Submitted:
18 July 2023
Posted:
21 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. SARS-CoV-2 Sample Collection and Selection
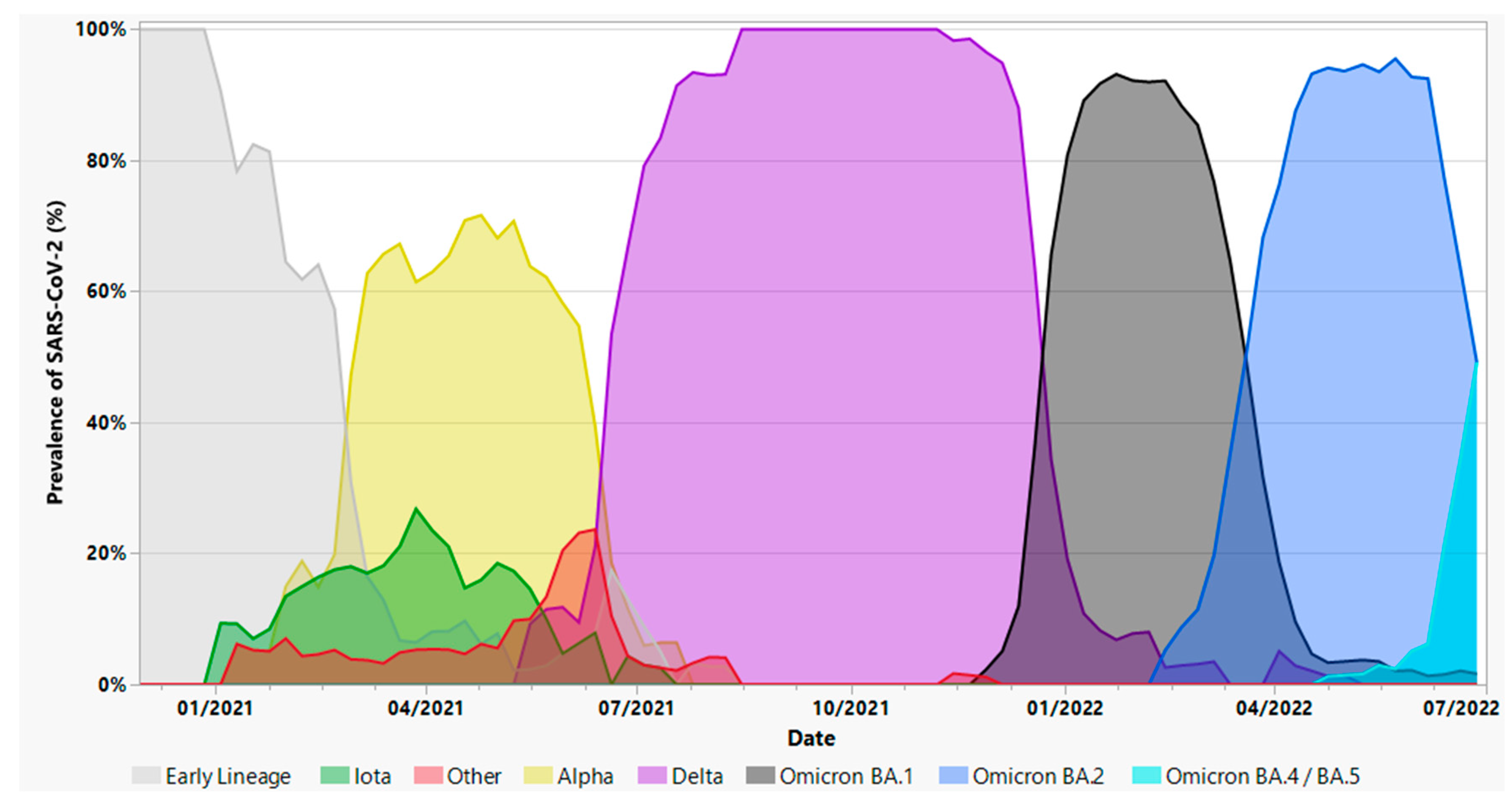
3.2. Variant Screening of SARS-CoV-2-Positive Nasopharyngeal Swabs
3.3. Whole Genome Sequencing of SARS-CoV-2-Positive Nasopharyngeal Swabs
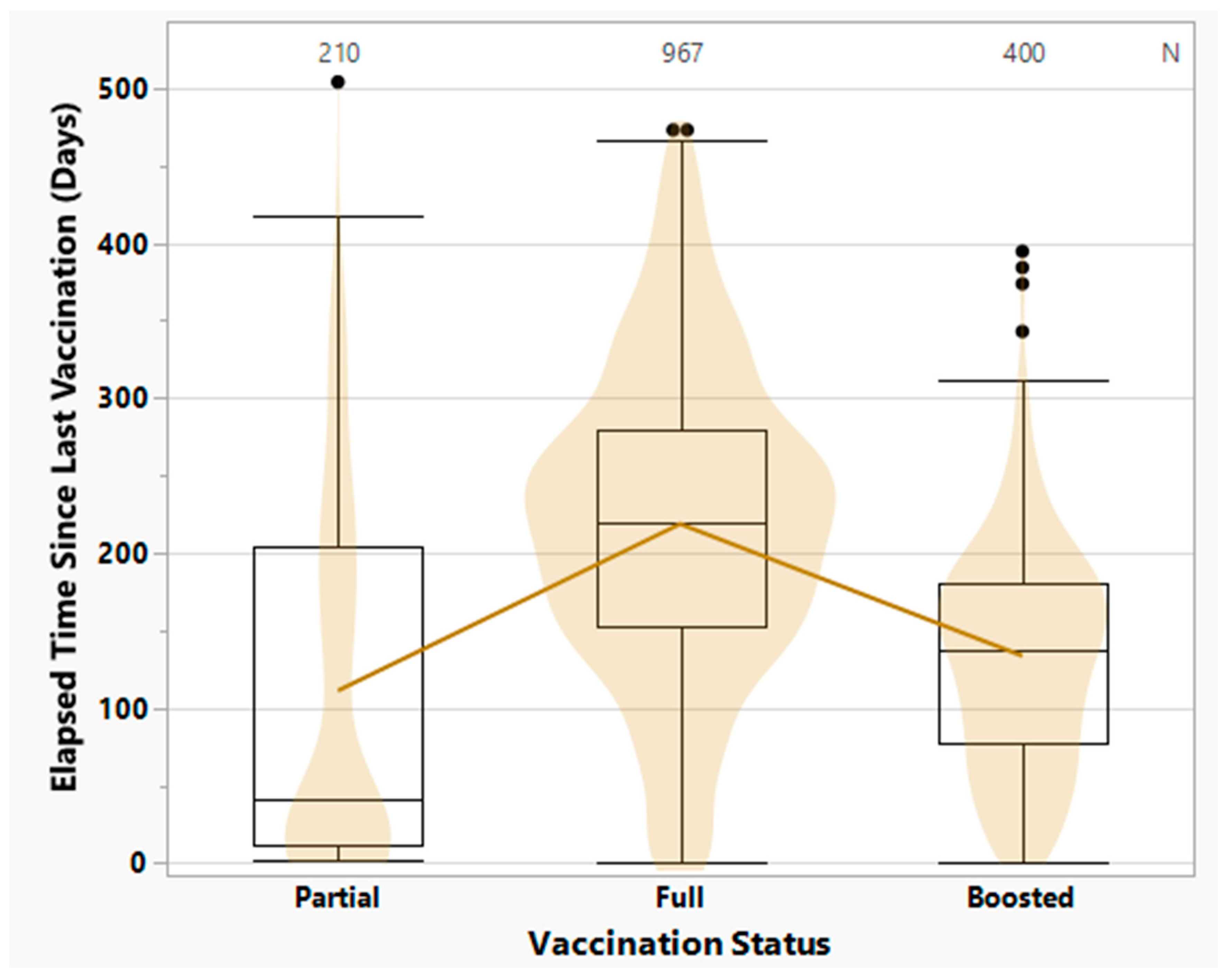
3.4. COVID-19 Vaccination Status and Vaccine-Breakthrough Infections
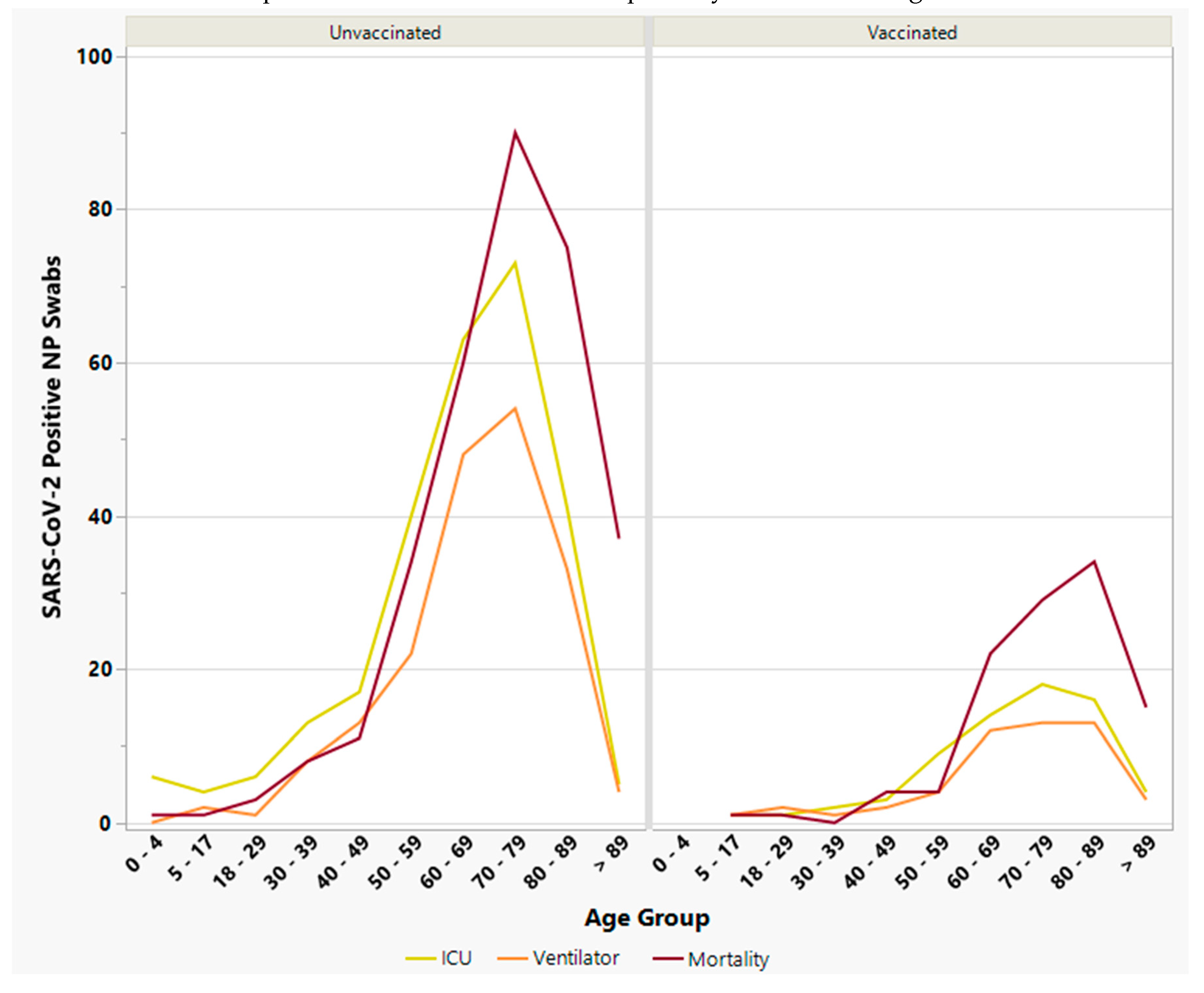
3.5. Severe Clinical Outcomes among Vaccinated vs. Unvaccinated Individuals
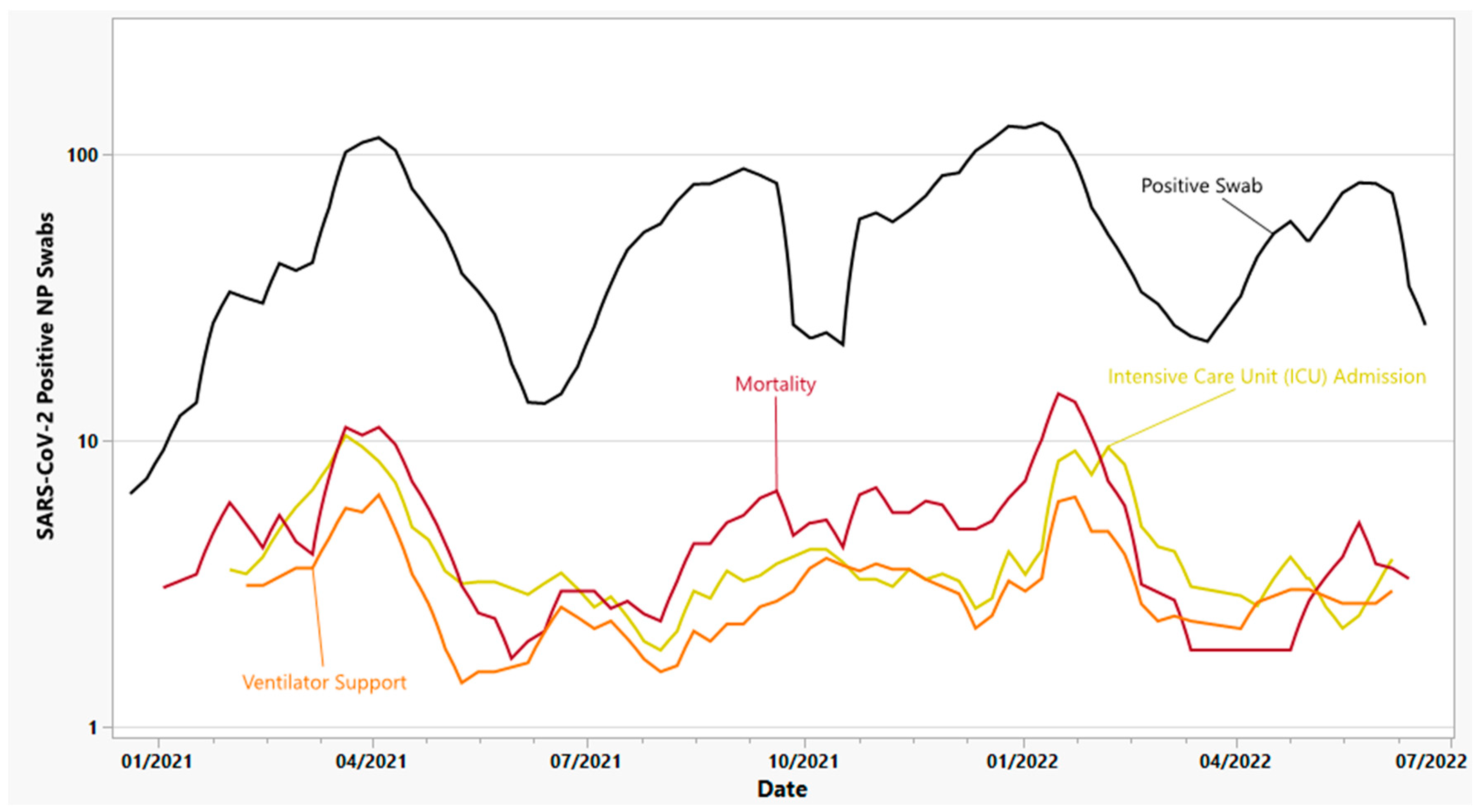
3.5.1. Intensive Care Unit (ICU) Admission Individuals
