Submitted:
18 July 2023
Posted:
20 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
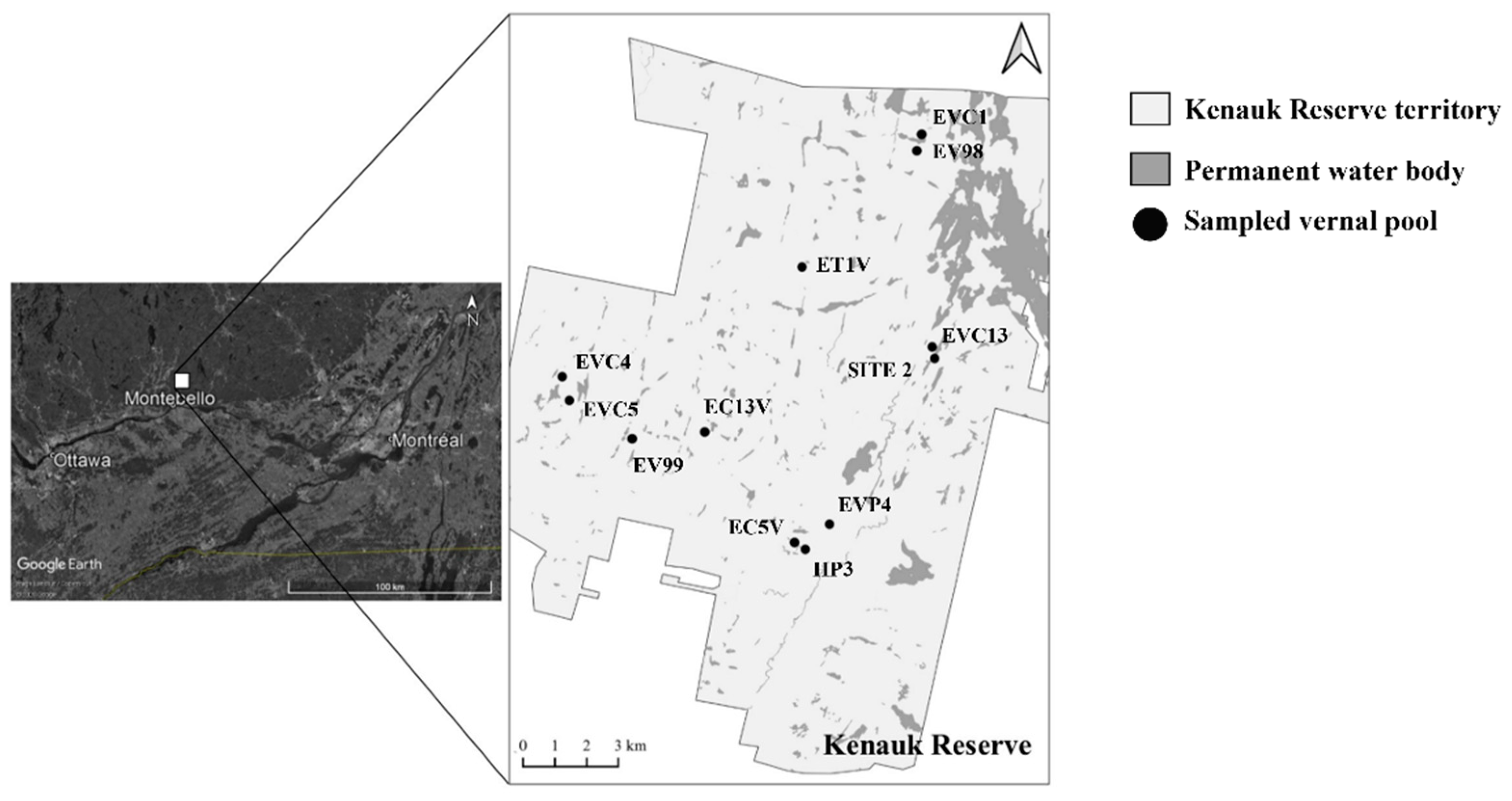
2.1. Sampling Site
2.2. Traditional Inventory of Amphibian Diversity
2.4. Amplicon Preparation and Sequencing
2.5. Contamination Control
2.6. Bioinformatics, Taxonomic Assignment
2.7. Statistical Analysis
3. Results
