Submitted:
18 July 2023
Posted:
19 July 2023
You are already at the latest version
Abstract
Keywords:
MSC: 90C20; 90C29; 90C90; 93E20
1. Introduction
2. The architecture of the proposed method
3. Density based support vector machine
2.1. Classical support vector machine
2.2. Density based support vector machine (DBSVM)
- A simple is called if .
- A simple is called if and
- A simple is called if and there exists a such as .
4. Recurrent neural network to optimal support vectors
3.1. Continuous Hopfield network based on the original energy function
- (1)
- Initialization: , and the step are randomly chosen;
- (2)
- Given and the step , the step is chosen such that is maximum and we calculate using: , then we calculate using the activation function , then the are given by: , where P is the projection operator on the set
- (3)
- Return to 1) until , where .
3.2. Continuous Hopfield network with optimal time step
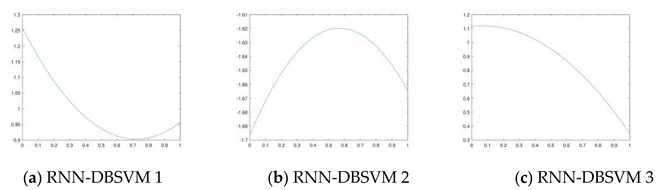
3.3. Opt-RNN-DBSVM algorithm
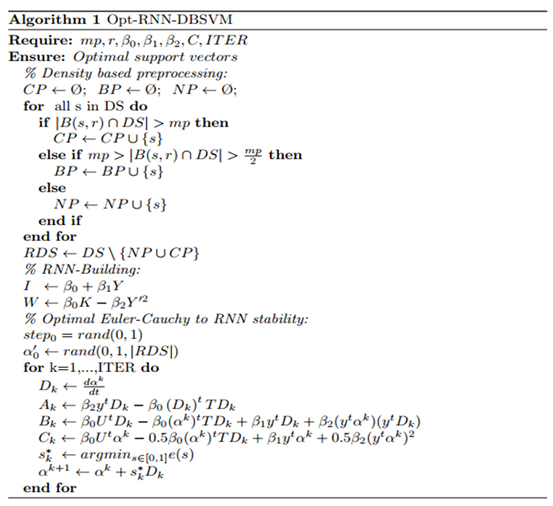
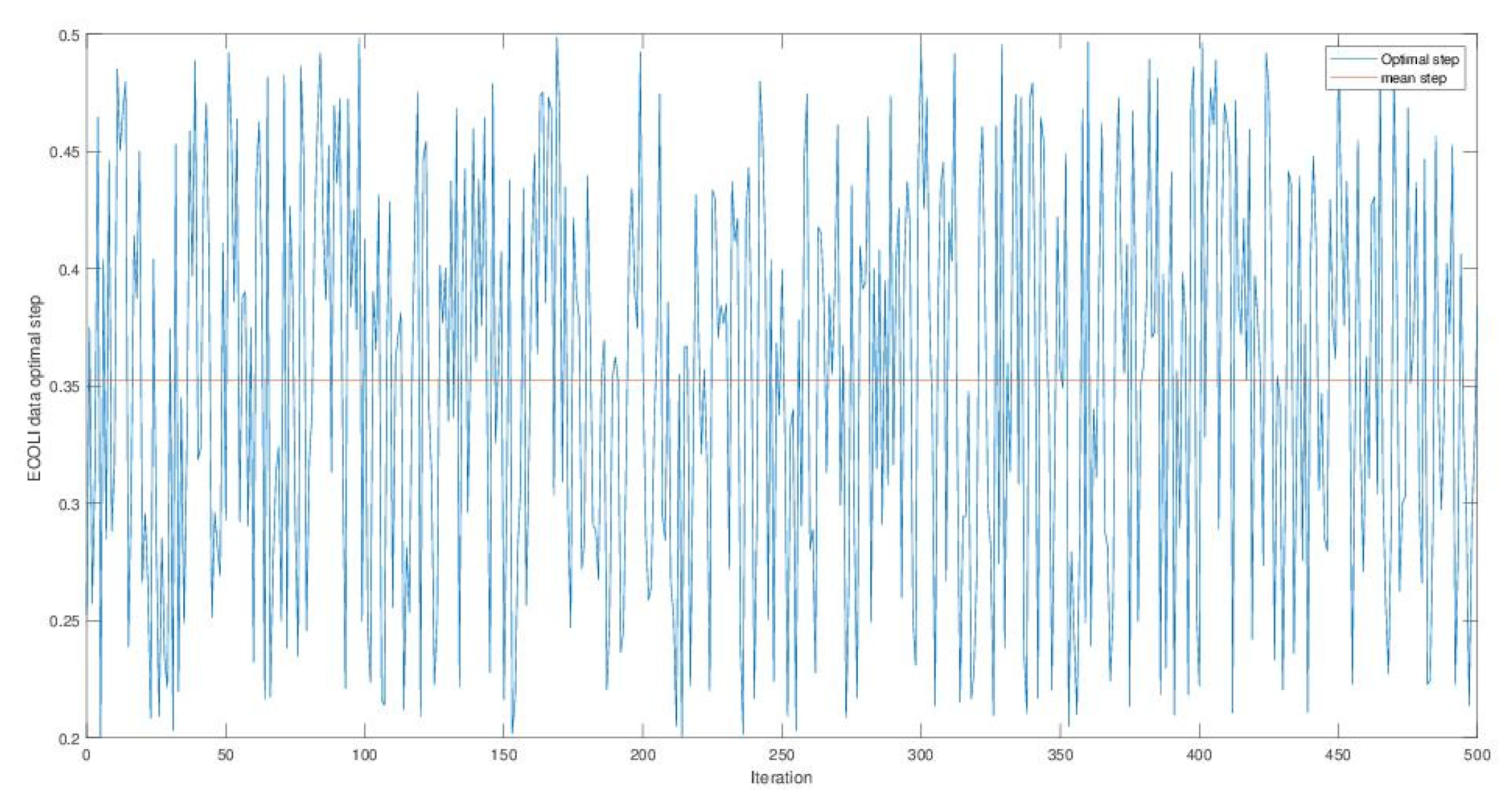
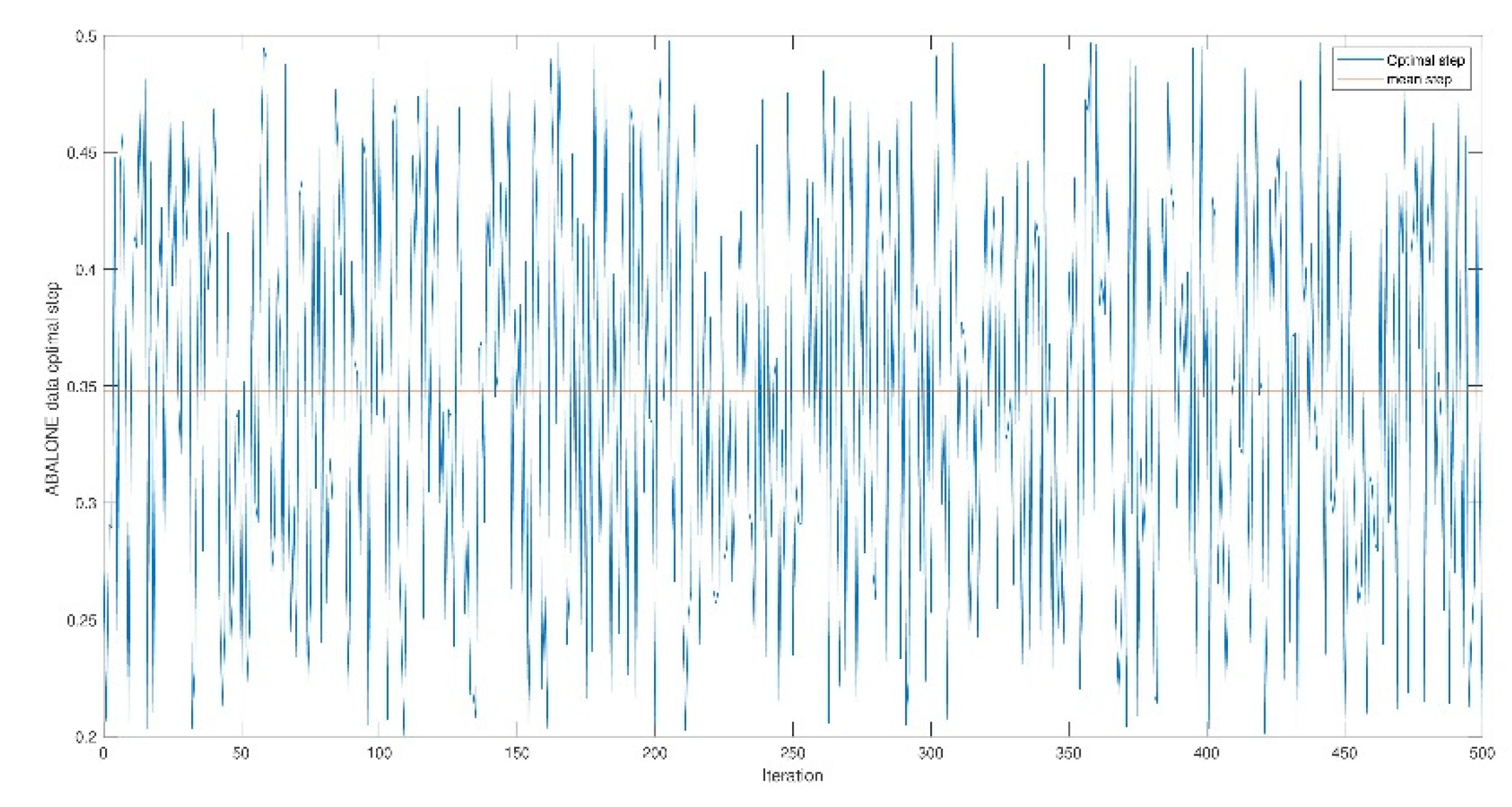
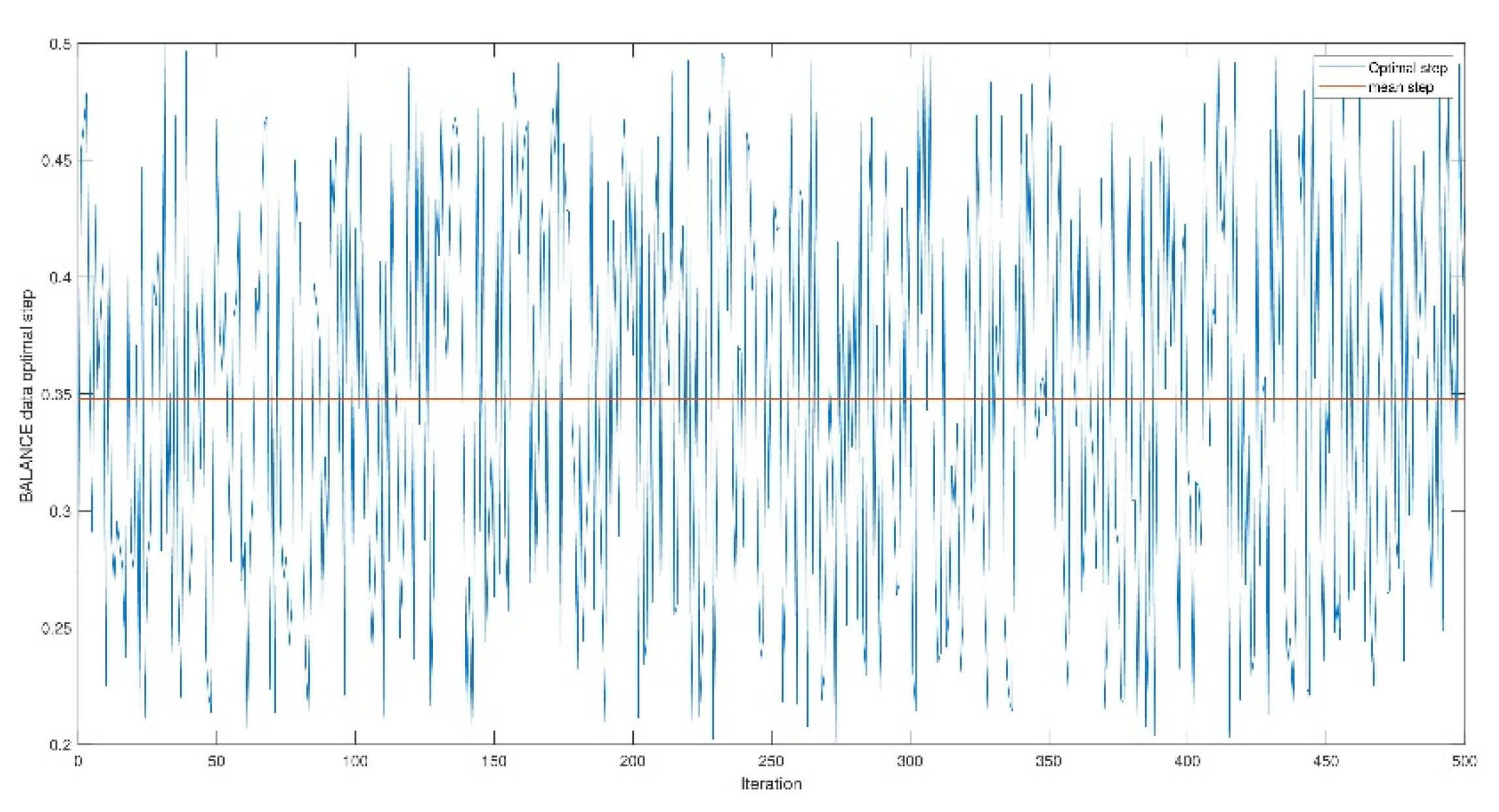
4. Experimentation
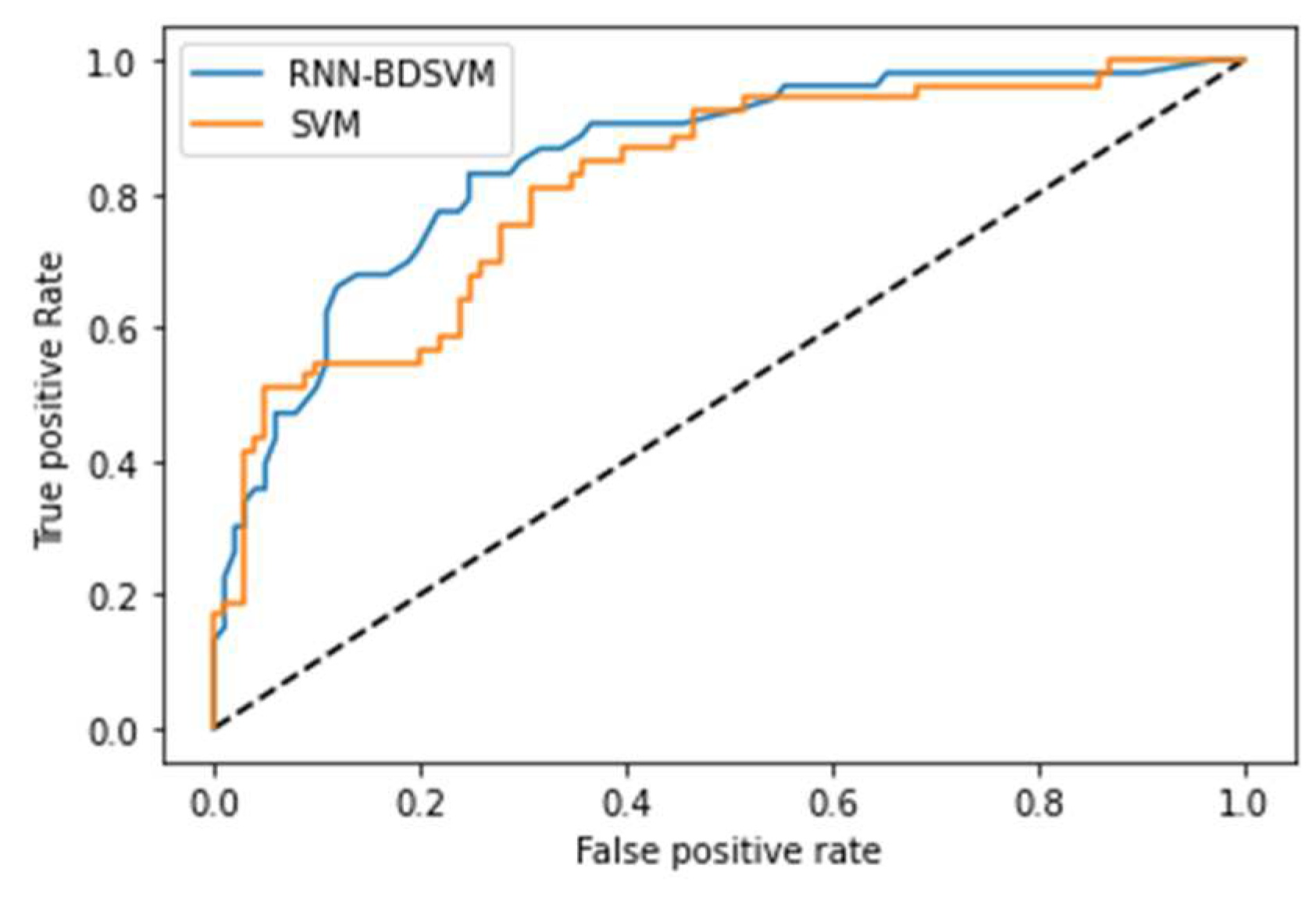
4.1. Opt-RNN-DBSVM vs Const-CHN-SVM
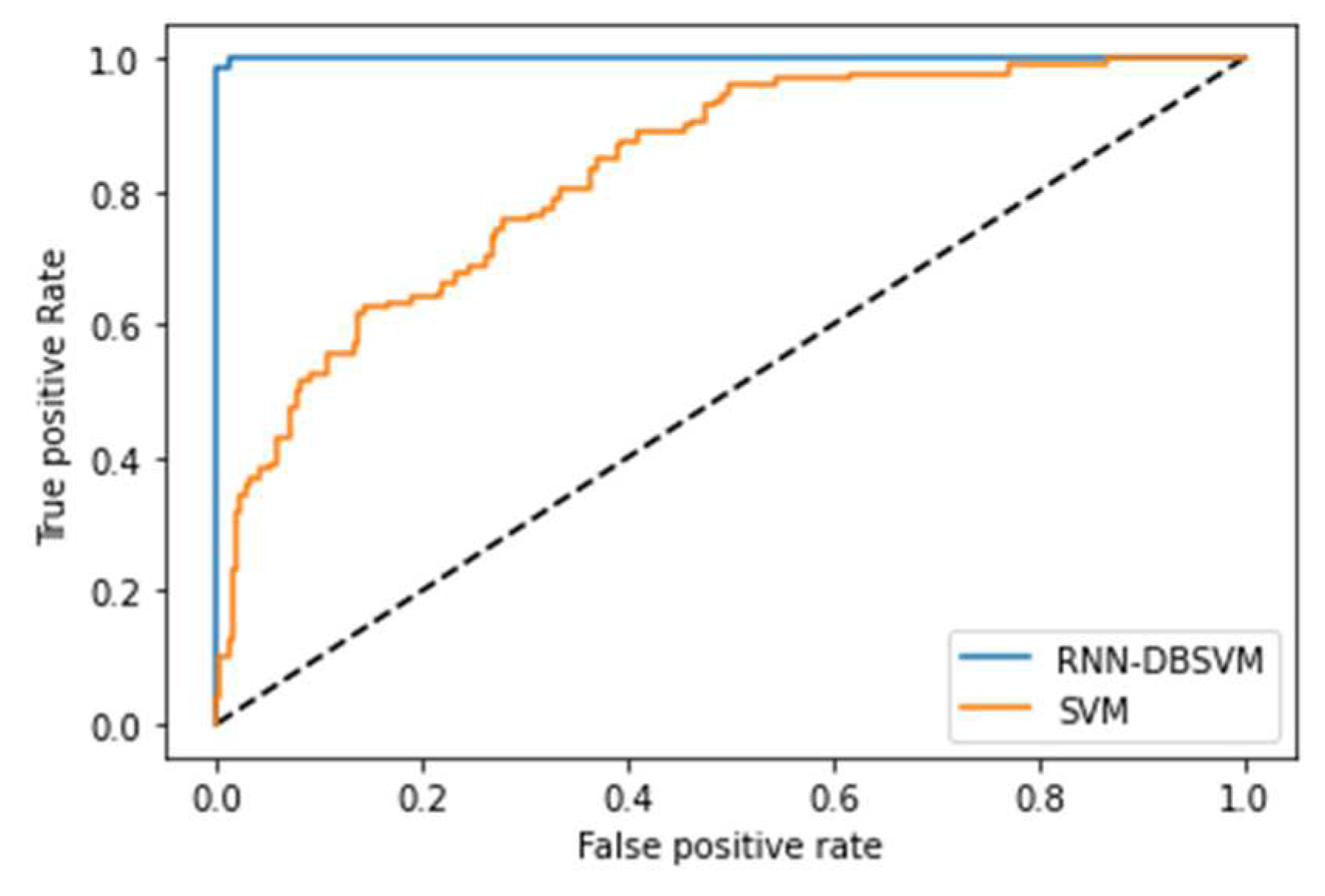