3.5.2. Ventilator Support
3.5.3. Mortality
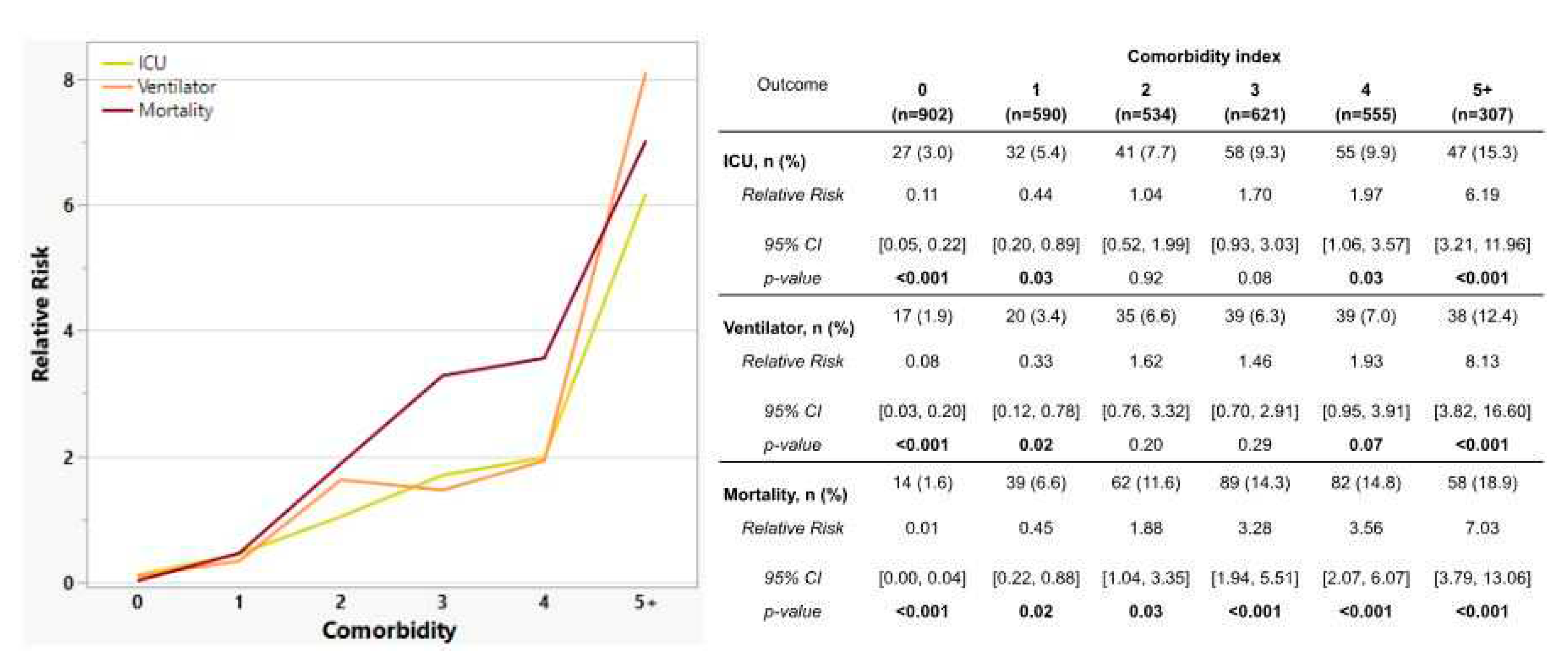
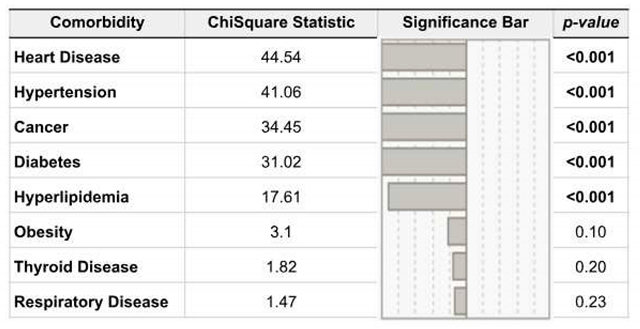
3.6. Risk Factors and Comorbidities among Vaccinated vs. Unvaccinated Individuals
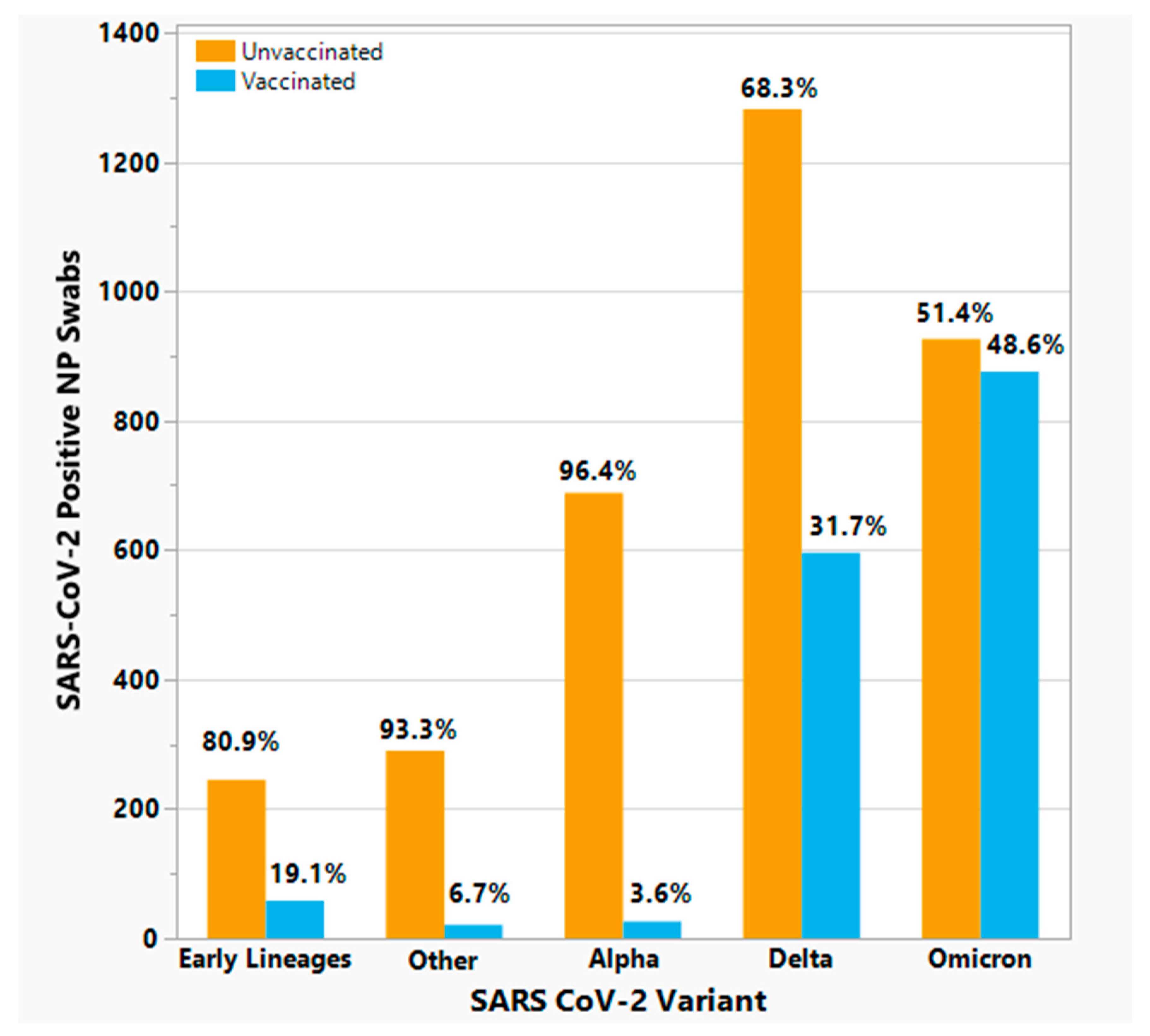
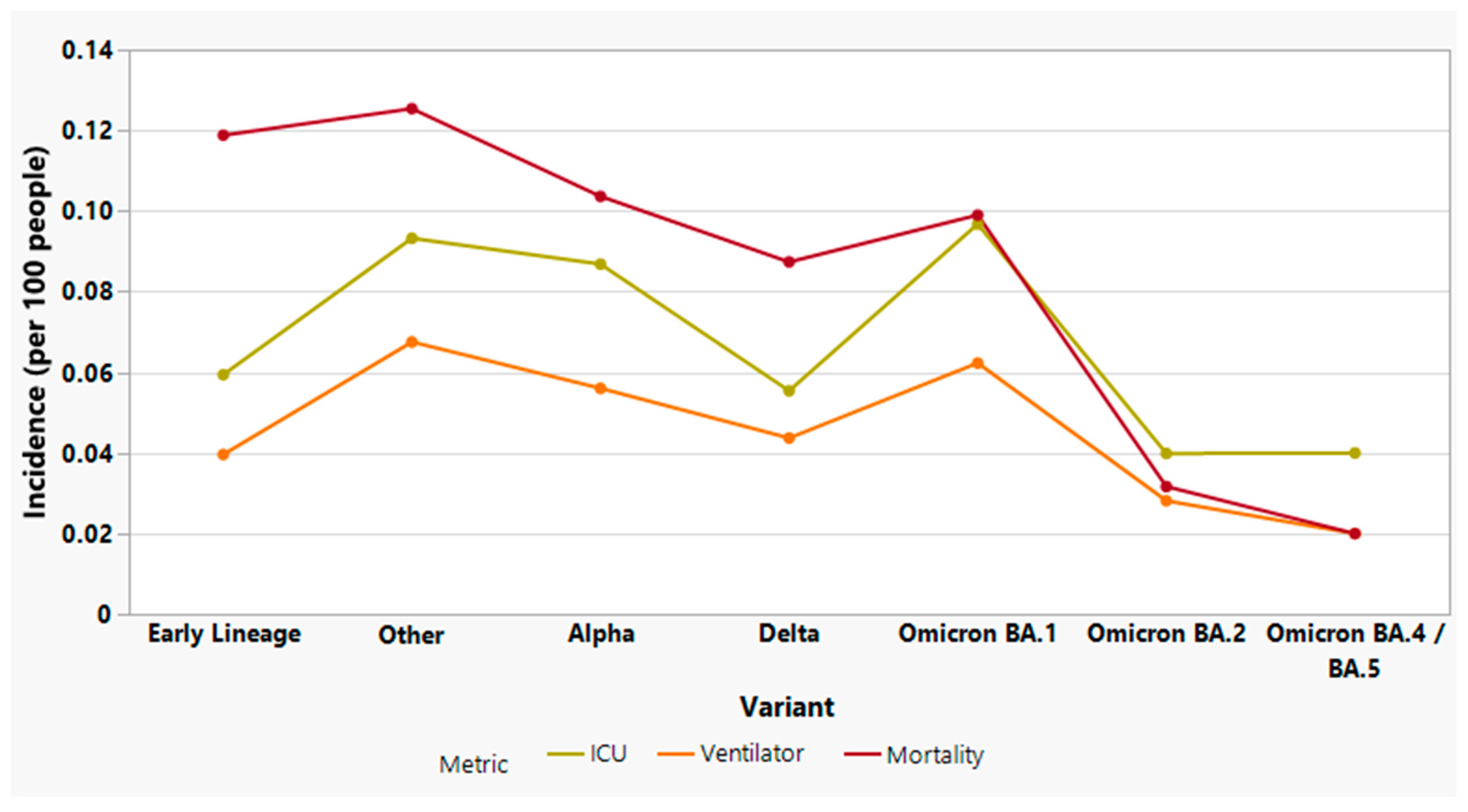
3.7. Analysis of SARS-CoV-2 variants and severe clinical
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Joseph, S.; Kutty Narayanan, A. , Covid 19-the 21st Century Pandemic: The Novel Coronavirus Outbreak and the Treatment Strategies. Adv Pharm Bull 2022, 12, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Agnihotri, K.; Tripathi, A.; Mukherjee, S.; Agnihotri, N.; Gupta, G. , COVID-19: A Worldwide, Zoonotic, Pandemic Outbreak. Altern Ther Health Med 2020, 26, 56–64. [Google Scholar] [PubMed]
- COVID-19 Mortality by State. https://www.cdc.gov/nchs/pressroom/sosmap/covid19_mortality_final/COVID19.htm.
- New Jersey COVID-19 Information Hub. https://covid19.nj.gov/.
- About Hackensack Meridian Health. https://www.hackensackmeridianhealth.org/en/about-us.
- Erminio, V. Coronavirus in New Jersey: A timeline of the outbreak. https://www.nj.com/coronavirus/2020/03/coronavirus-in-new-jersey-a-timeline-of-the-outbreak.html.
- Center for Discovery & Innovation. https://hmh-cdi.org/.
