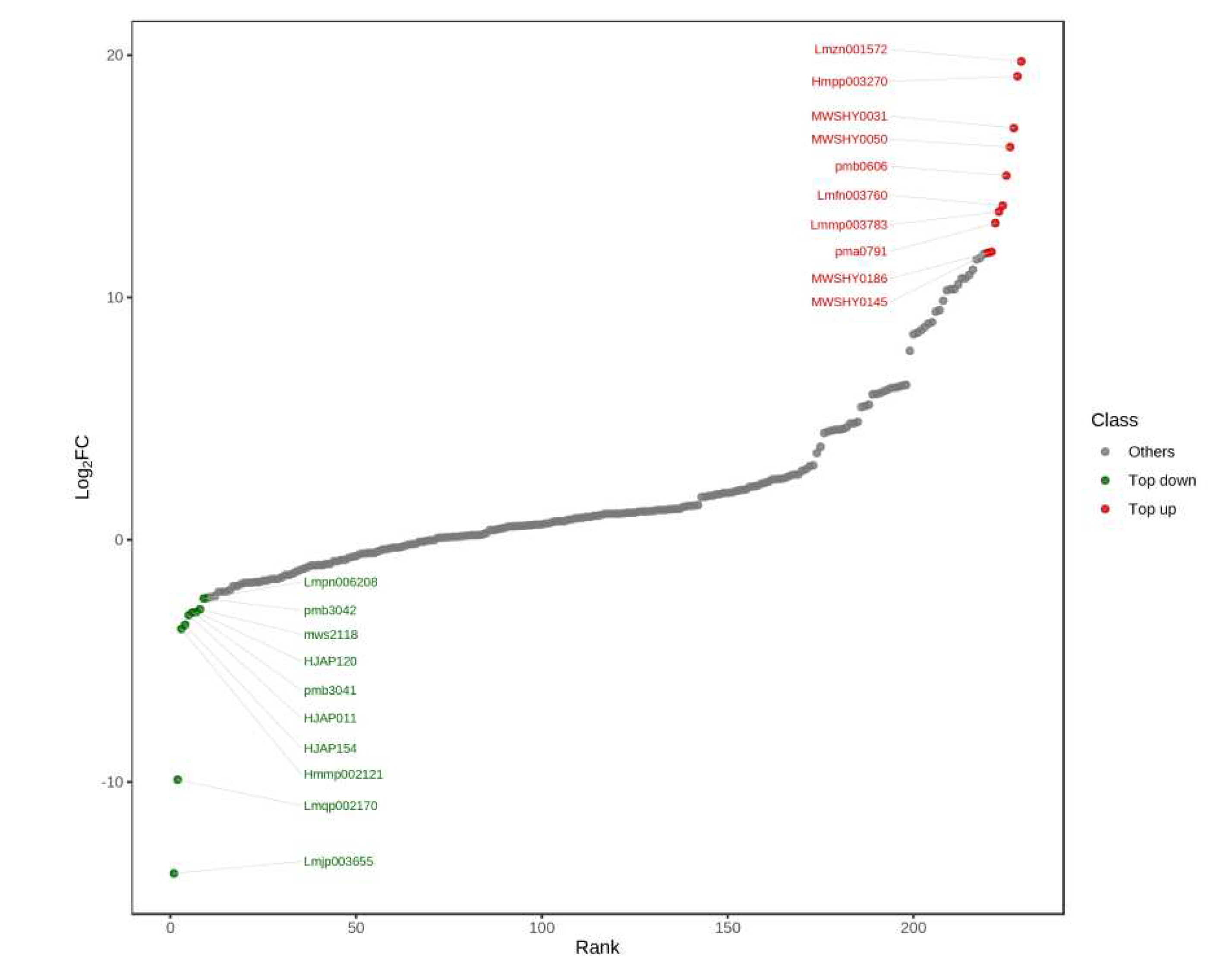
3.2.1. Differentially expressed metabolites
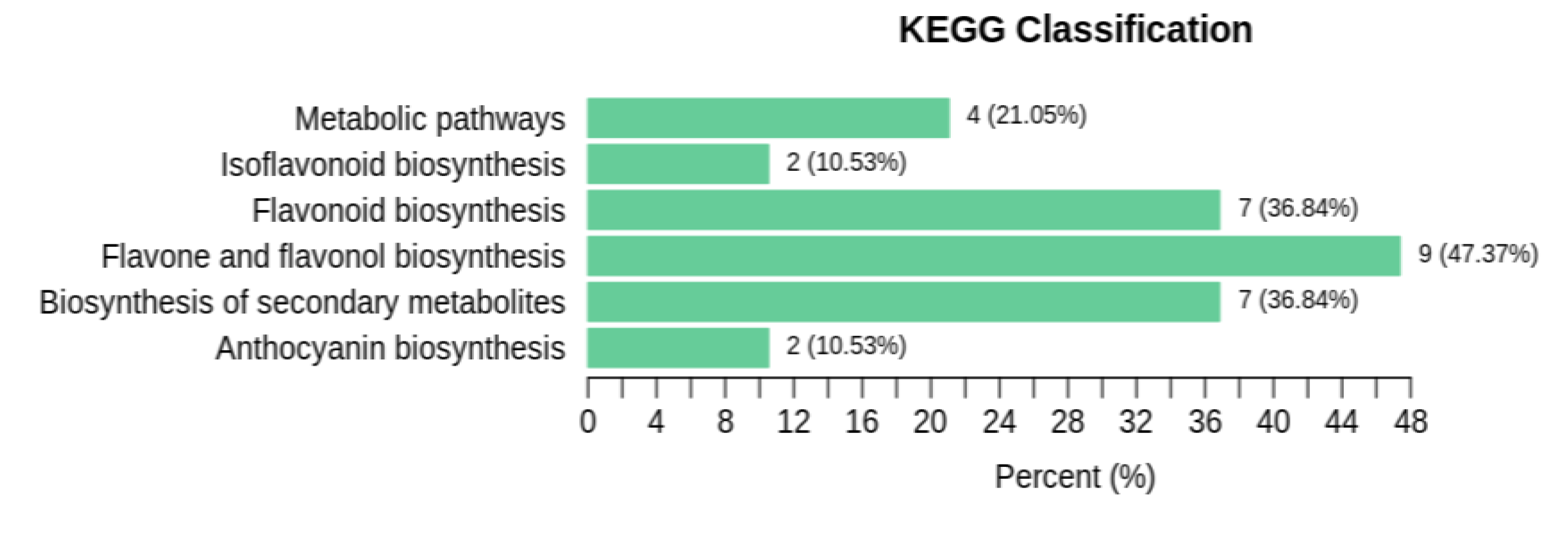
3.2.1. Enrichment analysis of DEMs
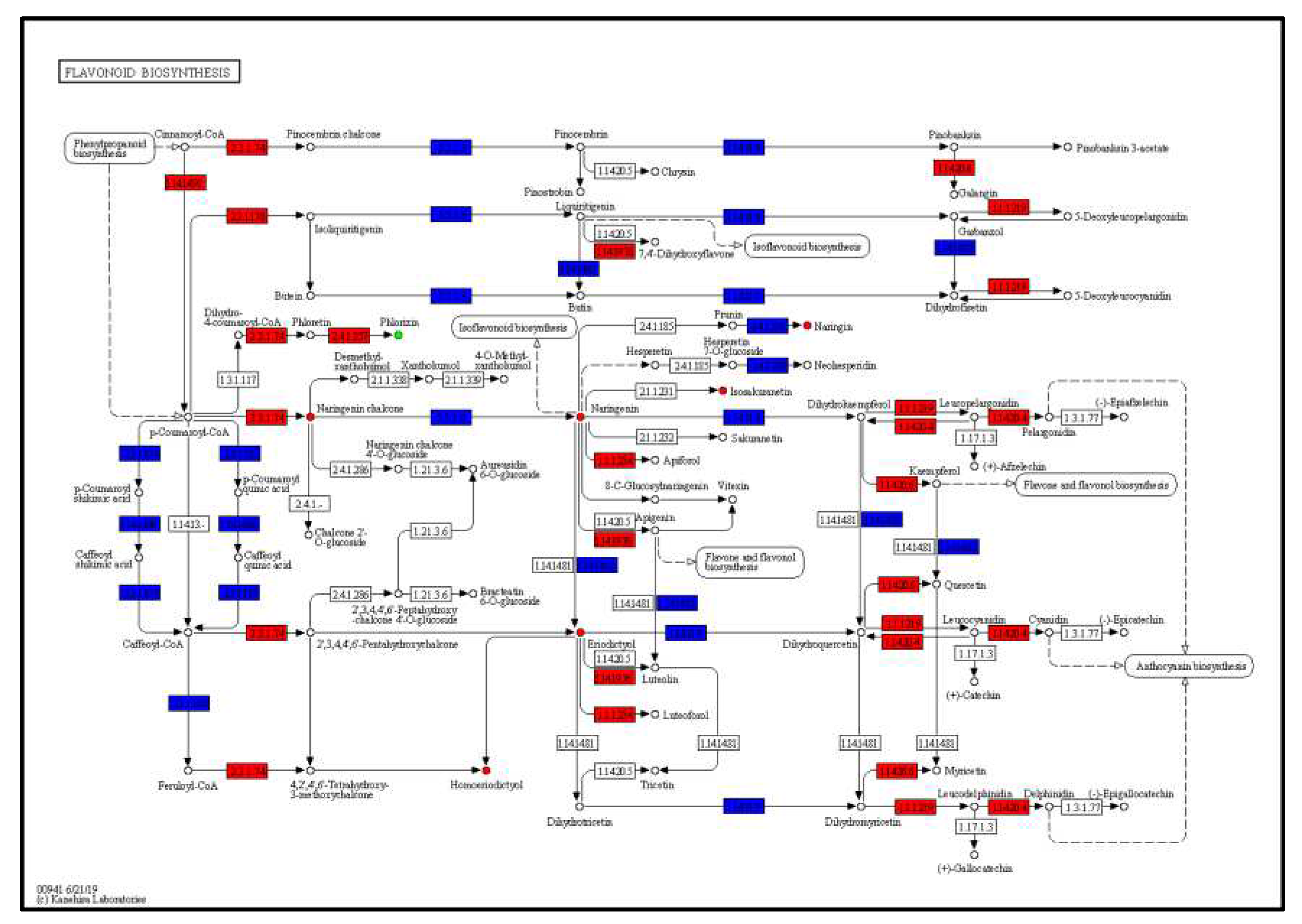
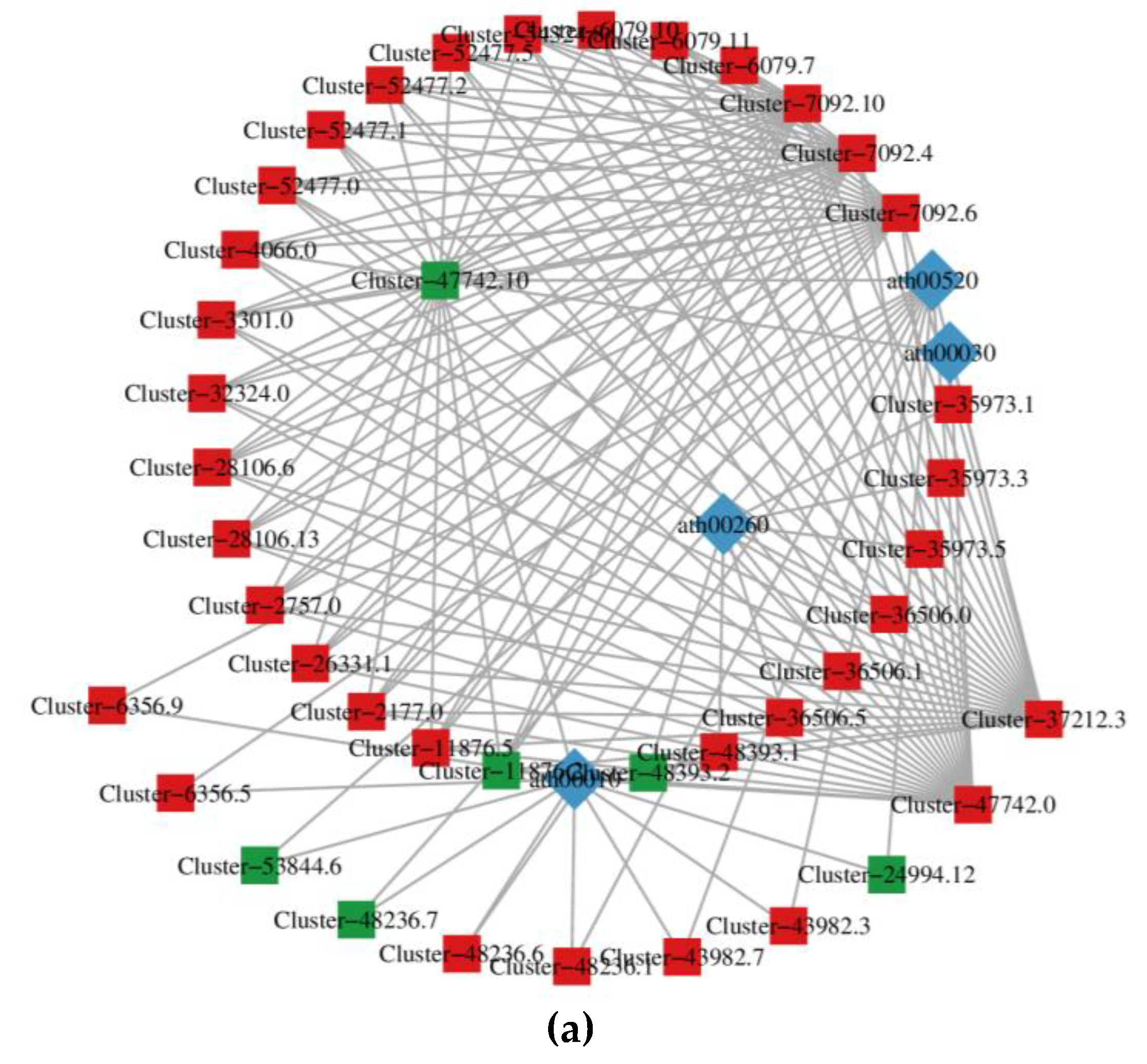
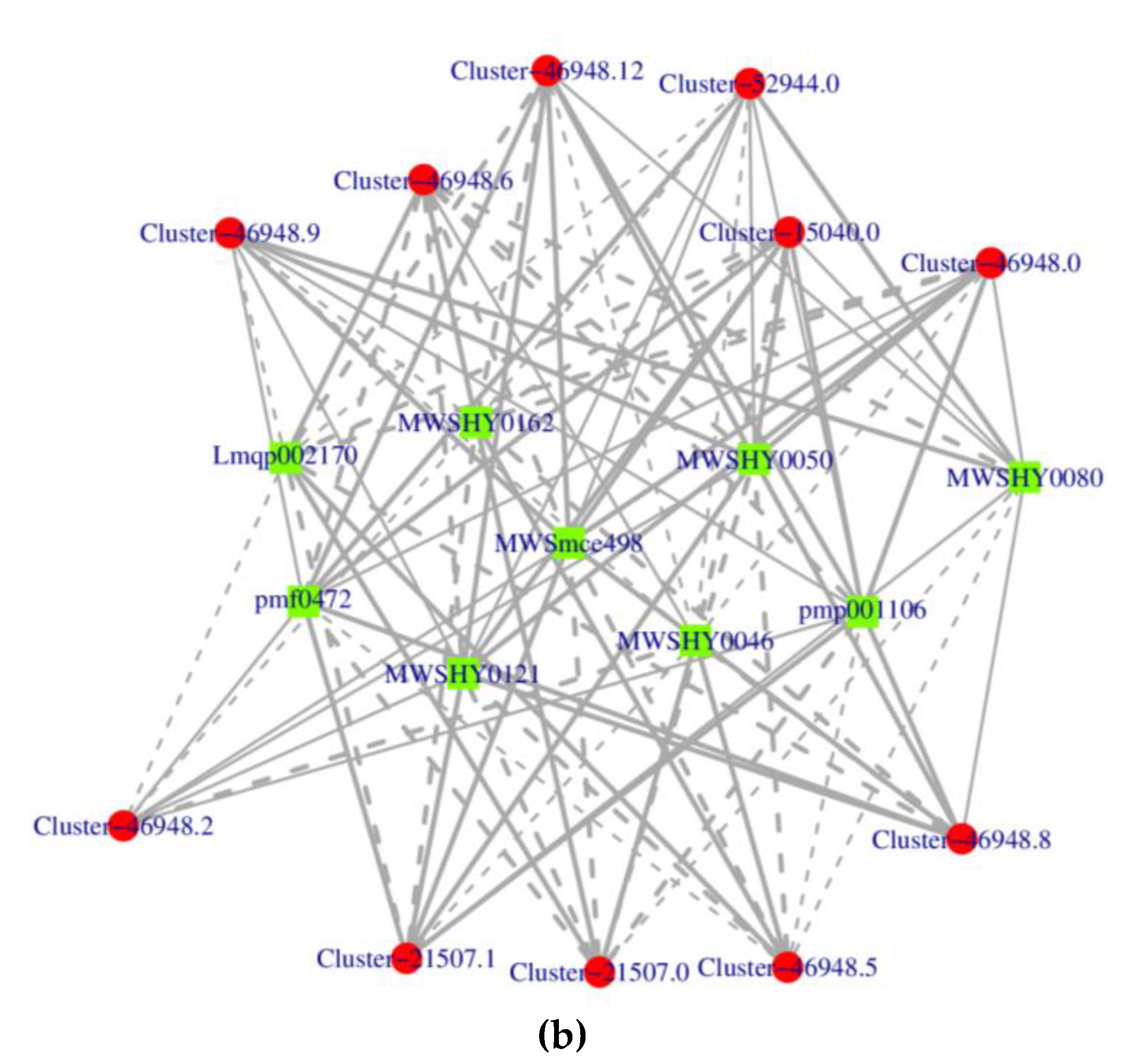
3.3. Joint analysis of transcriptome and metabolome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bahadori, M.B.; Zengin, G.; Bahadori, S.; Dinparast, L.; Movahhedin, N. Phenolic composition and functional properties of wild mint (Mentha longifolia var. calliantha (Stapf) Briq.). International journal of food properties. 2018, 21, 183–193. [Google Scholar] [CrossRef]
- Wani, S.A.; Naik, H.; Wagay, J.A.; Ganie, N.A.; Mulla, M.Z.; Dar, B. Mentha: A review on its bioactive compounds and potential health benefits. Quality Assurance and Safety of Crops & Foods. 2022, 14, 154–168. [Google Scholar] [CrossRef]
- Yilmaztekin, M.; Lević, S.; Kalušević, A.; Cam, M.; Bugarski, B.; Rakić, V.; Pavlović, V.; Nedović, V. Characterisation of peppermint (Mentha piperita L.) essential oil encapsulates. Journal of microencapsulation. 2019, 36, 109–119. [Google Scholar] [CrossRef]
- Shaikh, S.; Yaacob, H.; Rahim, Z. Prospectve role in treatment of major illnesses and potental benefts as a safe insectcide and natural food preservatve of mint (Mentha spp.): A review. Asian J Biomed Pharm. 2014, 4. [Google Scholar]