3.1. NGS Data Analyses
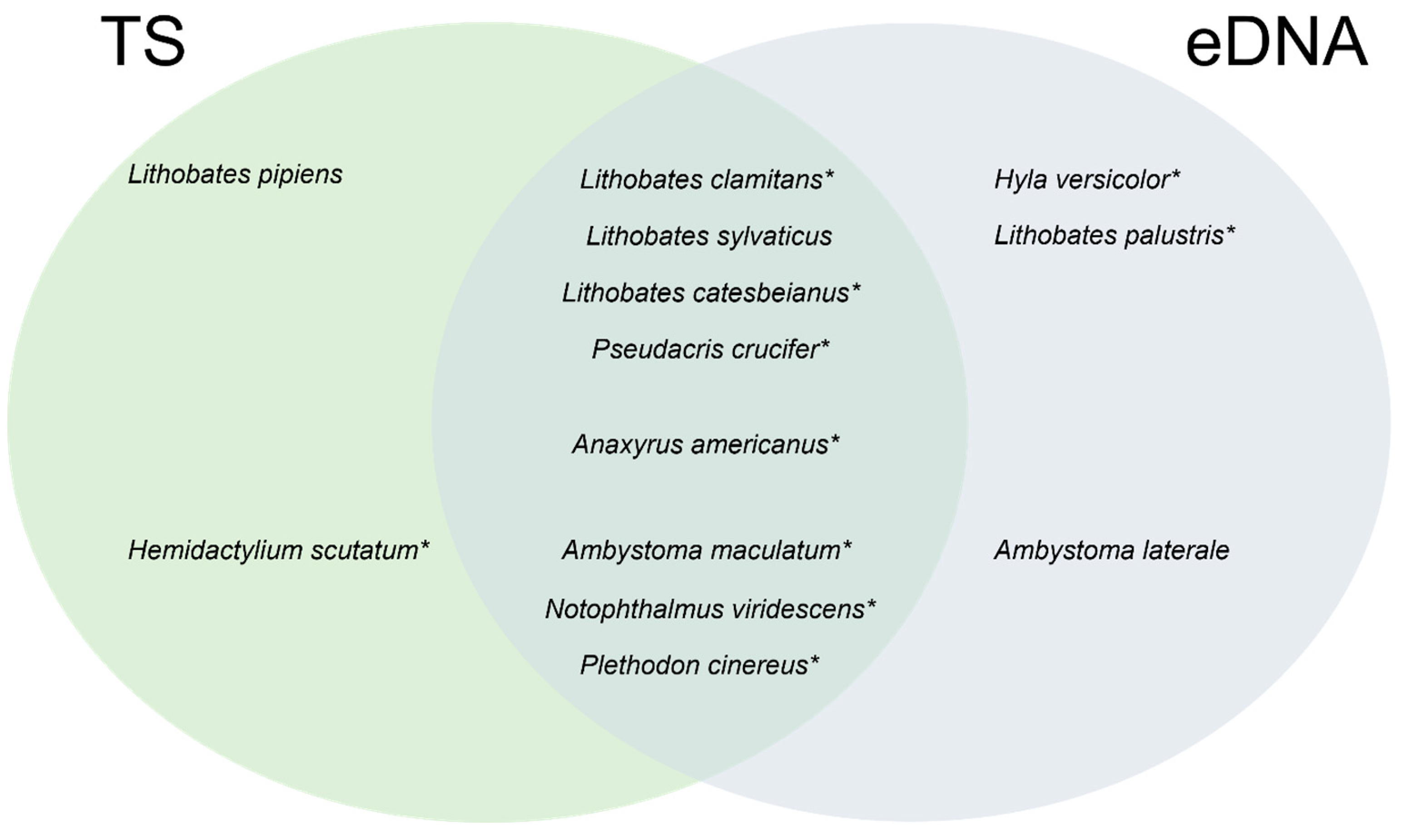
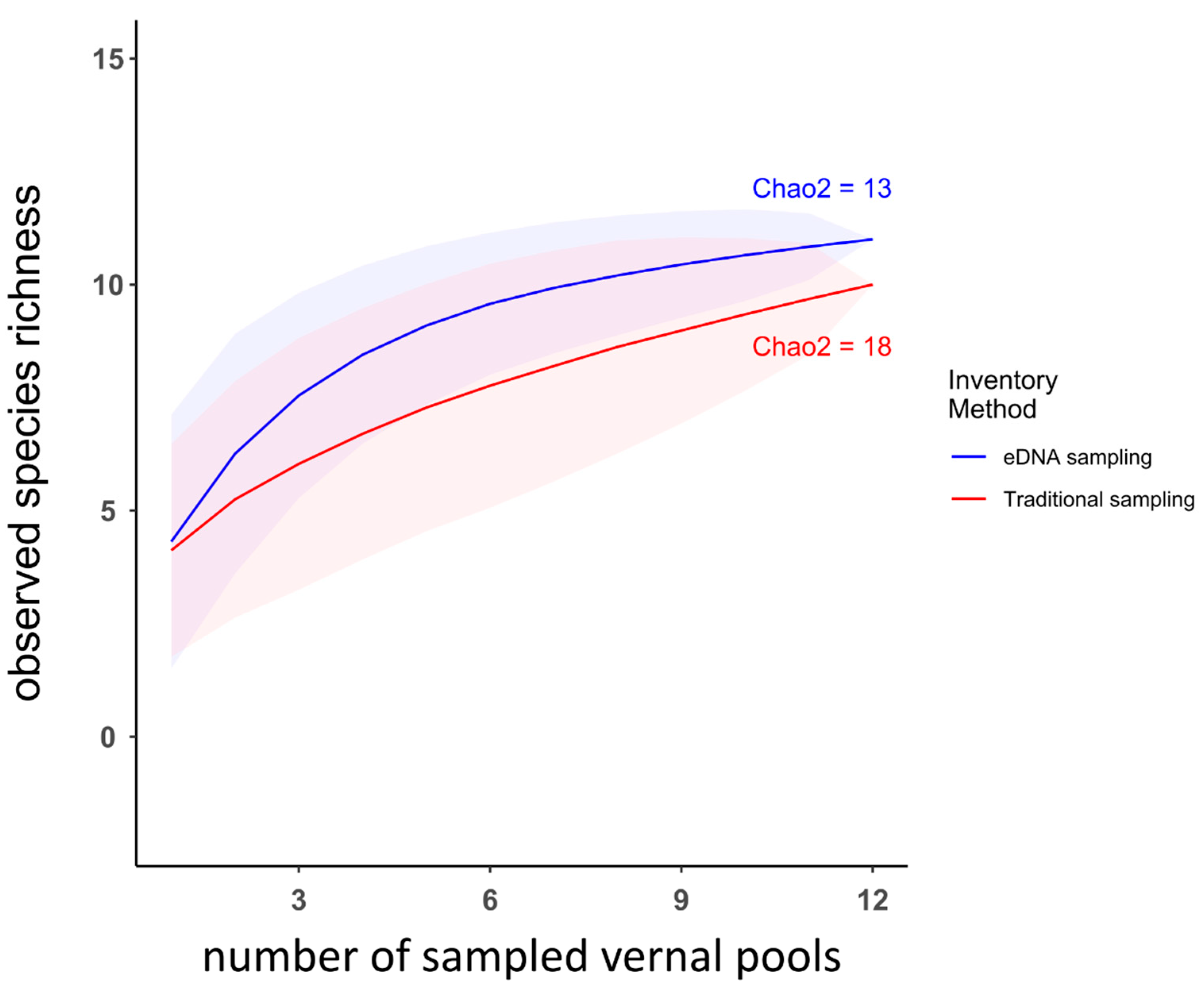
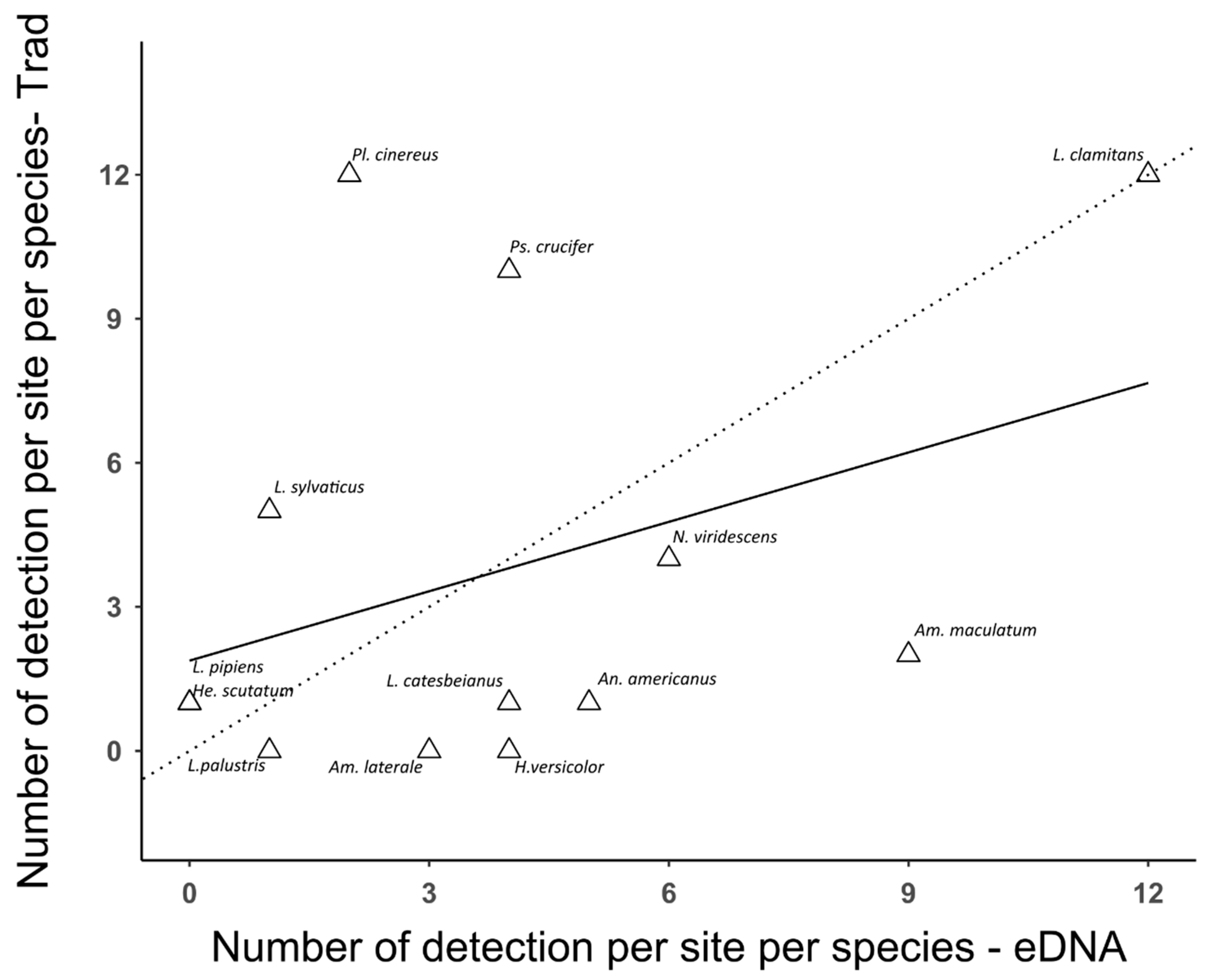
3.2. Comparison between Traditional Inventory Methods and eDNA Sampling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- J. P. Collins, “Amphibian decline and extinction: What we know and what we need to learn,” Dis. Aquat. Organ., vol. 92, no. 2–3, pp. 93–99, 2010. [CrossRef]
- D. M. Green, M. J. Lannoo, D. Lesbarrères, and E. Muths, “Amphibian population declines: 30 years of progress in confronting a complex problem,” Herpetologica, vol. 76, no. 2, pp. 97–100, 2020. [CrossRef]
- T. J. C. Beebee and R. A. Griffiths, “The amphibian decline crisis: A watershed for conservation biology?,” Biol. Conserv., vol. 125, no. 3, pp. 271–285, 2005. [CrossRef]
- J. R. Vonesh, J. C. Mitchell, K. M. Howell, and A. J. Crawford, “Rapid assessments of amphibian diversity,” in Amphibian Ecology and Conservation: A handbook of Techniques, C. K. Dodd, Ed. Oxford University Press, 2008, pp. 263–280.