- Zhao, Y.; Cunningham, M. H.; Mediavilla, J. R.; Park, S.; Fitzgerald, S.; Ahn, H. S.; Li, X.; Zhan, C.; Hong, T.; Munk, G.; Chow, K. F.; Perlin, D. S. , Diagnosis, clinical characteristics, and outcomes of COVID-19 patients from a large healthcare system in northern New Jersey. Scientific reports 2021, 11, 4389. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lee, A.; Composto, K.; Cunningham, M. H.; Mediavilla, J. R.; Fennessey, S.; Corvelo, A.; Chow, K. F.; Zody, M.; Chen, L.; Kreiswirth, B. N.; Perlin, D. S. , A novel diagnostic test to screen SARS-CoV-2 variants containing E484K and N501Y mutations. Emerging microbes & infections 2021, 10, 994–997. [Google Scholar]
- Polack, F. P.; Thomas, S. J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J. L.; Pérez Marc, G.; Moreira, E. D.; Zerbini, C.; Bailey, R.; Swanson, K. A.; Roychoudhury, S.; Koury, K.; Li, P.; Kalina, W. V.; Cooper, D.; Frenck, R. W., Jr.; Hammitt, L. L.; Türeci, Ö.; Nell, H.; Schaefer, A.; Ünal, S.; Tresnan, D. B.; Mather, S.; Dormitzer, P. R.; Şahin, U.; Jansen, K. U.; Gruber, W. C. , Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. The New England journal of medicine 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Anderson, E. J.; Rouphael, N. G.; Widge, A. T.; Jackson, L. A.; Roberts, P. C.; Makhene, M.; Chappell, J. D.; Denison, M. R.; Stevens, L. J.; Pruijssers, A. J.; McDermott, A. B.; Flach, B.; Lin, B. C.; Doria-Rose, N. A.; O'Dell, S.; Schmidt, S. D.; Corbett, K. S.; Swanson, P. A., 2nd; Padilla, M.; Neuzil, K. M.; Bennett, H.; Leav, B.; Makowski, M.; Albert, J.; Cross, K.; Edara, V. V.; Floyd, K.; Suthar, M. S.; Martinez, D. R.; Baric, R.; Buchanan, W.; Luke, C. J.; Phadke, V. K.; Rostad, C. A.; Ledgerwood, J. E.; Graham, B. S.; Beigel, J. H. , Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. The New England journal of medicine 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Stephenson, K. E.; Le Gars, M.; Sadoff, J.; de Groot, A. M.; Heerwegh, D.; Truyers, C.; Atyeo, C.; Loos, C.; Chandrashekar, A.; McMahan, K.; Tostanoski, L. H.; Yu, J.; Gebre, M. S.; Jacob-Dolan, C.; Li, Z.; Patel, S.; Peter, L.; Liu, J.; Borducchi, E. N.; Nkolola, J. P.; Souza, M.; Tan, C. S.; Zash, R.; Julg, B.; Nathavitharana, R. R.; Shapiro, R. L.; Azim, A. A.; Alonso, C. D.; Jaegle, K.; Ansel, J. L.; Kanjilal, D. G.; Guiney, C. J.; Bradshaw, C.; Tyler, A.; Makoni, T.; Yanosick, K. E.; Seaman, M. S.; Lauffenburger, D. A.; Alter, G.; Struyf, F.; Douoguih, M.; Van Hoof, J.; Schuitemaker, H.; Barouch, D. H. , Immunogenicity of the Ad26.COV2.S Vaccine for COVID-19. Jama 2021, 325, 1535–1544. [Google Scholar] [CrossRef]
- Mathema, B.; Chen, L.; Chow, K. F.; Zhao, Y.; Zody, M. C.; Mediavilla, J. R.; Cunningham, M. H.; Composto, K.; Lee, A.; Oschwald, D. M.; Germer, S.; Fennessey, S.; Patel, K.; Wilson, D.; Cassell, A.; Pascual, L.; Ip, A.; Corvelo, A.; Dar, S.; Kramer, Y.; Maniatis, T.; Perlin, D. S.; Kreiswirth, B. N. , Postvaccination SARS-COV-2 among Health Care Workers in New Jersey: A Genomic Epidemiological Study. Microbiology spectrum 2021, 9, e0188221. [Google Scholar] [CrossRef]
- González-Candelas, F.; Shaw, M. A.; Phan, T.; Kulkarni-Kale, U.; Paraskevis, D.; Luciani, F.; Kimura, H.; Sironi, M. , One year into the pandemic: Short-term evolution of SARS-CoV-2 and emergence of new lineages. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases 2021, 92, 104869. [Google Scholar] [CrossRef]
- Li, J.; Lai, S.; Gao, G. F.; Shi, W. , The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef]
- Tosta, S.; Moreno, K.; Schuab, G.; Fonseca, V.; Segovia, F. M. C.; Kashima, S.; Elias, M. C.; Sampaio, S. C.; Ciccozzi, M.; Alcantara, L. C. J.; Slavov, S. N.; Lourenço, J.; Cella, E.; Giovanetti, M. , Global SARS-CoV-2 genomic surveillance: What we have learned (so far). Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases 2023, 108, 105405. [Google Scholar] [CrossRef]
- Boehm, E.; Kronig, I.; Neher, R. A.; Eckerle, I.; Vetter, P.; Kaiser, L. , Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2021, 27, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Sharma, P.; Singh, M.; Kumar, M.; Ethayathulla, A. S.; Kaur, P. , Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cellular and molecular life sciences : CMLS 2021, 78, 7967–7989. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, I.; Pravica, V.; Miljanovic, D.; Cupic, M. , Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Bhola, S.; Thakur, P.; Patel, S. K. S.; Kulshrestha, S.; Ratho, R. K.; Kumar, P. , Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe. Infection 2022, 50, 309–325. [Google Scholar] [CrossRef]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Haider, N.; Abbasi, A. F.; Jaferi, U.; Prakash, S.; Balendra, V. , The emerging SARS-CoV-2 variants of concern. Therapeutic advances in infectious disease 2021, 8, 20499361211024372. [Google Scholar] [CrossRef]
- Tao, K.; Tzou, P. L.; Nouhin, J.; Gupta, R. K.; de Oliveira, T.; Kosakovsky Pond, S. L.; Fera, D.; Shafer, R. W. , The biological and clinical significance of emerging SARS-CoV-2 variants. Nature reviews. Genetics 2021, 22, 757–773. [Google Scholar] [CrossRef]
- Thye, A. Y.; Law, J. W.; Pusparajah, P.; Letchumanan, V.; Chan, K. G.; Lee, L. H. , Emerging SARS-CoV-2 Variants of Concern (VOCs): An Impending Global Crisis. Biomedicines 2021, 9. [Google Scholar] [CrossRef]
- Kannan, S.; Shaik Syed Ali, P.; Sheeza, A. , Omicron (B.1.1.529) - variant of concern - molecular profile and epidemiology: a mini review. European review for medical and pharmacological sciences 2021, 25, 8019–8022. [Google Scholar]
- Saxena, S. K.; Kumar, S.; Ansari, S.; Paweska, J. T.; Maurya, V. K.; Tripathi, A. K.; Abdel-Moneim, A. S. , Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. Journal of medical virology 2022, 94, 1738–1744. [Google Scholar] [CrossRef]
- Esper, F. P.; Adhikari, T. M.; Tu, Z. J.; Cheng, Y. W.; El-Haddad, K.; Farkas, D. H.; Bosler, D.; Rhoads, D.; Procop, G. W.; Ko, J. S.; Jehi, L.; Li, J.; Rubin, B. P. , Alpha to Omicron: Disease Severity and Clinical Outcomes of Major SARS-CoV-2 Variants. The Journal of infectious diseases 2023, 227, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Faust, J. S.; Du, C.; Liang, C.; Mayes, K. D.; Renton, B.; Panthagani, K.; Krumholz, H. M. , Excess Mortality in Massachusetts During the Delta and Omicron Waves of COVID-19. Jama 2022, 328, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Loucera, C.; Perez-Florido, J.; Casimiro-Soriguer, C. S.; Ortuño, F. M.; Carmona, R.; Bostelmann, G.; Martínez-González, L. J.; Muñoyerro-Muñiz, D.; Villegas, R.; Rodriguez-Baño, J.; Romero-Gomez, M.; Lorusso, N.; Garcia-León, J.; Navarro-Marí, J. M.; Camacho-Martinez, P.; Merino-Diaz, L.; Salazar, A.; Viñuela, L.; The Andalusian Covid-Sequencing, I.; Lepe, J. A.; Garcia, F.; Dopazo, J. , Assessing the Impact of SARS-CoV-2 Lineages and Mutations on Patient Survival. Viruses 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Wrenn, J. O.; Pakala, S. B.; Vestal, G.; Shilts, M. H.; Brown, H. M.; Bowen, S. M.; Strickland, B. A.; Williams, T.; Mallal, S. A.; Jones, I. D.; Schmitz, J. E.; Self, W. H.; Das, S. R. , COVID-19 severity from Omicron and Delta SARS-CoV-2 variants. Influenza and other respiratory viruses 2022, 16, 832–836. [Google Scholar] [CrossRef] [PubMed]