- Salehi, B.; Stojanović-Radić, Z.; Matejić, J.; Sharopov, F.; Antolak, H.; Kręgiel, D.; Sen, S.; Sharifi-Rad, M.; Acharya, K.; Sharifi-Rad, R. Plants of genus Mentha: From farm to food factory. Plants. 2018, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Šarić-Kundalić, B.; Fialová, S.; Dobeš, C.; Ölzant, S.; Tekeľová, D.; Grančai, D.; Reznicek, G.; Saukel, J. Multivariate numerical taxonomy of Mentha species, hybrids, varieties and cultivars. Scientia Pharmaceutica. 2009, 77, 851–876. [Google Scholar] [CrossRef]
- Arumugam, P.; Priya, N.G.; Subathra, M.; Ramesh, A. Anti-inflammatory activity of four solvent fractions of ethanol extract of Mentha spicata L. investigated on acute and chronic inflammation induced rats. Environmental toxicology and pharmacology. 2008, 26, 92–95. [Google Scholar] [CrossRef]
- Diop, S.M.; Guèye, M.T.; Ndiaye, I.; Ndiaye, E.H.B.; Diop, M.B.; Heuskin, S.; Fauconnier, M.-L.; Lognay, G. Chemical composition of essential oils and floral waters of Mentha longifolia (L.) Huds. from Senegal. American Journal of Essential Oils and Natural Products. 2016, 4, 46–49. [Google Scholar]