4.2. Opt-RNN-DBSVM vs Classical-Optimizer-SVM
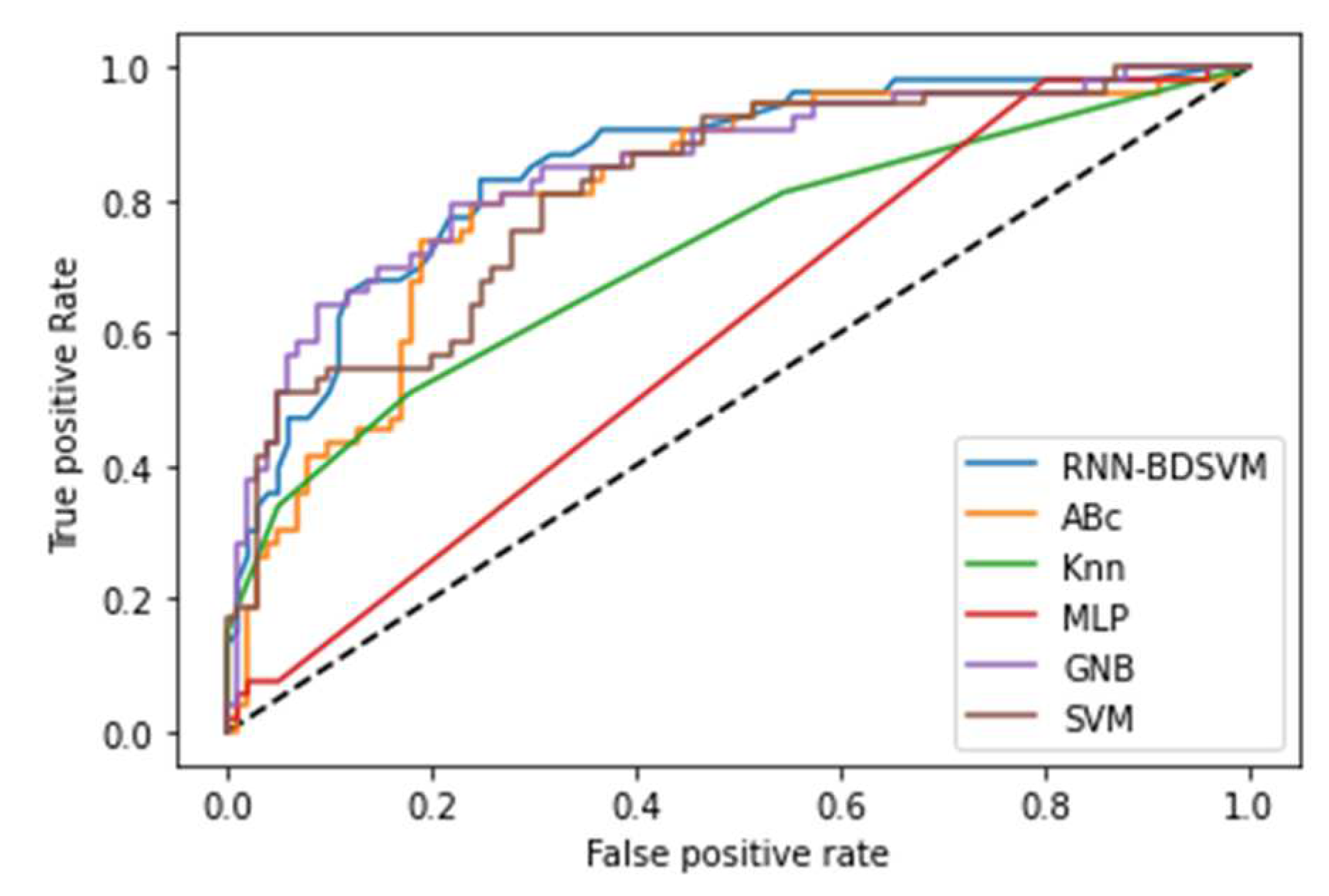
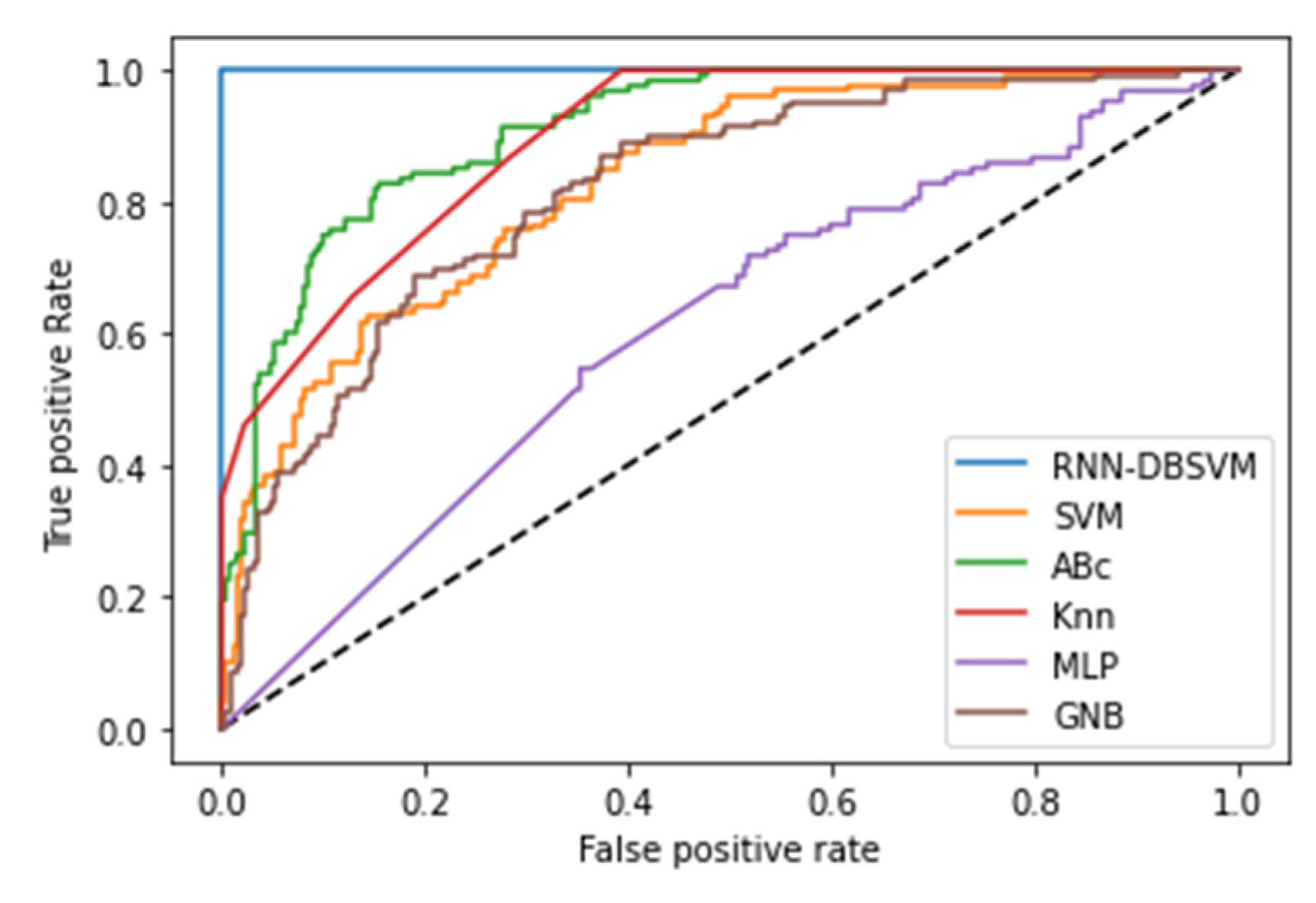
4.3. Opt-RNN-DBSVM vs non-kernel classifiers
5. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
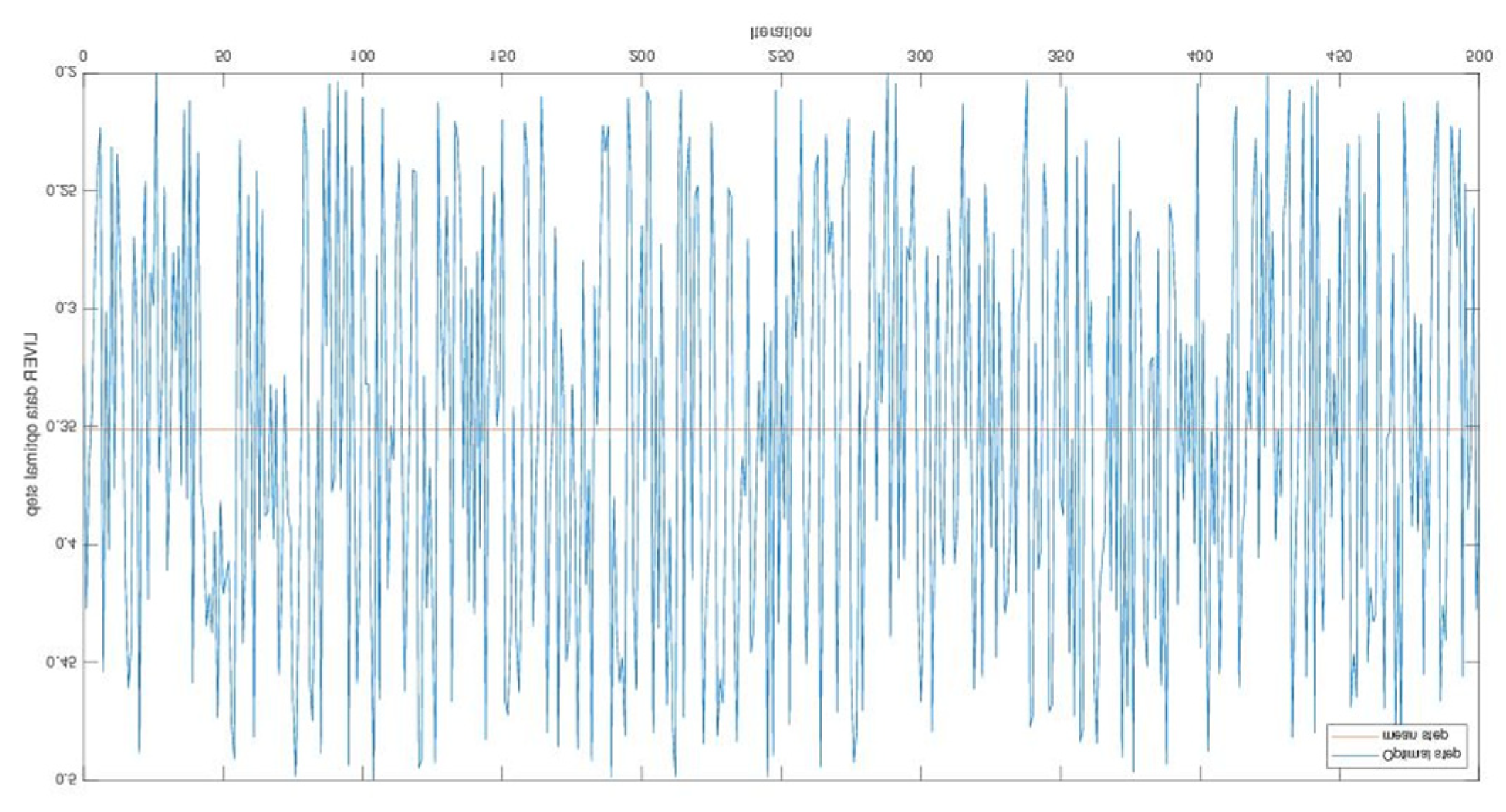
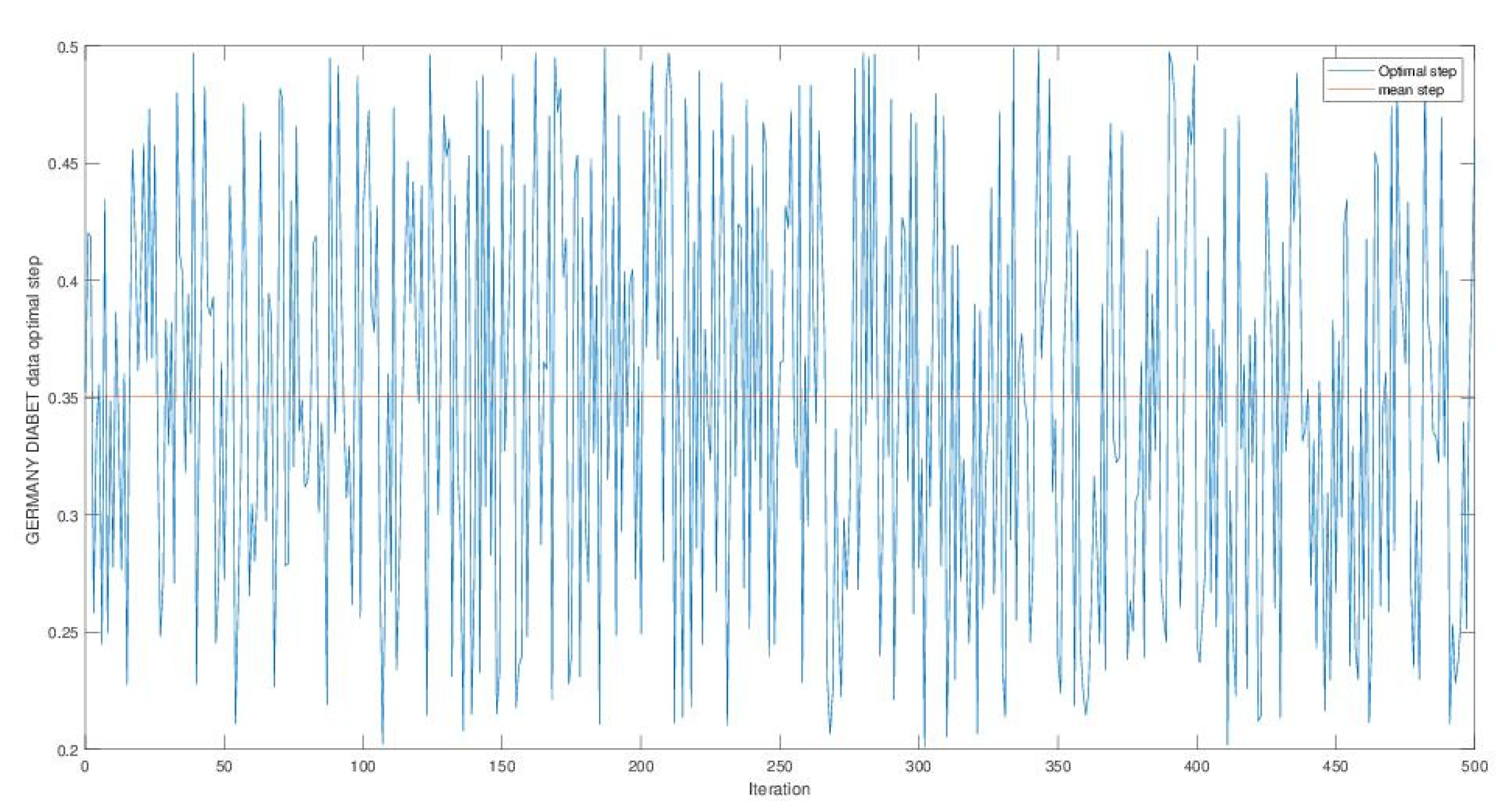
References
- V. Vapnik, The Nature of Statistical Learning Theory. Springer Science and Business Media (1999).