- DeCuir, J.; Surie, D.; Zhu, Y.; Gaglani, M.; Ginde, A. A.; Douin, D. J.; Talbot, H. K.; Casey, J. D.; Mohr, N. M.; McNeal, T.; Ghamande, S.; Gibbs, K. W.; Files, D. C.; Hager, D. N.; Phan, M.; Prekker, M. E.; Gong, M. N.; Mohamed, A.; Johnson, N. J.; Steingrub, J. S.; Peltan, I. D.; Brown, S. M.; Martin, E. T.; Monto, A. S.; Khan, A.; Bender, W. S.; Duggal, A.; Wilson, J. G.; Qadir, N.; Chang, S. Y.; Mallow, C.; Kwon, J. H.; Exline, M. C.; Lauring, A. S.; Shapiro, N. I.; Columbus, C.; Gottlieb, R.; Vaughn, I. A.; Ramesh, M.; Lamerato, L. E.; Safdar, B.; Halasa, N.; Chappell, J. D.; Grijalva, C. G.; Baughman, A.; Womack, K. N.; Rhoads, J. P.; Hart, K. W.; Swan, S. A.; Lewis, N.; McMorrow, M. L.; Self, W. H.; Network, I. V. Y. , Effectiveness of Monovalent mRNA COVID-19 Vaccination in Preventing COVID-19-Associated Invasive Mechanical Ventilation and Death Among Immunocompetent Adults During the Omicron Variant Period - IVY Network, 19 U.S. States, February 1, 2022-January 31, 2023. MMWR. Morbidity and mortality weekly report 2023, 72, 463–468. [Google Scholar]
- Berry, D. A.; Ip, A.; Lewis, B. E.; Berry, S. M.; Berry, N. S.; MrKulic, M.; Gadalla, V.; Sat, B.; Wright, K.; Serna, M.; Unawane, R.; Trpeski, K.; Koropsak, M.; Kaur, P.; Sica, Z.; McConnell, A.; Bednarz, U.; Marafelias, M.; Goy, A. H.; Pecora, A. L.; Sawczuk, I. S.; Goldberg, S. L. , Development and validation of a prognostic 40-day mortality risk model among hospitalized patients with COVID-19. PloS one 2021, 16, e0255228. [Google Scholar] [CrossRef]
- Fiore, M. C.; Smith, S. S.; Adsit, R. T.; Bolt, D. M.; Conner, K. L.; Bernstein, S. L.; Eng, O. D.; Lazuk, D.; Gonzalez, A.; Jorenby, D. E.; D'Angelo, H.; Kirsch, J. A.; Williams, B.; Nolan, M. B.; Hayes-Birchler, T.; Kent, S.; Kim, H.; Piasecki, T. M.; Slutske, W. S.; Lubanski, S.; Yu, M.; Suk, Y.; Cai, Y.; Kashyap, N.; Mathew, J. P.; McMahan, G.; Rolland, B.; Tindle, H. A.; Warren, G. W.; An, L. C.; Boyd, A. D.; Brunzell, D. H.; Carrillo, V.; Chen, L. S.; Davis, J. M.; Dilip, D.; Ellerbeck, E. F.; Iturrate, E.; Jose, T.; Khanna, N.; King, A.; Klass, E.; Newman, M.; Shoenbill, K. A.; Tong, E.; Tsoh, J. Y.; Wilson, K. M.; Theobald, W. E.; Baker, T. B. , The first 20 months of the COVID-19 pandemic: Mortality, intubation and ICU rates among 104,590 patients hospitalized at 21 United States health systems. PloS one 2022, 17, e0274571. [Google Scholar] [CrossRef]
- Gupta, S.; Hayek, S. S.; Wang, W.; Chan, L.; Mathews, K. S.; Melamed, M. L.; Brenner, S. K.; Leonberg-Yoo, A.; Schenck, E. J.; Radbel, J.; Reiser, J.; Bansal, A.; Srivastava, A.; Zhou, Y.; Sutherland, A.; Green, A.; Shehata, A. M.; Goyal, N.; Vijayan, A.; Velez, J. C. Q.; Shaefi, S.; Parikh, C. R.; Arunthamakun, J.; Athavale, A. M.; Friedman, A. N.; Short, S. A. P.; Kibbelaar, Z. A.; Abu Omar, S.; Admon, A. J.; Donnelly, J. P.; Gershengorn, H. B.; Hernán, M. A.; Semler, M. W.; Leaf, D. E. , Factors Associated With Death in Critically Ill Patients With Coronavirus Disease 2019 in the US. JAMA internal medicine 2020, 180, 1436–1447. [Google Scholar] [CrossRef]
- Lipsitch, M.; Krammer, F.; Regev-Yochay, G.; Lustig, Y.; Balicer, R. D. , SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nature reviews. Immunology 2022, 22, 57–65. [Google Scholar] [CrossRef]
- Vet, J. A.; Van der Rijt, B. J.; Blom, H. J. , Molecular beacons: colorful analysis of nucleic acids. Expert review of molecular diagnostics 2002, 2, 77–86. [Google Scholar] [CrossRef]
- Mathema, B.; Chen, L.; Wang, P.; Cunningham, M. H.; Mediavilla, J. R.; Chow, K. F.; Luo, Y.; Zhao, Y.; Composto, K.; Zuckerman, J.; Zody, M. C.; Wilson, N.; Lee, A.; Oschwald, D. M.; Liu, L.; Iketani, S.; Germer, S.; Fennessey, S.; Wang, M.; Kramer, Y.; Toole, P.; Maniatis, T.; Ho, D. D.; Perlin, D. S.; Kreiswirth, B. N. , Genomic Epidemiology and Serology Associated with a SARS-CoV-2 R.1 Variant Outbreak in New Jersey. mBio 2022, 13, e0214122. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Megill, C.; Bell, S. M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R. A. , Nextstrain: real-time tracking of pathogen evolution. Bioinformatics (Oxford, England) 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- O'Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J. T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; Yeats, C.; du Plessis, L.; Maloney, D.; Medd, N.; Attwood, S. W.; Aanensen, D. M.; Holmes, E. C.; Pybus, O. G.; Rambaut, A. , Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus evolution 2021, 7, veab064. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. , GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin, 2017; 22. [Google Scholar]
- Data Definitions for COVID-19 Vaccinations in the United States. https://www.cdc.gov/coronavirus/2019- 184 ncov/vaccines/reporting-vaccinations.html.
- Washington, N. L.; White, S.; Schiabor-Barrett, K. M.; E.T.; Bolze, A.; Lu, J. T., S gene dropout patterns in SARS-CoV-2 tests suggest spread of the H69del/V70del mutation in the US. medRxiv : the preprint server for health sciences 2020, 2020.12.24.20248814. [CrossRef]
- Metzger, C.; Lienhard, R.; Seth-Smith, H. M. B.; Roloff, T.; Wegner, F.; Sieber, J.; Bel, M.; Greub, G.; Egli, A. , PCR performance in the SARS-CoV-2 Omicron variant of concern? Swiss medical weekly 2021, 151, w30120. [Google Scholar] [CrossRef] [PubMed]
- Robishaw, J. D.; Alter, S. M.; Solano, J. J.; Shih, R. D.; DeMets, D. L.; Maki, D. G.; Hennekens, C. H. , Genomic surveillance to combat COVID-19: challenges and opportunities. The Lancet. Microbe 2021, 2, e481–e484. [Google Scholar] [CrossRef]
- van Dorp, L.; Houldcroft, C. J.; Richard, D.; Balloux, F. , COVID-19, the first pandemic in the post-genomic era. Current opinion in virology 2021, 50, 40–48. [Google Scholar] [CrossRef]
- Steele, M. K.; Couture, A.; Reed, C.; Iuliano, D.; Whitaker, M.; Fast, H.; Hall, A. J.; MacNeil, A.; Cadwell, B.; Marks, K. J.; Silk, B. J. , Estimated Number of COVID-19 Infections, Hospitalizations, and Deaths Prevented Among Vaccinated Persons in the US, December 2020 to September 2021. JAMA network open 2022, 5, e2220385. [Google Scholar] [CrossRef]
- Tatsi, E. B.; Filippatos, F.; Michos, A. , SARS-CoV-2 variants and effectiveness of vaccines: a review of current evidence. Epidemiology and infection 2021, 149, e237. [Google Scholar] [CrossRef]
- Yaqinuddin, A.; Shafqat, A.; Kashir, J.; Alkattan, K. , Effect of SARS-CoV-2 Mutations on the Efficacy of Antibody Therapy and Response to Vaccines. Vaccines 2021, 9. [Google Scholar] [CrossRef]
- Adams, K.; Rhoads, J. P.; Surie, D.; Gaglani, M.; Ginde, A. A.; McNeal, T.; Ghamande, S.; Huynh, D.; Talbot, H. K.; Casey, J. D.; Mohr, N. M.; Zepeski, A.; Shapiro, N. I.; Gibbs, K. W.; Files, D. C.; Hicks, M.; Hager, D. N.; Ali, H.; Prekker, M. E.; Frosch, A. E.; Exline, M. C.; Gong, M. N.; Mohamed, A.; Johnson, N. J.; Srinivasan, V.; Steingrub, J. S.; Peltan, I. D.; Brown, S. M.; Martin, E. T.; Monto, A. S.; Lauring, A. S.; Khan, A.; Hough, C. L.; Busse, L. W.; Ten Lohuis, C. C.; Duggal, A.; Wilson, J. G.; Gordon, A. J.; Qadir, N.; Chang, S. Y.; Mallow, C.; Rivas, C.; Babcock, H. M.; Kwon, J. H.; Chappell, J. D.; Halasa, N.; Grijalva, C. G.; Rice, T. W.; Stubblefield, W. B.; Baughman, A.; Lindsell, C. J.; Hart, K. W.; Lester, S. N.; Thornburg, N. J.; Park, S.; McMorrow, M. L.; Patel, M. M.; Tenforde, M. W.; Self, W. H., Vaccine Effectiveness of Primary Series and Booster Doses against Omicron Variant COVID-19-Associated Hospitalization in the United States. medRxiv : the preprint server for health sciences 2022. [CrossRef]