- Peixoto, I.; Furlanetti, V.; Anibal, P.; Duarte, M.; Höfling, J. Potential pharmacological and toxicological basis of the essential oil from Mentha spp. Revista de Ciências Farmacêuticas Básica e Aplicada. 2009, 30. [Google Scholar]
- Tiwari, P. Recent advances and challenges in trichome research and essential oil biosynthesis in Mentha arvensis L. Industrial Crops and Products. 2016, 82, 141–148. [Google Scholar] [CrossRef]
- Dimandja, J.M.D.; Stanfill, S.B.; Grainger, J.; Patterson Jr, D.G. Application of comprehensive two-dimensional gas chromatography (GC× GC) to the qualitative analysis of essential oils. Journal of High Resolution Chromatography. 2000, 23, 208–214. [Google Scholar] [CrossRef]
- Gherman, C.; Culea, M.; Cozar, O. Comparative analysis of some active principles of herb plants by GC/MS. Talanta. 2000, 53, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Carrera, J.; Rodrigo, G.; Jaramillo, A.; Elena, S.F. Reverse-engineering the Arabidopsis thaliana transcriptional network under changing environmental conditions. Genome biology. 2009, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Vining, K.J.; Johnson, S.R.; Ahkami, A.; Lange, I.; Parrish, A.N.; Trapp, S.C.; Croteau, R.B.; Straub, S.C.; Pandelova, I.; Lange, B.M. Draft genome sequence of Mentha longifolia and development of resources for mint cultivar improvement. Molecular plant. 2017, 10, 323–339. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: enabling differential gene expression analysis for de novoassembled transcriptomes. Genome biology. 2014, 15, 1–14. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014, 15, 550. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nature methods. 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Bouhaddani, S.E.; Houwing-Duistermaat, J.; Salo, P.; Perola, M.; Jongbloed, G.; Uh, H.W. Evaluation of O2PLS in Omics data integration. BMC Bioinformatics. 2016, 17, 117–132. [Google Scholar] [CrossRef] [PubMed]
- López-Hernández, F.; Cortés, A.J. Whole transcriptome sequencing unveils the genomic determinants of putative somaclonal variation in Mint (Mentha L.). International Journal of Molecular Sciences. 2022, 23, 5291. [Google Scholar] [CrossRef] [PubMed]
- Boachon, B.; Buell, C.R.; Crisovan, E.; Dudareva, N.; Garcia, N.; Godden, G.; Henry, L.; Kamileen, M.O.; Kates, H.R.; Kilgore, M.B. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Molecular plant. 2018, 11, 1084–1096. [Google Scholar] [CrossRef]
- Yu, X.; Qi, X.; Li, S.; Fang, H.; Bai, Y.; Li, L.; Liu, D.; Chen, Z.; Li, W.; Liang, C. Transcriptome analysis of light-regulated Monoterpenes biosynthesis in leaves of Mentha canadensis L. Plants. 2021, 10, 930. [Google Scholar] [CrossRef]