- G. Boussarie et al., “Environmental DNA illuminates the dark diversity of sharks,” Sci. Adv., vol. 4, no. 5, 2018. [CrossRef]
- B. Hänfling et al., “Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods,” Mol. Ecol., vol. 25, no. 13, pp. 3101–3119, 2016. [CrossRef]
- K. Deiner et al., “Environmental DNA metabarcoding: Transforming how we survey animal and plant communities,” Mol. Ecol., vol. 26, no. 21, pp. 5872–5895, 2017. [CrossRef]
- G. F. Ficetola, C. Miaud, F. Pompanon, and P. Taberlet, “Species detection using environmental DNA from water samples,” Biol. Lett., vol. 4, no. 4, pp. 423–425, 2008. [CrossRef]
- W. E. Moss, L. R. Harper, M. A. Davis, C. S. Goldberg, M. M. Smith, and P. T. J. Johnson, “Navigating the trade-offs between environmental DNA and conventional field surveys for improved amphibian monitoring,” Ecosphere, vol. 13, no. 2, pp. 1–17, 2022. [CrossRef]
- Valentini et al., “Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding,” Mol. Ecol., vol. 25, no. 4, pp. 929–942, 2016. [CrossRef]
- Kačergytė et al., “Environmental DNA metabarcoding elucidates patterns of fish colonisation and co-occurrences with amphibians in temperate wetlands created for biodiversity,” Freshw. Biol., vol. 66, no. 10, pp. 1915–1929, 2021. [CrossRef]
- R. A. Griffiths, “Temporary ponds as amphibian habitats,” Aquat. Conserv. Mar. Freshw. Ecosyst., vol. 7, no. 2, pp. 119–126, 1997.
- J. K. Calhoun, J. Arrigoni, R. P. Brooks, M. L. Hunter, and S. C. Richter, “Creating Successful Vernal Pools: A Literature Review and Advice for Practitioners,” Wetlands, vol. 34, no. 5, pp. 1027–1038, 2014. [CrossRef]
- P. W. C. Paton, “Vernal pool,” in Wetlands and Habitats, 2nd Editio., Y. Wang, Ed. Boca Raton, FL: CRC Press, 2020, p. 5.
- L. D. Nagel, S. A. McNulty, M. D. Schlesinger, and J. P. Gibbs, “Breeding Effort and Hydroperiod Indicate Habitat Quality of Small, Isolated Wetlands for Amphibians Under Climate Extremes,” Wetlands, vol. 41, no. 2, 2021.
- D. K. Skelly, E. E. Werner, and S. A. Cortwright, “Long-term distributional dynamics of a Michigan amphibian assemblage,” Ecology, vol. 80, no. 7, pp. 2326–2337, 1999. [CrossRef]
- R. D. Semlitsch, “Principles for Management of Aquatic-Breeding Amphibians,” J. Wildl. Manage., vol. 64, no. 3, pp. 615–631, 2000. [CrossRef]
- B. Windmiller and A. J. K. Calhoun, “Conserving Vernal Pool Wildlife in Urbanizing Landscapes,” in Science and conservation of vernal pools in northeastern North America., 1st., A. J. K. Calhoun and P. G. DeMaynadier, Eds. Boca Raton, FL: CRC Press, 2007, pp. 233–252.
- P. Evans et al., “Widespread Degradation of a Vernal Pool Network in the Southeastern United States: Challenges to Current and Future Management,” Wetlands, vol. 37, no. 6, pp. 1093–1103, 2017. [CrossRef]
- Société d’histoire naturelle de la vallée du Saint-Laurent, “L’Atlas des amphibiens et reptiles du Québec.” [Online]. Available: https://www.atlasamphibiensreptiles.qc.ca/wp/. [Accessed: 10-Jun-2023].
- R. D. Semlitsch and D. K. Skelly, “Ecology and conservation of amphibians,” in Science and conservation of vernal pools in northeastern North America., A. J. K. Calhoun and P. G. DeMaynadier, Eds. Boca Raton, FL: CRC Press, 2007, pp. 127–148.
- C. R. Troth, M. J. Sweet, J. Nightingale, and A. Burian, “Seasonality, DNA degradation and spatial heterogeneity as drivers of eDNA detection dynamics,” Sci. Total Environ., vol. 768, p. 144466, 2021. [CrossRef]
- M. E. Cristescu and P. D. N. Hebert, “Uses and Misuses of Environmental DNA in Biodiversity Science and Conservation,” Annu. Rev. Ecol. Evol. Syst., vol. 49, no. 1, pp. 209–230, 2018. [CrossRef]
- T. Jo, H. Murakami, S. Yamamoto, R. Masuda, and T. Minamoto, “Effect of water temperature and fish biomass on environmental DNA shedding, degradation, and size distribution,” Ecol. Evol., vol. 9, no. 3, pp. 1135–1146, 2019.