- DeSilva, M. B.; Mitchell, P. K.; Klein, N. P.; Dixon, B. E.; Tenforde, M. W.; Thompson, M. G.; Naleway, A. L.; Grannis, S. J.; Ong, T. C.; Natarajan, K.; Reese, S. E.; Zerbo, O.; Kharbanda, A. B.; Patel, P.; Stenehjem, E.; Raiyani, C.; Irving, S. A.; Fadel, W. F.; Rao, S.; Han, J.; Reynolds, S.; Davis, J. M.; Lewis, N.; McEvoy, C.; Dickerson, M.; Dascomb, K.; Valvi, N. R.; Barron, M. A.; Goddard, K.; Vazquez-Benitez, G.; Grisel, N.; Mamawala, M.; Embi, P. J.; Fireman, B.; Essien, I. J.; Griggs, E. P.; Arndorfer, J.; Gaglani, M. , Protection of Two and Three mRNA Vaccine Doses Against Severe Outcomes Among Adults Hospitalized With COVID-19-VISION Network, August 2021 to March 2022. The Journal of infectious diseases 2023, 227, 961–969. [Google Scholar] [CrossRef]
- Ferdinands, J. M.; Rao, S.; Dixon, B. E.; Mitchell, P. K.; DeSilva, M. B.; Irving, S. A.; Lewis, N.; Natarajan, K.; Stenehjem, E.; Grannis, S. J.; Han, J.; McEvoy, C.; Ong, T. C.; Naleway, A. L.; Reese, S. E.; Embi, P. J.; Dascomb, K.; Klein, N. P.; Griggs, E. P.; Konatham, D.; Kharbanda, A. B.; Yang, D. H.; Fadel, W. F.; Grisel, N.; Goddard, K.; Patel, P.; Liao, I. C.; Birch, R.; Valvi, N. R.; Reynolds, S.; Arndorfer, J.; Zerbo, O.; Dickerson, M.; Murthy, K.; Williams, J.; Bozio, C. H.; Blanton, L.; Verani, J. R.; Schrag, S. J.; Dalton, A. F.; Wondimu, M. H.; Link-Gelles, R.; Azziz-Baumgartner, E.; Barron, M. A.; Gaglani, M.; Thompson, M. G.; Fireman, B. , Waning 2-Dose and 3-Dose Effectiveness of mRNA Vaccines Against COVID-19-Associated Emergency Department and Urgent Care Encounters and Hospitalizations Among Adults During Periods of Delta and Omicron Variant Predominance - VISION Network, 10 States, August 2021-January 2022. MMWR. Morbidity and mortality weekly report 2022, 71, 255–263. [Google Scholar] [PubMed]
- Flacco, M. E.; Acuti Martellucci, C.; Baccolini, V.; De Vito, C.; Renzi, E.; Villari, P.; Manzoli, L. , COVID-19 vaccines reduce the risk of SARS-CoV-2 reinfection and hospitalization: Meta-analysis. Frontiers in medicine 2022, 9, 1023507. [Google Scholar] [CrossRef] [PubMed]
- Havers, F. P.; Pham, H.; Taylor, C. A.; Whitaker, M.; Patel, K.; Anglin, O.; Kambhampati, A. K.; Milucky, J.; Zell, E.; Moline, H. L.; Chai, S. J.; Kirley, P. D.; Alden, N. B.; Armistead, I.; Yousey-Hindes, K.; Meek, J.; Openo, K. P.; Anderson, E. J.; Reeg, L.; Kohrman, A.; Lynfield, R.; Como-Sabetti, K.; Davis, E. M.; Cline, C.; Muse, A.; Barney, G.; Bushey, S.; Felsen, C. B.; Billing, L. M.; Shiltz, E.; Sutton, M.; Abdullah, N.; Talbot, H. K.; Schaffner, W.; Hill, M.; George, A.; Hall, A. J.; Bialek, S. R.; Murthy, N. C.; Murthy, B. P.; McMorrow, M. , COVID-19-Associated Hospitalizations Among Vaccinated and Unvaccinated Adults 18 Years or Older in 13 US States, January 2021 to April 2022. JAMA internal medicine 2022, 182, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Surie, D.; Bonnell, L.; Adams, K.; Gaglani, M.; Ginde, A. A.; Douin, D. J.; Talbot, H. K.; Casey, J. D.; Mohr, N. M.; Zepeski, A.; McNeal, T.; Ghamande, S.; Gibbs, K. W.; Files, D. C.; Hager, D. N.; Shehu, A.; Frosch, A. P.; Erickson, H. L.; Gong, M. N.; Mohamed, A.; Johnson, N. J.; Srinivasan, V.; Steingrub, J. S.; Peltan, I. D.; Brown, S. M.; Martin, E. T.; Khan, A.; Bender, W. S.; Duggal, A.; Wilson, J. G.; Qadir, N.; Chang, S. Y.; Mallow, C.; Rivas, C.; Kwon, J. H.; Exline, M. C.; Lauring, A. S.; Shapiro, N. I.; Halasa, N.; Chappell, J. D.; Grijalva, C. G.; Rice, T. W.; Stubblefield, W. B.; Baughman, A.; Womack, K. N.; Hart, K. W.; Swan, S. A.; Zhu, Y.; DeCuir, J.; Tenforde, M. W.; Patel, M. M.; McMorrow, M. L.; Self, W. H. , Effectiveness of Monovalent mRNA Vaccines Against COVID-19-Associated Hospitalization Among Immunocompetent Adults During BA.1/BA.2 and BA.4/BA.5 Predominant Periods of SARS-CoV-2 Omicron Variant in the United States - IVY Network, 18 States, December 26, 2021-August 31, 2022. MMWR. Morbidity and mortality weekly report 2022, 71, 1327–1334. [Google Scholar]
- Thompson, M. G.; Natarajan, K.; Irving, S. A.; Rowley, E. A.; Griggs, E. P.; Gaglani, M.; Klein, N. P.; Grannis, S. J.; DeSilva, M. B.; Stenehjem, E.; Reese, S. E.; Dickerson, M.; Naleway, A. L.; Han, J.; Konatham, D.; McEvoy, C.; Rao, S.; Dixon, B. E.; Dascomb, K.; Lewis, N.; Levy, M. E.; Patel, P.; Liao, I. C.; Kharbanda, A. B.; Barron, M. A.; Fadel, W. F.; Grisel, N.; Goddard, K.; Yang, D. H.; Wondimu, M. H.; Murthy, K.; Valvi, N. R.; Arndorfer, J.; Fireman, B.; Dunne, M. M.; Embi, P.; Azziz-Baumgartner, E.; Zerbo, O.; Bozio, C. H.; Reynolds, S.; Ferdinands, J.; Williams, J.; Link-Gelles, R.; Schrag, S. J.; Verani, J. R.; Ball, S.; Ong, T. C. , Effectiveness of a Third Dose of mRNA Vaccines Against COVID-19-Associated Emergency Department and Urgent Care Encounters and Hospitalizations Among Adults During Periods of Delta and Omicron Variant Predominance - VISION Network, 10 States, August 2021-January 2022. MMWR. Morbidity and mortality weekly report 2022, 71, 139–145. [Google Scholar]
- Worldometers.info United States COVID - Coronavirus Statistics. https://www.worldometers.info/coronavirus/country/us/.
- New Jersey COVID-19 Dashboard. https://www.nj.gov/health/cd/topics/covid2019_dashboard.shtml.
- Sullivan, S. P. 1 in 500 New Jerseyans have died from COVID-19 since pandemic began, data shows. https://www.nj.com/coronavirus/2020/12/1-in-500-new-jerseyans-have-died-from-covid-19-since-pandemic-began-data-shows.
- Barrett, E. S.; Horton, D. B.; Roy, J.; Gennaro, M. L.; Brooks, A.; Tischfield, J.; Greenberg, P.; Andrews, T.; Jagpal, S.; Reilly, N.; Carson, J. L.; Blaser, M. J.; Panettieri, R. A., Jr. , Prevalence of SARS-CoV-2 infection in previously undiagnosed health care workers in New Jersey, at the onset of the U.S. COVID-19 pandemic. BMC infectious diseases 2020, 20, 853. [Google Scholar] [CrossRef]
- Bhavsar, S. M.; Clouser, K. N.; Gadhavi, J.; Anene, O.; Kaur, R.; Lewis, R.; Naganathan, S.; Michalak, Z.; Chen, C. Q.; Shah, P.; Siu, A.; Ballance, C. , COVID-19 in Pediatrics: Characteristics of Hospitalized Children in New Jersey. Hospital pediatrics 2021, 11, 79–87. [Google Scholar] [CrossRef]
- Donato, M. L.; Park, S.; Baker, M.; Korngold, R.; Morawski, A.; Geng, X.; Tan, M.; Ip, A.; Goldberg, S.; Rowley, S.; Chow, K.; Brown, E.; Zenreich, J.; McKiernan, P.; Buttner, K.; Ullrich, A.; Long, L.; Feinman, R.; Ricourt, A.; Kemp, M.; Vendivil, M.; Suh, H.; Balani, B.; Cicogna, C.; Sebti, R.; Al-Khan, A.; Sperber, S.; Desai, S.; Fanning, S.; Arad, D.; Go, R.; Tam, E.; Rose, K.; Sadikot, S.; Siegel, D.; Gutierrez, M.; Feldman, T.; Goy, A.; Pecora, A.; Biran, N.; Leslie, L.; Gillio, A.; Timmapuri, S.; Boonstra, M.; Singer, S.; Kaur, S.; Richards, E.; Perlin, D. S. , Clinical and laboratory evaluation of patients with SARS-CoV-2 pneumonia treated with high-titer convalescent plasma. JCI insight 2021, 6. [Google Scholar] [CrossRef]
- Samuel, A.; Mechineni, A.; Aronow, W. S.; Ismail, M.; Manickam, R. , A review of the characteristics and outcomes of 900 COVID-19 patients hospitalized at a Tertiary Care Medical Center in New Jersey, USA. Archives of medical sciences. Atherosclerotic diseases 2020, 5, e306–e312. [Google Scholar] [CrossRef]
- Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu/region/us/new-jersey.