- Godden, G.T.; Kinser, T.J.; Soltis, P.S.; Soltis, D.E. Phylotranscriptomic analyses reveal asymmetrical gene duplication dynamics and signatures of ancient polyploidy in mints. Genome Biology and Evolution. 2019, 11, 3393–3408. [Google Scholar] [CrossRef]
- Vining, K.J.; Pandelova, I. Dynamic tissue-specific transcriptome changes in response to Verticillium dahliae in wild mint species Mentha longifolia. Plants. 2022, 11, 674. [Google Scholar] [CrossRef] [PubMed]
- Aminfar, Z.; Rabiei, B.; Tohidfar, M.; Mirjalili, M.H. Identification of key genes involved in the biosynthesis of triterpenic acids in the mint family. Scientific Reports. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Current opinion in biotechnology. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Rencoret, J.; Rosado, M.J.; Kim, H.; Timokhin, V.I.; Gutiérrez, A.; Bausch, F.; Rosenau, T.; Potthast, A.; Ralph, J.; Del Río, J.C. Flavonoids naringenin chalcone, naringenin, dihydrotricin, and tricin are lignin monomers in papyrus. Plant Physiology. 2022, 188, 208–219. [Google Scholar] [CrossRef]
- Lam, P.Y.; Wang, L.; Lui, A.C.; Liu, H.; Takeda-Kimura, Y.; Chen, M.-X.; Zhu, F.-Y.; Zhang, J.; Umezawa, T.; Tobimatsu, Y. Deficiency in flavonoid biosynthesis genes CHS, CHI, and CHIL alters rice flavonoid and lignin profiles. Plant Physiology. 