- Schölkopf, B.; Smola, A.J.; Williamson, R.C.; Bartlett, P.L. New Support Vector Algorithms. Neural Comput. 2000, 12, 1207–1245, . [CrossRef]
- J.J. Hopfield, D.W. Tank, Neural computation of decisions in optimization problems. Biol. Cybern. 52 (1985) 1–25.
- K. Haddouch, & K. El Moutaouakil, New Starting Point of the Continuous Hopfield Network. In Int. Conf. on Big Data, Cloud Appl. Springer, Cham (2018, April) 379-389.
- W. Steyerberg, Clinical prediction models, Cham: Springer International Publishing (2019) 309-328.
- M. Law. How to build valid and credible simulation models. In 2019 Winter Simulation Conference (WSC) IEEE. (2019, December) 1402-1414.
- E. L. Glaeser, S. D. Kominers, M. Luca, & N. Naik. Big data and big cities: The promises and limitations of improved measures of urban life. Eco. Inqui. 56(1) (2018) 114-137.
- Cortes, V. Vapnik, Support-vector networks. Machine learning, 20(3) (1995) 273-297.
- J. Mercer, Functions of positive and negative type, and their connection the theory of integral equations. Philosophical transactions of the royal society of London. Ser. A, Cont. Pap. Math. Phy. Char., 209(441-458) (1909) 415-446.
- Ahmadi, M.; Khashei, M. Generalized support vector machines (GSVMs) model for real-world time series forecasting. Soft Comput. 2021, 25, 14139–14154, . [CrossRef]
- F. Lin, S. D.Wang, Fuzzy support vector machines. IEEE Trans. Neur. Netw. 13(2) (2002) 464-471.
- Aghbashlo, M.; Peng, W.; Tabatabaei, M.; Kalogirou, S.A.; Soltanian, S.; Hosseinzadeh-Bandbafha, H.; Mahian, O.; Lam, S.S. Machine learning technology in biodiesel research: A review. Prog. Energy Combust. Sci. 2021, 85, 100904, . [CrossRef]
- Schölkopf, B.; Platt, J.C.; Shawe-Taylor, J.; Smola, A.J.; Williamson, R.C. Estimating the Support of a High-Dimensional Distribution. Neural Comput. 2001, 13, 1443–1471, . [CrossRef]
- Bi, T. Zhang, Support vector classification with input data uncertainty. In Adv. Neur. Inf. Proc. syst. (2005) 161-168.
- Hazarika, B.B.; Gupta, D. Density-weighted support vector machines for binary class imbalance learning. Neural Comput. Appl. 2020, 33, 4243–4261, . [CrossRef]
- Guo, H.; Wang, W. Granular support vector machine: a review. Artif. Intell. Rev. 2019, 51, 19–32, . [CrossRef]
- Lee, Y.-J.; Mangasarian, O. SSVM: A Smooth Support Vector Machine for Classification. Comput. Optim. Appl. 2001, 20, 5–22, . [CrossRef]
- B. Schölkopf, AJ. Smola , F. Bach, Learning with kernels: support vector machines, regularization, optimization, and beyond. MIT press (2002).
- Xie, X.; Xiong, Y. Generalized multi-view learning based on generalized eigenvalues proximal support vector machines. Expert Syst. Appl. 2022, 194, 116491, . [CrossRef]
- Tanveer, M.; Rajani, T.; Rastogi, R.; Shao, Y.H.; Ganaie, M.A. Comprehensive review on twin support vector machines. Ann. Oper. Res. 2022, 1–46, . [CrossRef]
- Laxmi, S.; Gupta, S. Multi-category intuitionistic fuzzy twin support vector machines with an application to plant leaf recognition. Eng. Appl. Artif. Intell. 2022, 110, 104687, . [CrossRef]
- Ettaouil, M.; Elmoutaouakil, K.; Ghanou, Y. The Placement of Electronic Circuits Problem: A Neural Network Approach. Math. Model. Nat. Phenom. 2010, 5, 109–115, . [CrossRef]
- M. Minoux, Mathematical programming: theories and algorithms, Duond (1983).
- S. Russell and P. Norvig, Artificial Intelligence A Modern Approach, 3rd edition, by Pearson Education (2010).
- Hopfield, J.J. Neurons with graded response have collective computational properties like those of two-state neurons. Proc. Natl. Acad. Sci. USA 1984, 81, 3088–3092, doi:10.1073/pnas.81.10.3088.
- K. El Moutaouakil M. Ettaouil, Reduction of the continuous Hopfield architecture, J. Compu. Volume 4, Issue 8 (2012) pp. 64-70.
- K. El Moutaouakil, A. El Ouissari, A. Touhafi and N. Aharrane, "An Improved Density Based Support Vector Machine (DBSVM)," 5th Inter. Confe. Cloud Compu. Artif. Intel.: Techn. Appl. (CloudTech) (2020) pp. 1-7.
- El Ouissari, A.; El Moutaouakil, K. Density based fuzzy support vector machine: application to diabetes dataset. Math. Model. Comput. 2021, 8, 747–760, . [CrossRef]
- Dua, D. and Graff, C. UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science (2019).
- Wickramasinghe, I.; Kalutarage, H. Naive Bayes: applications, variations and vulnerabilities: a review of literature with code snippets for implementation. Soft Comput. 2021, 25, 2277–2293, . [CrossRef]
- O. Tolstikhin, N. Houlsby, A. Kolesnikov, L. Beyer, X. Zhai, T. Unterthiner, &A.Dosovitskiy, Mlp-mixer: An all-mlp architecture for vision. Adv. Neur. Inf. Proc. Sys. 34 (2021).
- Shokrzade, A.; Ramezani, M.; Tab, F.A.; Mohammad, M.A. A novel extreme learning machine based kNN classification method for dealing with big data. Expert Syst. Appl. 2021, 183, 115293, . [CrossRef]
- Chen, P.; Pan, C. Diabetes classification model based on boosting algorithms. BMC Bioinform. 2018, 19, 109, doi:10.1186/s12859-018-2090-9.
- Charbuty, B.; Abdulazeez, A. Classification Based on Decision Tree Algorithm for Machine Learning. J. Appl. Sci. Technol. Trends 2021, 2, 20–28, . [CrossRef]
- Almustafa, K.M. Classification of epileptic seizure dataset using different machine learning algorithms. Informatics Med. Unlocked 2020, 21, 100444, . [CrossRef]
- K E. Moutaouakil, & A. Touhafi, A New Recurrent Neural Network Fuzzy Mean Square Clustering Method. In 2020 5th International Conference on Cloud Computing and Artificial Intelligence: Technologies and Applications (CloudTech) IEEE (2020) 1-5.