- Focosi, D.; Quiroga, R.; McConnell, S.; Johnson, M. C.; Casadevall, A. , Convergent Evolution in SARS-CoV-2 Spike Creates a Variant Soup from Which New COVID-19 Waves Emerge. International journal of molecular sciences 2023, 24. [Google Scholar] [CrossRef]
- Chrysostomou, A. C.; Aristokleous, A.; Rodosthenous, J. H.; Christodoulou, C.; Stathi, G.; Kostrikis, L. G., Detection of Circulating SARS-CoV-2 Variants of Concern (VOCs) Using a Multiallelic Spectral Genotyping Assay. Life (Basel, Switzerland) 2023, 13. [CrossRef]
- Jiang, W.; Ji, W.; Zhang, Y.; Xie, Y.; Chen, S.; Jin, Y.; Duan, G. , An Update on Detection Technologies for SARS-CoV-2 Variants of Concern. Viruses 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Xu, Y.; Zheng, R.; Zeng, X.; Chen, Z.; Lin, S.; Xia, Z.; Liao, Y.; Zhang, Y.; Li, Q. , Accessible and Adaptable Multiplexed Real-Time PCR Approaches to Identify SARS-CoV-2 Variants of Concern. Microbiology spectrum 2022, 10, e0322222. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.; Peacock, T. P.; Harvey, W. T.; Hughes, J.; Wright, D. W.; Willett, B. J.; Thomson, E.; Gupta, R. K.; Peacock, S. J.; Robertson, D. L.; Carabelli, A. M. , SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nature reviews. Microbiology 2023, 21, 112–124. [Google Scholar] [CrossRef]
- Murano, K.; Guo, Y.; Siomi, H. , The emergence of SARS-CoV-2 variants threatens to decrease the efficacy of neutralizing antibodies and vaccines. Biochemical Society transactions 2021, 49, 2879–2890. [Google Scholar] [CrossRef] [PubMed]
- Sabbatucci, M.; Vitiello, A.; Clemente, S.; Zovi, A.; Boccellino, M.; Ferrara, F.; Cimmino, C.; Langella, R.; Ponzo, A.; Stefanelli, P.; Rezza, G. , Omicron variant evolution on vaccines and monoclonal antibodies. Inflammopharmacology 2023, 1–10. [Google Scholar] [CrossRef]
- Wahid, M.; Jawed, A.; Mandal, R. K.; Dailah, H. G.; Janahi, E. M.; Dhama, K.; Somvanshi, P.; Haque, S. , Variants of SARS-CoV-2, their effects on infection, transmission and neutralization by vaccine-induced antibodies. European review for medical and pharmacological sciences 2021, 25, 5857–5864. [Google Scholar]
- Banada, P.; Green, R.; Banik, S.; Chopoorian, A.; Streck, D.; Jones, R.; Chakravorty, S.; Alland, D. , A Simple Reverse Transcriptase PCR Melting-Temperature Assay To Rapidly Screen for Widely Circulating SARS-CoV-2 Variants. Journal of clinical microbiology 2021, 59, e0084521. [Google Scholar] [CrossRef]
- Chung, H. Y.; Jian, M. J.; Chang, C. K.; Lin, J. C.; Yeh, K. M.; Chen, C. W.; Hsieh, S. S.; Hung, K. S.; Tang, S. H.; Perng, C. L.; Chang, F. Y.; Wang, C. H.; Shang, H. S. , Emergency SARS-CoV-2 Variants of Concern: Novel Multiplex Real-Time RT-PCR Assay for Rapid Detection and Surveillance. Microbiology spectrum 2022, 10, e0251321. [Google Scholar] [CrossRef]
- Dikdan, R. J.; Marras, S. A. E.; Field, A. P.; Brownlee, A.; Cironi, A.; Hill, D. A.; Tyagi, S. , Multiplex PCR Assays for Identifying all Major Severe Acute Respiratory Syndrome Coronavirus 2 Variants. The Journal of molecular diagnostics : JMD 2022, 24, 309–319. [Google Scholar] [CrossRef]
- Moisan, A.; Soares, A.; De Oliveira, F.; Alessandri-Gradt, E.; Lecoquierre, F.; Fourneaux, S.; Plantier, J. C.; Gueudin, M. , Evaluation of Analytical and Clinical Performance and Usefulness in a Real-Life Hospital Setting of Two in-House Real-Time RT-PCR Assays to Track SARS-CoV-2 Variants of Concern. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Bouzid, D.; Visseaux, B.; Kassasseya, C.; Daoud, A.; Fémy, F.; Hermand, C.; Truchot, J.; Beaune, S.; Javaud, N.; Peyrony, O.; Chauvin, A.; Vaittinada Ayar, P.; Bourg, A.; Riou, B.; Marot, S.; Bloom, B.; Cachanado, M.; Simon, T.; Freund, Y. , Comparison of Patients Infected With Delta Versus Omicron COVID-19 Variants Presenting to Paris Emergency Departments : A Retrospective Cohort Study. Annals of internal medicine 2022, 175, 831–837. [Google Scholar] [CrossRef]
- Hause, A. M.; Marquez, P.; Zhang, B.; Myers, T. R.; Gee, J.; Su, J. R.; Parker, C.; Thompson, D.; Panchanathan, S. S.; Shimabukuro, T. T.; Shay, D. K. , COVID-19 mRNA Vaccine Safety Among Children Aged 6 Months-5 Years - United States, June 18, 2022-August 21, 2022. MMWR. Morbidity and mortality weekly report 2022, 71, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Roy, A. , Determinants of Covid-19 vaccination: Evidence from the US pulse survey. PLOS global public health 2023, 3, e0001927. [Google Scholar] [CrossRef] [PubMed]
- Fisher, K. A.; Nguyen, N.; Fouayzi, H.; Crawford, S.; Singh, S.; Dong, M.; Wittenberg, R.; Mazor, K. M. , From COVID-19 Vaccine Hesitancy to Vaccine Acceptance: Results of a Longitudinal Survey. Public health reports (Washington, D.C. : 1974) 2023, 138, 681–690. [Google Scholar] [CrossRef]
- Zintel, S.; Flock, C.; Arbogast, A. L.; Forster, A.; von Wagner, C.; Sieverding, M., Gender differences in the intention to get vaccinated against COVID-19: a systematic review and meta-analysis. Zeitschrift fur Gesundheitswissenschaften = Journal of public health 2022, 1-25.
- Zhou, Z.; Zhu, Y.; Chu, M. , Role of COVID-19 Vaccines in SARS-CoV-2 Variants. Frontiers in immunology 2022, 13, 898192. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Gao, L.; Zhou, Q.; Yu, K.; Sun, F. , Effectiveness of COVID-19 vaccines against SARS-CoV-2 variants of concern: a systematic review and meta-analysis. BMC medicine 2022, 20, 200. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, Y.; Yu, D.; Du, B.; Cheng, W.; Li, L.; Yu, Z.; Luo, S.; Zhang, Y.; Wang, H.; Zhang, X.; Zhang, W. , A systematic review of Vaccine Breakthrough Infections by SARS-CoV-2 Delta Variant. International journal of biological sciences 2022, 18, 889–900. [Google Scholar] [CrossRef]
- Klann, J. G.; Strasser, Z. H.; Hutch, M. R.; Kennedy, C. J.; Marwaha, J. S.; Morris, M.; Samayamuthu, M. J.; Pfaff, A. C.; Estiri, H.; South, A. M.; Weber, G. M.; Yuan, W.; Avillach, P.; Wagholikar, K. B.; Luo, Y.; Omenn, G. S.; Visweswaran, S.; Holmes, J. H.; Xia, Z.; Brat, G. A.; Murphy, S. N. , Distinguishing Admissions Specifically for COVID-19 From Incidental SARS-CoV-2 Admissions: National Retrospective Electronic Health Record Study. Journal of medical Internet research 2022, 24, e37931. [Google Scholar] [CrossRef]
- Peckham, H.; de Gruijter, N. M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L. R.; Rosser, E. C.; Webb, K.; Deakin, C. T. , Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nature communications 2020, 11, 6317. [Google Scholar] [CrossRef]
- Pradhan, A.; Olsson, P. E. , Sex differences in severity and mortality from COVID-19: are males more vulnerable? Biology of sex differences 2020, 11, 53. [Google Scholar] [CrossRef]
- Chanana, N.; Palmo, T.; Sharma, K.; Kumar, R.; Graham, B. B.; Pasha, Q. , Sex-derived attributes contributing to SARS-CoV-2 mortality. American journal of physiology. Endocrinology and metabolism 2020, 319, E562–e567. [Google Scholar] [CrossRef] [PubMed]
- Thangavel, H.; Dhanyalayam, D.; Lizardo, K.; Oswal, N.; Dolgov, E.; Perlin, D. S.; Nagajyothi, J. F. , Susceptibility of Fat Tissue to SARS-CoV-2 Infection in Female hACE2 Mouse Model. International journal of molecular sciences 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Gadi, N.; Wu, S. C.; Spihlman, A. P.; Moulton, V. R. , What's Sex Got to Do With COVID-19? Gender-Based Differences in the Host Immune Response to Coronaviruses. Frontiers in immunology 2020, 11, 2147. [Google Scholar] [CrossRef] [PubMed]
- Bwire, G. M. , Coronavirus: Why Men are More Vulnerable to Covid-19 Than Women? SN comprehensive clinical medicine 2020, 2, 874–876. [Google Scholar] [CrossRef] [PubMed]
- Janssen (Johnson & Johnson) COVID-19 Vaccine. https://www.cdc.gov/vaccines/covid-19/info-by-product/janssen/index.