2022, 188, 1993–2011. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst. 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Georgii, E.; Jin, M.; Zhao, J.; Kanawati, B.; Schmitt-Kopplin, P.; Albert, A.; Winkler, J.B.; Schäffner, A.R. Relationships between drought, heat and air humidity responses revealed by transcriptome-metabolome co-analysis. BMC plant biology. 2017, 17, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Cho, K.-S.; Sohn, H.-B.; Ha, I.J.; Hong, S.-Y.; Lee, H.; Kim, Y.-M.; Nam, M.H. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. Journal of experimental botany. 2016, 67, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
| Materials | Chlorophyll (mg/g) | Anthocyanin (mg/g) | Total flavone (mg/g) | Soluble sugar (mg/g) | Soluble protein (mg/g) |
| Purple mint | 0.215±0.008 a | 8.493±0.481 a | 8.531±0.758 a | 60.861±10.027 a | 4.126±0.306 a |
| Green mint | 0.152±0.009 b | 0.363±0.035 b | 5.457±0.276 b | 70.407±0.439 a | 1.773±0.104 b |
| Index | Compounds | Class | Log2FC | Type |
| Lmzn001572 | Scutellarein-7-O-glucuronosyl-(1->2)-glucuronide | flavones | 1.97E+01 | up |
| Hmpp003270 | Luteolin-4'-O-glucoside | flavones | 1.91E+01 | up |
| MWSHY0031 | Apigenin-7-O-glucuronide | flavones | 1.70E+01 | up |
| MWSHY0050 | Kaempferol-3-O-rutinoside | flavonols | 1.62E+01 | up |