- El Moutaouakil, K.; Yahyaouy, A.; Chellak, S.; Baizri, H. An Optimized Gradient Dynamic-Neuro-Weighted-Fuzzy Clustering Method: Application in the Nutrition Field. Int. J. Fuzzy Syst. 2022, 24, 3731–3744, . [CrossRef]
- El Moutaouakil, K.; Roudani, M.; El Ouissari, A. Optimal Entropy Genetic Fuzzy-C-Means SMOTE (OEGFCM-SMOTE). Knowledge-Based Syst. 2023, 262, . [CrossRef]
- Anlauf, J.K.; Biehl, M. The AdaTron: An Adaptive Perceptron Algorithm. EPL (Europhysics Lett. 1989, 10, 687–692, . [CrossRef]
- T.-T. Frieß, , Cristianini, N., Campbell, I. C. G., The Kernel-Adatron: a Fast and Simple Learning Procedure for Support Vector Machines. In Shavlik, J., editor, Proceedings of the 15th International Conference on Machine Learning, Morgan Kaufmann, San Francisco, CA (1998) 188–196.
- T.-M. Huang, V. Kecman, Bias Term b in SVMs Again, Proc. of ESANN 2004, 12th European Symposium on Artificial Neural Networks Bruges, Belgium, 2004.
- T. Joachims, Making large-scale svm learning practical. advances in kernel methods-support vector learning. http://svmlight. joachims.org/ (1999).
- V. Kecman, M. Vogt, T.-M. Huang, On the Equality of Kernel AdaTron and Sequential Minimal Optimization in Classification and Regression Tasks and Alike Algorithms for Kernel Machines, Proc. of the 11 th European Symposium on Artificial Neural Networks, ESANN, , Bruges, Belgium (2003) 215–222.
- E. Osuna, R. Freund, F. Girosi, An Improved Training Algorithm for Support Vector Machines. In Neural Networks for Signal Processing VII, Proceedings of the 1997 Signal Processing Society Workshop, (1997) 276–285.
- J. Platt, Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines, Microsoft Research Technical Report MSR-TR-98-14, (1998).
- Veropoulos, K., Machine Learning Approaches to Medical Decision Making, PhD Thesis, The University of Bristol, Bristol, UK (2001).
- M. Vogt, SMO Algorithms for Support Vector Machines without Bias, Institute Report, Institute of Automatic Control, TU Darmstadt, Darmstadt, Germany, (Available at http://www.iat.tu-darmstadt.de/vogt) (2002).
- V. Kecman, T. M.Huang, & M. Vogt, Iterative single data algorithm for training kernel machines from huge data sets: Theory andperformance. Support vector machines: Theory and Applications, (2005) 255-274.
- V. Kecman, Iterative k data algorithm for solving both the least squares SVM and the system of linear equations. In SoutheastCon IEEE (2015) 1-6.


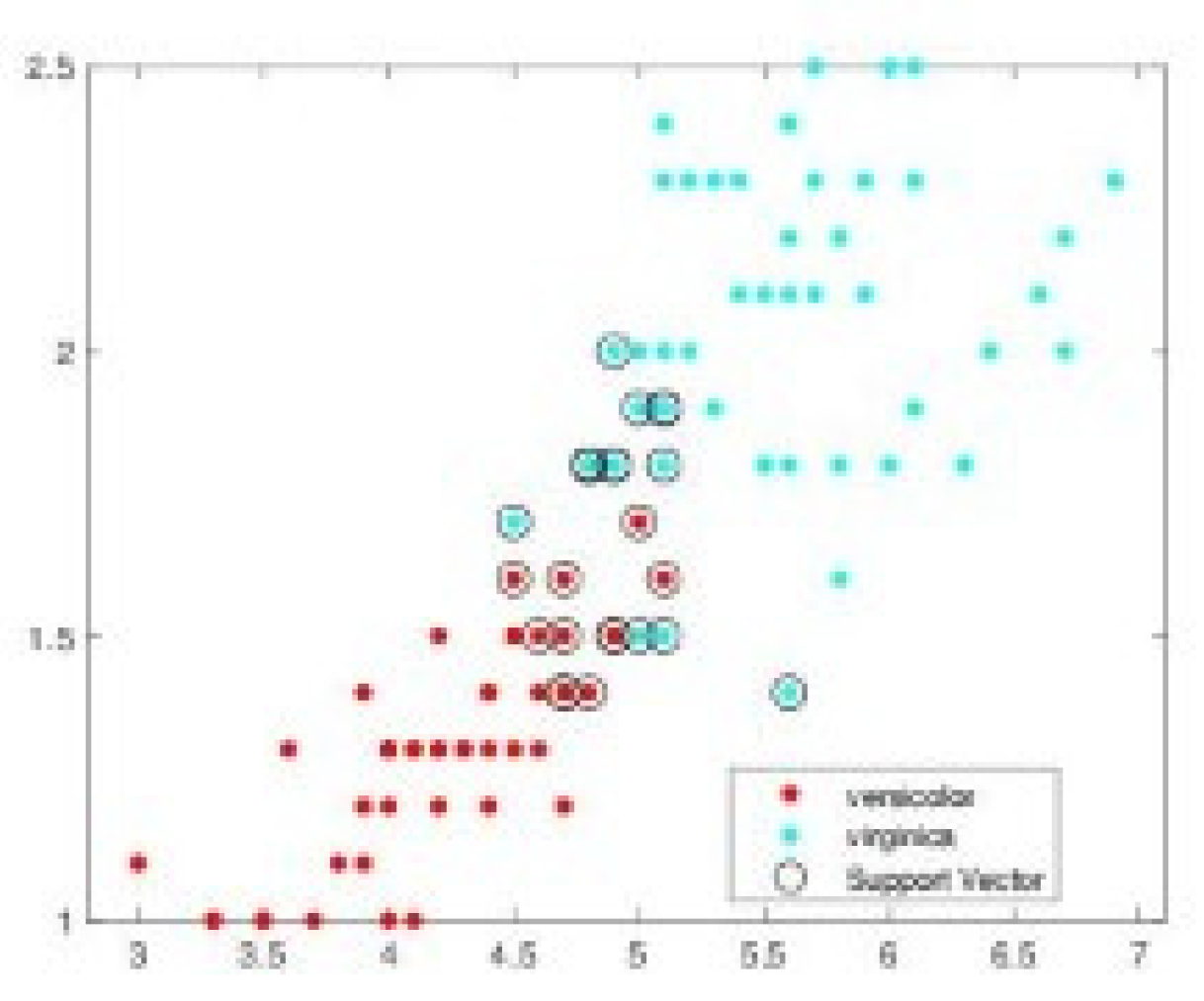
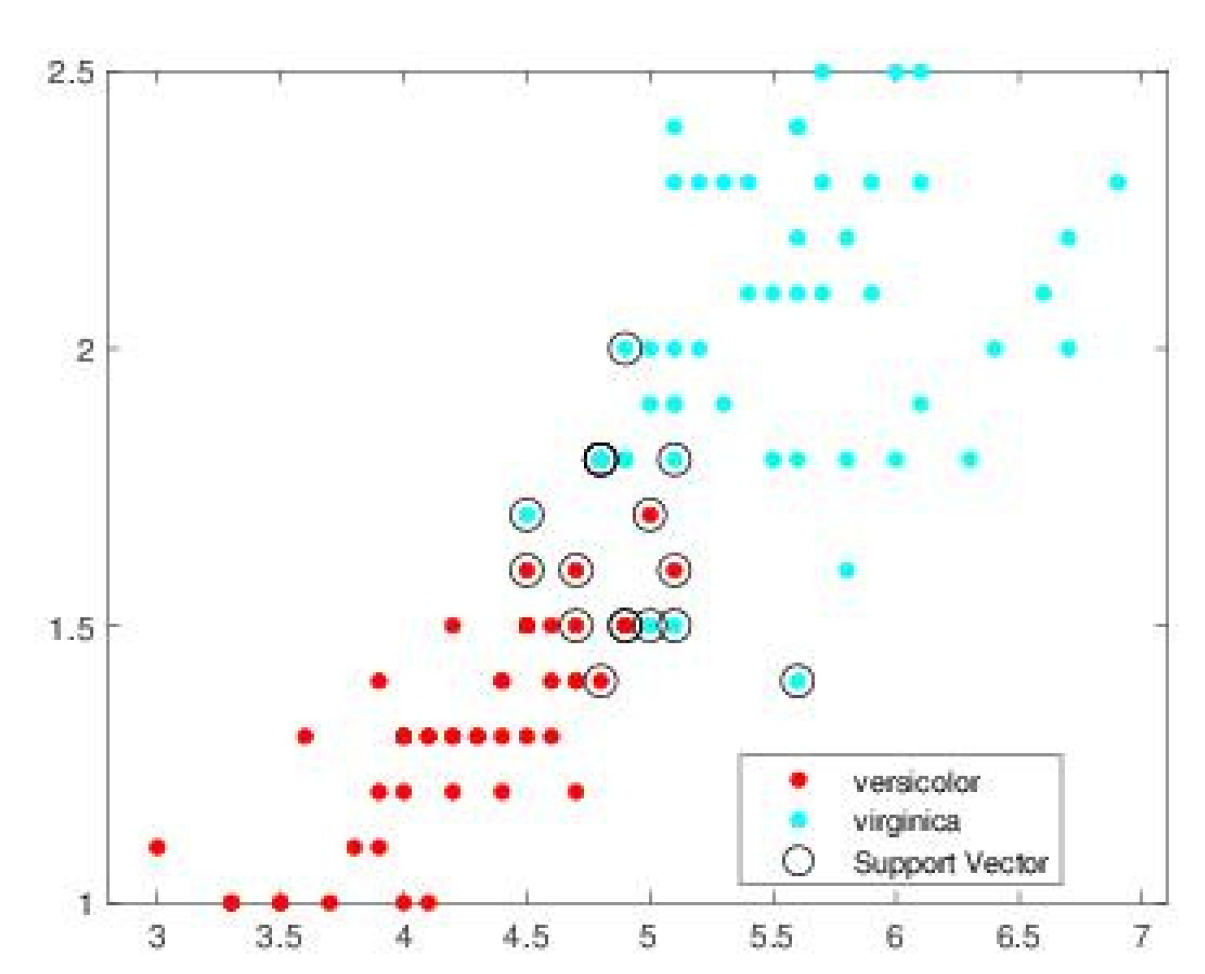
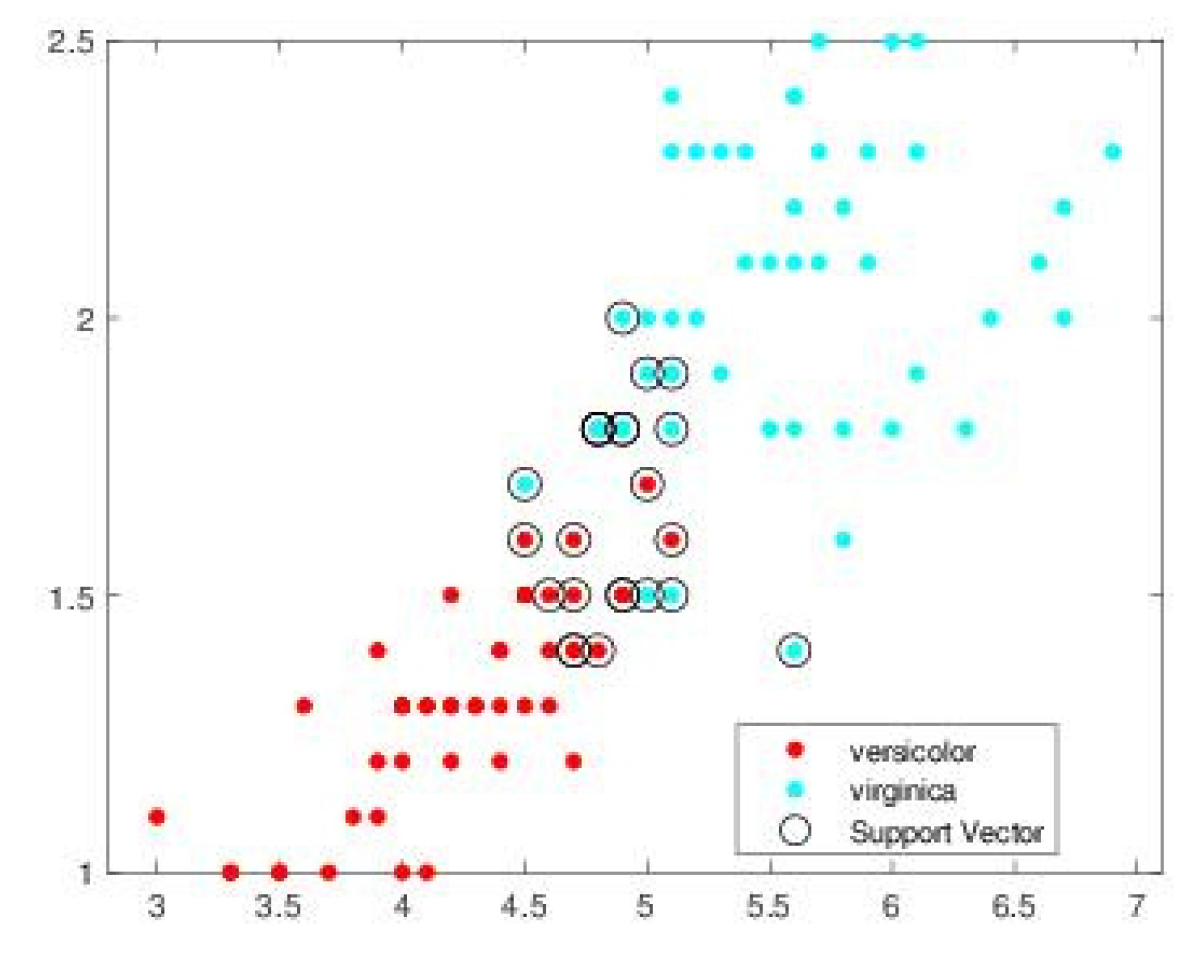
| SVM-CHN s = .1 | SVM-CHN s = .2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | F1-score | Precision | Recall | Accuracy | F1-score | Precision | Recall | |
| Iris | 95.98 | 96.66 | 90.52 | 92.00 | 96.66 | 95.98 | 91.62 | 92.00 |
| Abalone | 80.98 | 40.38 | 82.00 | 27.65 | 80.66 | 40.38 | 81.98 | 27.65 |
| Wine | 79.49 | 78.26 | 73.52 | 74.97 | 79.49 | 78.26 | 73.52 | 74.97 |
| Ecoli | 88.05 | 96.77 | 97.83 | 97.33 | 88.05 | 97.77 | 97.83 | 97.99 |
| Balance | 79.70 | 70.7 | 55.60 | 62.70 | 79.70 | 70.7 | 55.60 | 62.70 |
| Liver | 80.40 | 77.67 | 77.90 | 70.08 | 80.40 | 77.67 | 77.90 | 70.08 |