- Gupta, A.; Marzook, H.; Ahmad, F. , Comorbidities and clinical complications associated with SARS-CoV-2 infection: an overview. Clinical and experimental medicine 2023, 23, 313–331. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; El Hosseiny, A.; Siam, R. , Assessing and Reassessing the Association of Comorbidities and Coinfections in COVID-19 Patients. Cureus 2023, 15, e36683. [Google Scholar] [CrossRef]
- Darvishi, M.; Nazer, M. R.; Shahali, H.; Nouri, M. , Association of thyroid dysfunction and COVID-19: A systematic review and meta-analysis. Frontiers in endocrinology 2022, 13, 947594. [Google Scholar] [CrossRef]
- Almonte, M.; Au, X. Y.; Ali, M.; Rajabalee, N.; Hasan, S.; Shibre, T.; Li, Y.; Kaplan, A. C. , Association Between COVID-19 Outcomes and Patient Characteristics: A Study in an Inner-City Community Hospital. Cureus 2021, 13, e17255. [Google Scholar] [CrossRef]
- Duerr, R.; Dimartino, D.; Marier, C.; Zappile, P.; Wang, G.; Lighter, J.; Elbel, B.; Troxel, A. B.; Heguy, A. , Dominance of Alpha and Iota variants in SARS-CoV-2 vaccine breakthrough infections in New York City. The Journal of clinical investigation 2021, 131. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, E. J.; Lee, S. W.; Kwon, D. , Review of the early reports of the epidemiological characteristics of the B.1.1.7 variant of SARS-CoV-2 and its spread worldwide. Osong public health and research perspectives 2021, 12, 139–148. [Google Scholar] [CrossRef]
- Manjunath, R.; Gaonkar, S. L.; Saleh, E. A. M.; Husain, K. , A comprehensive review on Covid-19 Omicron (B.1.1.529) variant. Saudi journal of biological sciences 2022, 29, 103372. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Bhattacharya, M.; Nag, S.; Dhama, K.; Chakraborty, C. , A Detailed Overview of SARS-CoV-2 Omicron: Its Sub-Variants, Mutations and Pathophysiology, Clinical Characteristics, Immunological Landscape, Immune Escape, and Therapies. Viruses 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, L. B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R. A. , Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Reviews in medical virology 2022, 32, e2381. [Google Scholar] [CrossRef] [PubMed]
- Alderson, J.; Batchelor, V.; O'Hanlon, M.; Cifuentes, L.; Richter, F. C.; Kopycinski, J. , Overview of approved and upcoming vaccines for SARS-CoV-2: a living review. Oxford open immunology 2021, 2, iqab010. [Google Scholar] [CrossRef]
- Lai, A.; Bergna, A.; Della Ventura, C.; Zehender, G. , Genomic epidemiology and phylogenetics applied to the study of SARS-CoV-2 pandemic. The new microbiologica 2023, 46, 1–8. [Google Scholar]
- Oude Munnink, B. B.; Worp, N.; Nieuwenhuijse, D. F.; Sikkema, R. S.; Haagmans, B.; Fouchier, R. A. M.; Koopmans, M. , The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nature medicine 2021, 27, 1518–1524. [Google Scholar] [CrossRef]
| Variant of Concern | Pango lineage | Spike protein mutations |
|---|---|---|
| Alpha | B.1.1.7 | ΔH69/V70, N501Y |
| Betaa | B.1.351 | E484K, N501Y |
| Gammaa | P.1 (B.1.1.28.1) | E484K, N501Y |
| Delta | B.1.617.2 | L452R, T478K |
| Omicron: | B.1.1.529 | T478K, E484A, N501Y, Y505H |
| BA.1 | B.1.1.529.1 | A67V, ΔH69/V70, T478K, E484A, N501Y, Y505H |
| BA.2 | B.1.1.529.2 | T478K, E484A, N501Y, Y505H |
| BA.2.12.1 | B.1.1.529.2.12.1 | L452Q, T478K, E484A, N501Y, Y505H |
| BA.4 / BA.5 b | B.1.1.529.4 / 5 | ΔH69/V70, L452R, T478K, E484A, F486V, N501Y, Y505H |
| Outcome | Total | Unvaccinated | Vaccinated | RRCRUDE | 95% CI | p-value | RRADJUSTED | 95% CI | p-value |
|---|---|---|---|---|---|---|---|---|---|
| ICU Admissions, n (%) | 336 (6.7) | 268 (7.8) | 68 (4.3) | 2.07 | [1.51, 2.83] | <.001 | |||
| Age, median (IQR) | 68 (57 - 78) | 67 (56 - 76) | 72 (60 - 83) | 1.07 | [1.06, 1.08] | <.001 | 1.07 | [1.06, 1.09] | <.001 |
| Gender, n (%) | |||||||||
| Male | 197 (58.6) | 154 (57.5) | 43 (63.2) | 1.82 | [1.41, 2.36] | <.001 | 0.79 | [0.57, 1.09] | 0.16 |
| Female | 139 (41.4) | 114 (42.5) | 25 (36.8) | 0.55 | [0.42, 0.71] | <.001 | 1.26 | [0.91, 1.75] | 0.16 |
| Race / Ethnicity, n (%) | |||||||||
| Hispanic | 59 (17.6) | 49 (18.3) | 10 (14.7) | 0.95 | [0.51, 1.74] | 0.87 | 0.52 | [0.20, 1.26] | 0.15 |
| Asian | 15 (4.5) | 12 (4.5) | 3 (4.4) | 2.49 | [0.37, 1.70] | 0.08 | 1.05 | [0.29, 4.56] | 0.94 |
| Black | 22 (6.5) | 21 (7.8) | 1 (1.5) | 0.35 | [0.14, 0.79] | 0.02 | 2.84 | [0.61, 41.66] | 0.28 |
| White | 210 (62.5) | 161 (60.1) | 49 (72.1) | 1.48 | [0.94, 2.36] | 0.09 | 0.72 | [0.32, 1.36] | 0.35 |
| Ventilator Support, n (%) | 236 (4.7) | 185 (5.4) | 51 (3.2) | 1.85 | [1.30, 2.69] | <.001 | |||
| Age, median (IQR) | 70 (60 - 79) | 69 (60 - 78) | 72 (64 - 83) | 1.09 | [1.07, 1.10] | <.001 | 1.09 | [1.07, 1.10] | <.001 |
| Gender, n (%) | |||||||||
| Male | 141 (59.7) | 109 (58.9) | 32 (62.7) | 1.90 | [1.40, 2.56] | <.001 | 0.83 | [0.57, 1.21] | 0.34 |
| Female | 95 (40.3) | 76 (41.1) | 19 (37.3) | 0.53 | [0.39, 0.71] | <.001 | 1.20 | [0.83, 1.76] | 0.34 |
| Race / Ethnicity, n (%) | |||||||||
| Hispanic | 49 (20.8) | 39 (21.1) | 10 (19.6) | 1.31 | [0.66, 2.57] | 0.44 | 1.04 | [0.48, 2.21] | 0.91 |
| Asian | 10 (4.2) | 8 (4.3) | 2 (3.9) | 1.89 | [0.50, 5.90] | 0.31 | 2.23 | [0.49, 7.93] | 0.25 |
| Black | 17 (7.2) | 17 (9.2) | 0 (0.0) | 0.41 | [0.15, 1.03] | 0.07 | 0.56 | [0.20, 1.46] | 0.26 |
| White | 137 (58.1) | 104 (56.2) | 33 (64.7) | 1.89 | [0.64, 1.89] | 0.75 | 1.06 | [0.58, 1.96] | 0.86 |
| Mortality, n (%) | 430 (8.6) | 320 (9.3) | 110 (7.0) | 1.44 | [1.11, 1.87] | 0.01 | |||
| Age, median (IQR) | 75 (65 - 85) | 74 (63 - 85) | 79 (68 - 87) | 1.14 | [1.13, 1.16] | <.001 | 1.14 | [1.13, 1.16] | <.001 |
| Gender, n (%) | |||||||||
| Male | 245 (57.0) | 174 (54.4) | 71 (64.5) | 2.03 | [1.56, 2.66] | <.001 | 0.7 | [0.53, 0.92] | 0.01 |
| Female | 185 (43.0) | 146 (45.6) | 39 (35.5) | 0.59 | [0.46, 0.74] | <.001 | 1.43 | [1.09, 1.87] | 0.01 |
| Race / Ethnicity, n (%) | |||||||||
| Hispanic | 58 (13.5) | 44 (13.8) | 14 (12.7) | 1.23 | [0.57, 2.65] | 0.60 | 0.46 | [0.21, 0.99] | 0.05 |
| Asian | 16 (3.7) | 13 (4.1) | 3 (2.7) | 1.76 | [0.42, 5.63] | 0.38 | 1.76 | [0.54, 7.40] | 0.38 |
| Black | 25 (5.8) | 23 (7.2) | 2 (1.8) | 0.22 | [0.04, 0.74] | 0.03 | 2.14 | [0.63, 12.07] | 0.29 |
| White | 304 (70.7) | 220 (68.8) | 84 (76.4) | 3.33 | [1.94, 6.15] | <.001 | 0.84 | [0.46, 1.44] | 0.55 |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