| pmb0606 | Chrysoeriol-7-O-glucuronide | flavones | 1.50E+01 | up |
| Lmfn003760 | Quercetin-4'-O-glucuronide | flavonols | 1.38E+01 | up |
| Lmmp003783 | Quercetin-3-O-glucuronide | flavonols | 1.35E+01 | up |
| pma0791 | Naringenin-7-O-(6''-malonyl) glucoside | flavanones | 1.31E+01 | up |
| MWSHY0186 | Isosakuranetin (5,7-Dihydroxy-4'-methoxy flavanone) | flavanones | 1.19E+01 | up |
| MWSHY0145 | Eriodictyol (5,7,3',4'-Tetrahydroxy flavanone) | flavanones | 1.18E+01 | up |
| Lmpn006208 | 8-Methoxykaempferol-7-O-rhamnoside | flavonols | -2.39E+00 | down |
| pmb3042 | Tricin-5-O-glucoside | flavones | -2.42E+00 | down |
| mws2118 | Phloretin-2'-O-glucoside | chalcones | -2.88E+00 | down |
| HJAP120 | Rhamnetin-3-O-rutinoside | flavonols | -2.98E+00 | down |
| pmb3041 | Tricin-7-O-saccharic acid | flavones | -3.00E+00 | down |
| HJAP011 | Chrysoeriol-8-C-glucoside | flavones | -3.11E+00 | down |
| HJAP154 | Galloylisorhamnetin | flavonols | -3.51E+00 | down |
| Hmmp002121 | Isorhamnetin-3-O-gallate | flavonols | -3.68E+00 | down |
| Lmqp002170 | Kaempferol-3-O-sophorotrioside | flavonols | -9.91E+00 | down |
| Lmjp003655 | 6-C-Methyl Kaempferol-3-glucoside | Flavones | -1.38E+01 | down |
| Ko_ID | Sig_compound | All_compound |
| ko00941 | 7 | 12 |
| ko01100 | 7 | 9 |