| Spect | 92.12 | 90.86 | 91.33 | 90.00 | 97.36 | 99.60 | 97.77 | 1.00 |
| Seed | 85.71 | 83.43 | 92.70 | 75.04 | 85.71 | 83.43 | 92.70 | 75.04 |
| PIMA | 79.22 | 61.90 | 84.7 | 49.6 | 79.22 | 61.90 | 83.97 | 49.6 |
| SVM-CHN s = .3 | SVM-CHN s = .4 | ||||||||||
| Accuracy | F1-score | Precision | Recall | Accuracy | F1-score | Precision | Recall | ||||
| Iris | 94.53 | 95.86 | 89.66 | 98.23 | 95.96 | 93.88 | 89.33 | 95.32 | |||
| Abalone | 77.99 | 51.68 | 83.85 | 30.88 | 81.98 | 41.66 | 83.56 | 33.33 | |||
| Wine | 80.23 | 77.66 | 74.89 | 74.97 | 81.33 | 77.65 | 73.11 | 74.43 | |||
| Ecoli | 88.86 | 95.65 | 96.88 | 97.33 | 86.77 | 97.66 | 97.83 | 97.95 | |||
| Balance | 79.75 | 70.89 | 55.96 | 62.32 | 79.66 | 70.45 | 56.1 | 66.23 | |||
| Liver | 80.51 | 78.33 | 77.9 | 70.56 | 80.40 | 77.67 | 77.90 | 70.08 | |||
| Spect | 97.63 | 98.99 | 97.81 | 98.56 | 96.40 | 98.71 | 96.83 | 97.79 | |||
| Seed | 85.71 | 83.43 | 92.70 | 75.88 | 85.71 | 83.43 | 92.70 | 75.61 | |||
| PIMA | 79.22 | 61.93 | 84.82 | 49.86 | 79.22 | 61.90 | 84.98 | 49.89 | |||
| SVM-CHN s = .5 | SVM-CHN s = .6 | ||||||||||
| Accuracy | F1-score | Precision | Recall | Accuracy | F1-score | Precision | Recall | ||||
| Iris | 94.53 | 95.86 | 89.66 | 98.23 | 95.96 | 93.88 | 89.33 | 95.32 | |||
| Abalone | 78.06 | 51.83 | 83.96 | 40.45 | 82.1 | 42.15 | 83.88 | 38.26 | |||
| Wine | 80.84 | 78.26 | 74.91 | 75.20 | 81.39 | 77.86 | 73.66 | 74.47 | |||
| Ecoli | 88.97 | 95.7 | 96.91 | 97.43 | 86.77 | 97.66 | 97.83 | 97.95 | |||
| Balance | 79.89 | 71.00 | 56.11 | 62.72 | 79.71 | 70.64 | 56.33 | 66.44 | |||
| Liver | 80.66 | 78.33 | 77.9 | 70.56 | 80.40 | 77.67 | 77.90 | 70.08 | |||
| Spect | 91.36 | 92.60 | 91.77 | 84.33 | 91.36 | 92.60 | 91.77 | 84.33 | |||
| Seed | 84.67 | 82.96 | 92.23 | 74.18 | 84.11 | 83.08 | 92.63 | 75.48 | |||
| PIMA | 79.12 | 61.75 | 84.62 | 49.86 | 79.12 | 61.33 | 84.68 | 48.55 | |||
| SVM-CHN s = .7 | SVM-CHN s = .8 | ||||||||||
| Accuracy | F1-score | Precision | Recall | Accuracy | F1-score | Precision | Recall | ||||
| Iris | 94.53 | 95.86 | 89.66 | 98.23 | 95.96 | 93.88 | 89.33 | 95.32 | |||
| Abalone | 77.99 | 51.68 | 83.85 | 30.88 | 81.98 | 41.66 | 83.56 | 33.33 | |||
| Wine | 80.23 | 77.66 | 74.89 | 74.97 | 81.33 | 77.65 | 73.11 | 74.43 | |||
| Ecoli | 88.86 | 95.65 | 96.88 | 97.33 | 86.77 | 97.66 | 97.83 | 97.95 | |||
| Balance | 79.75 | 70.89 | 55.96 | 62.32 | 79.66 | 70.45 | 56.1 | 66.23 | |||
| Liver | 80.51 | 78.33 | 77.9 | 70.56 | 80.40 | 77.67 | 77.90 | 70.08 | |||
| Spect | 94.36 | 84.60 | 83.77 | 85.99 | 94.36 | 84.60 | 83.77 | 85.99 | |||
| Seed | 85.71 | 83.43 | 92.70 | 75.88 | 85.71 | 83.43 | 92.70 | 75.61 | |||
| PIMA | 79.22 | 61.93 | 84.82 | 49.86 | 79.22 | 61.90 | 84.98 | 49.89 | |||
| SVM-CHN s = .9 | DBSVM optimal value of s | ||||||||||
| Accuracy | F1-score | Precision | Recall | Accuracy | F1-score | Precision | Recall | ||||
| Iris | 95.96 | 93.88 | 89.33 | 95.32 | 97.96 | 96.19 | 95.85 | 98.5 | |||
| Abalone | 81.98 | 41.66 | 83.56 | 33.33 | 98.38 | 96.07 | 96.18 | 93.19 | |||
| Wine | 81.33 | 77.65 | 73.11 | 74.43 | 96.47 | 95.96 | 96.08 | 96.02 | |||
| Ecoli | 86.77 | 88.46 | 90.77 | 91.19 | 91.82 | 97.66 | 97.83 | 97.95 | |||