- Y. Chen, O. Tournayre, H. Tian, and S. C. Lougheed, “Assessing the breeding phenology of a threatened frog species using eDNA and automatic acoustic monitoring,” PeerJ, vol. 11, 2023. [CrossRef]
- Zanovello et al., “A validated protocol for eDNA-based monitoring of within-species genetic diversity in a pond-breeding amphibian,” Sci. Rep., pp. 1–10, 123AD.
- K. N. Svenningsen, C. Pertoldi, and D. Bruhn, “eDNA Metabarcoding Benchmarked towards Conventional Survey Methods in Amphibian Monitoring,” Animals, vol. 12, no. 6, pp. 1–15, 2022. [CrossRef]
- P. Bournival, M. Varin, and J. Fink, “Validation d’une méthode semi-automatisée de détection des milieux humides à partir du lidar aéroporté,” 2017.
- Varin, P. Bournival, M. Dupuis, and J. Fink, “DÉVELOPPEMENT D’UNE MÉTHODE DE CARTOGRAPHIE D’ÉTANGS VERNAUX À L’AIDE DU LIDAR ET D’IMAGES MULTISPECTRALES,” 2016.
- The Kenauk Insitute and Nature Conservancy Canada, “Kenauk Amphibians, Reptiles and Mollusc Species List,” 2020. [Online]. Available: https://institutkenauk.wpengine.com/wp-content/uploads/2020/03/Amphibians-Reptiles-Molluscs.pdf. [Accessed: 10-Jun-2023].
- P. F. Thomsen and E. Willerslev, “Environmental DNA - An emerging tool in conservation for monitoring past and present biodiversity,” Biol. Conserv., vol. 183, pp. 4–18, 2015. [CrossRef]
- B. Duhaime and R. Cable, “DNA Extraction from Filters using QIAgen DNeasy and QIAshredder,” 2019.
- K. C. Beng et al., “The utility of DNA metabarcoding for studying the response of arthropod diversity and composition to land-use change in the tropics,” Sci. Rep., vol. 6, no. October 2015, pp. 1–13, 2016. [CrossRef]
- E. Robert, “Usearch.” Berkeley, CA, 2010.
- S. Ratnasingham and P. D. N. Hebert, “BOLD: The Barcode of Life Data System (www.barcodinglife.org),” Mol. Ecol. Notes, vol. 7, pp. 355–364, 2007.
- J. Oksanen and G. L. Simpson, “Package ‘ vegan ,’” no. April, 2022.
- H. Whickham, ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016.
- R. C. Team, “R: A language and environment for statistical computing,” 2013.
- Chao, R. K. Colwell, C. W. Lin, and N. J. Gotelli, “Sufficient sampling for asymptotic minimum species richness estimators,” Ecology, vol. 90, no. 4, pp. 1125–1133, 2009. [CrossRef]
- J. Vavrek, “fossil: palaeoecological and palaeogeographical analysis tools,” Palaeontol. Electron., vol. 14, no. 1, p. 1T, 2011.
- T. Sørensen, “A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons,” K. Danske Vidensk. Selsk., vol. 5, no. 4, pp. 1–34, 1948.
- S. H. Hurlbert, “The Breeding Migrations and Interhabitat Wandering of the Vermilion-Spotted Newt Notophthalmus viridescens (Rafinesque),” Ecol. Monogr., vol. 39, no. 4, pp. 465–488, 1969. [CrossRef]
- B. Stoler and R. A. Relyea, “Leaf litter quality induces morphological and developmental changes in larval amphibians,” Ecology, vol. 94, no. 7, pp. 1594–1603, 2013. [CrossRef]
- R. A. Relyea, “Competitor-induced plasticity in tadpoles: Consequences, cues, and connections to predator-induced plasticity,” Ecol. Monogr., vol. 72, no. 4, pp. 523–540, 2002.
- J. Merilä, A. Laurila, A. T. Laugen, and K. Räsänen, “Heads or tails? Variation in tadpole body proportions in response to temperature and food stress,” Evol. Ecol. Res., vol. 6, no. 5, pp. 727–738, 2004.