| ko01100 | 4 | 5 |
| ko00944 | 9 | 16 |
| ko00942 | 2 | 2 |
| ko00943 | 2 | 2 |
| KEGG_map | Description | P-value_meta | Count_meta | P-value_gene | Count_gene |
|---|---|---|---|---|---|
| ko01100 | Metabolic pathways | 0.3808 | 4 | 0.0000 | 4708 |
| ko01110 | Biosynthesis of secondary metabolites | 0.2583 | 7 | 0.0000 | 2625 |
| ko00941 | Flavonoid biosynthesis | 0.8027 | 7 | 0.0000 | 141 |
| ko00942 | Anthocyanin biosynthesis | 0.3931 | 2 | 0.0031 | 36 |
| ko00943 | Isoflavonoid biosynthesis | 0.3931 | 2 | 0.0140 | 64 |
| ko00944 | Flavone and flavonol biosynthesis | 0.8931 | 9 | 0.2014 | 11 |
| Genes | Metabolites | Description_meta |
| Cluster-51396.2 | Lmmp003783 | Quercetin-3-O-glucuronide |
| Cluster-7658.8 | Lmmn004625 | Dihydrokaempferol-7-O-glucoside |
| Cluster-16235.0 | HJN086 | Eriodictyol-3'-O-glucoside |
| Cluster-52100.1 | pmb2976 | Chrysoeriol-8-C-arabinoside-7-O-sophoroside |
| Cluster-25073.0 | Hmpp003270 | Luteolin-4'-O-glucoside |
| Cluster-42022.2 | MWSHY0031 | Apigenin-7-O-glucuronide |
| Cluster-53874.0 | Lmzn001572 | Scutellarein-7-O-glucuronosyl-(1->2)-glucuronide |
| Cluster-1116.5 | pma0791 | Naringenin-7-O-(6''-malonyl) glucoside |
| Cluster-1676.2 | MWSHY0050 | Kaempferol-3-O-rutinoside (Nicotiflorin) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