| Balance | 79.66 | 70.45 | 56.1 | 66.23 | 91.31 | 90.33 | 89.54 | 90.89 | |||
| Liver | 80.40 | 77.67 | 77.90 | 70.08 | 88.10 | 85.95 | 86.00 | 85.50 | |||
| Spect | 94.36 | 84.60 | 83.77 | 85.99 | 95.55 | 86.20 | 85.28 | 86.31 | |||
| Seed | 85.71 | 83.43 | 86.18 | 75.61 | 88.90 | 84.31 | 92.70 | 84.40 | |||
| PIMA | 79.22 | 61.90 | 75.90 | 49.89 | 79.87 | 68.04 | 84.98 | 62.05 | |||
| L1QP-SVM | ISDA-SVM | |||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | F1-score | Precision | Recall | Accuracy | F1-score | Precision | Recal | |
| Iris | 71.59 | 62.02 | 70.80 | 55.17 | 82.00 | 83.64 | 76.67 | 92.00 |
| Abalone | 74.15 | 70.80 | 71.48 | 59.30 | 83.70 | 68.22 | 70.00 | 80.66 |
| Wine | 72.90 | 65.79 | 75.53 | 60.11 | 66.08 | 65.80 | 66.00 | 70.02 |
| Ecoli | 66.15 | 55.89 | 61.33 | 41.30 | 51.60 | 48.30 | 33.33 | 51.39 |
| Balance | 65.20 | 53.01 | 60.51 | 41.22 | 50.44 | 58.36 | 68.32 | 60.20 |
| Liver | 64.66 | 52.06 | 60.77 | 40.44 | 50.00 | 48.00 | 62.31 | 51.22 |
| Spect | 70.66 | 62.02 | 67.48 | 50.11 | 77.60 | 71.20 | 75.33 | 70.11 |
| Seed | 70.51 | 58.98 | 67.30 | 45.30 | 80.66 | 81.25 | 79.80 | 79.30 |
| PIMA | 65.18 | 53.23 | 60.88 | 39.48 | 49.32 | 44.33 | 48.90 | 50.27 |
| SMO- | SVM | CHN- | DBSVM optimal value of s | |||||
| Accuracy | F1-score | Precision | Recall | Accuracy | F1-score | Precision | Recal | |
| Iris | 71.59 | 62.02 | 70.80 | 55.17 | 97.96 | 96.19 | 95.85 | 98.5 |
| Abalone | 74.15 | 70.80 | 71.48 | 59.30 | 98.38 | 96.07 | 96.18 | 93.19 |
| Wine | 72.90 | 65.79 | 75.53 | 60.11 | 96.47 | 95.96 | 96.08 | 96.02 |
| Ecoli | 66.15 | 55.89 | 61.33 | 41.30 | 91.82 | 88.46 | 90.77 | 91.19 |
| Balance | 65.20 | 53.01 | 60.51 | 41.22 | 91.31 | 90.33 | 89.54 | 90.89 |
| Liver | 64.66 | 52.06 | 60.77 | 40.44 | 88.10 | 85.95 | 86.00 | 85.50 |
| Spect | 70.66 | 62.02 | 67.48 | 50.11 | 95.55 | 86.20 | 85.28 | 86.31 |
| Seed | 70.51 | 58.98 | 67.30 | 45.30 | 88.90 | 84.31 | 86.18 | 84.40 |
| PIMA | 65.18 | 53.23 | 60.88 | 39.48 | 79.87 | 68.04 | 75.90 | 62.05 |
| Iris | ||||
| Method | Accuracy | F1-score | Precision | Recall |
| Niave Bayes | 90.00 | 87.99 | 77.66 | 1.00 |
| MLP | 26.66 | 0.00 | 0.00 | 0.00 |
| Knn | 96.66 | 95.98 | 91.62 | 1.00 |
| AdaBoostM1 | 86.66 | 83.66 | 71.77 | 1.00 |
| Dicision Tree | 69.25 | 76.12 | 70.01 | 69.55 |
| SGDClassifier | 76.66 | 46.80 | 1.00 | 30.10 |
| Random Forest Classifier | 90.00 | 87.99 | 77.66 | 1.00 |
| Nearest Centroid Classifier | 96.66 | 95.98 | 91.62 | 1.00 |
| Classical SVM | 96.66 | 95.98 | 91.62 | 1.00 |
| Opt-RNN-DBSVM | 97.96 | 92.19 | 95.85 | 96.05 |
| Abalone | ||||
| Method | Accuracy | F1-score | Precision | Recall |
| Niave Bayes | 68.89 | 51.19 | 41.37 | 67.33 |
| MLP | 62.91 | 47.63 | 36.32 | 47.63 |
| Knn | 81.93 | 53.74 | 70.23 | 43.02 |
| AdaBoostM1 | 82.29 | 55.99 | 70.56 | 55.06 |
| Dicision Tree | 76.79 | 51.33 | 52.06 | 49.63 |
| SGDClassifier | 80.86 | 64.74 | 58.08 | 70.57 |
| Nearest Centroid Classifier | 76.07 | 64.79 | 62.60 | 61.15 |
| RandomForestClassifier | 82.28 | 57.56 | 71.11 | 48.34 |
| Classical SVM | 80.98 | 40.38 | 82.00 | 27.65 |
| Opt-RNN-DBSVM | 98.38 | 96.07 | 96.18 | 93.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