- J.-F. Desroches and D. Rodrigue, Amphibiens et reptiles du Québec et des Maritimes. Montreal: Michel Quintin, 2018.
- S. R. Goldberg, “Notes on Reproduction of Spring Peepers , Pseudacris crucifer ( Anura : Hylidae ), from Oklahoma,” Bull. Chicago Herpetol. Soc., vol. 58, no. 4, pp. 58–62, 2023.
- T. Dejean, “Déclin et inventaire de la biodiversité : les maladies des amphibiens et la méthode de l’ADN environnemental,” 2011.
- M. A. Barnes, C. R. Turner, C. L. Jerde, M. A. Renshaw, W. L. Chadderton, and D. M. Lodge, “Environmental conditions influence eDNA persistence in aquatic systems,” Environ. Sci. Technol., vol. 48, no. 3, pp. 1819–1827, 2014. [CrossRef]
- R. N. Homan, C. D. Wright, G. L. White, L. F. Michael, B. S. Slaby, and S. A. Edwards, “Multiyear Study of the Migration Orientation of Ambystoma maculatum (Spotted Salamanders) among Varying Terrestrial Habitats,” J. Herpetol., vol. 42, no. 4, pp. 600–607, 2008. [CrossRef]
- D. L. Chambers, “Logging road effects on breeding-site selection in Notophthalmus viridescens (Red-spotted Newt) and three ambystomatid salamanders in South-central Pennsylvania,” Northeast. Nat., vol. 15, no. 1, pp. 123–130, 2008. [CrossRef]
- K. J. Ryan and A. J. K. Calhoun, “Postbreeding habitat use of the rare, pure-diploid blue-spotted salamander (ambystoma laterale),” J. Herpetol., vol. 48, no. 4, pp. 556–566, 2014.
- Ruiz-Garciá, Á. S. Roco, and M. Bullejos, “Sex Differentiation in Amphibians: Effect of Temperature and Its Influence on Sex Reversal,” Sex. Dev., vol. 15, no. 1, pp. 157–167, 2021. [CrossRef]
- E. Jorde and N. Ryman, “Temporal allele frequency change and estimation of effective size in populations with overlapping generations,” Genetics, vol. 139, no. 2, pp. 1077–1090, 1995. [CrossRef]
- Dournon, C. Houillon, and C. Pieau, “Temperature sex-reversal in amphibians and reptiles,” Int. J. Dev. Biol., vol. 34, no. 1, pp. 81–92, 1990.
| Order | Family | Species | eDNA | Acco | Trap | Enco |
|---|---|---|---|---|---|---|
| Anura | Bufonidae | Anaxyrus americanus* | 5 | 1 | 0 | 0 |
| Hylidae | Hyla versicolor* | 4 | 0 | 0 | 0 | |
| Pseudacris crucifer* | 4 | 10 | 1 | 0 | ||
| Ps. maculata | 0 | 0 | 0 | 0 | ||
| Ps. triseriata | 0 | 0 | 0 | 0 | ||
| Ranidae | Lithobates catesbeianus* | 4 | 0 | 0 | 1 | |
| L. clamitans* | 12 | 3 | 8 | 7 | ||
| L. palustris* | 1 | 0 | 0 | 0 | ||
| L. pipiens | 0 | 0 | 0 | 1 | ||
| L. septentrionalis* | 0 | 0 | 0 | 0 | ||
| L. sylvaticus | 1 | 4 | 0 | 1 | ||
| Urodela | Ambystomatidae | Ambystoma laterale | 3 | 0 | 0 | 0 |
| Am. maculatum* | 9 | 0 | 0 | 2 | ||
| Plethodontidae | Desmognathus fuscus | 0 | 0 | 0 | 0 | |
| D. ochrophaeus | 0 | 0 | 0 | 0 | ||
| Eurycea bislineata* | 0 | 0 | 0 | 0 | ||
| Gyrinophilus porphyriticus | 0 | 0 | 0 | 0 | ||
| Hemidactylium scutatum* | 0 | 0 | 0 | 1 | ||
| Plethodon cinereus* | 2 | 0 | 0 | 12 | ||
| Proteidae | Necturus maculosus | 0 | 0 | 0 | 0 | |
| Salamandridae | Notophthalmus viridescens* | 6 | 0 | 0 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





