Submitted:
19 July 2023
Posted:
19 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
2.1. Plasmid preparation
2.2. Cell lines
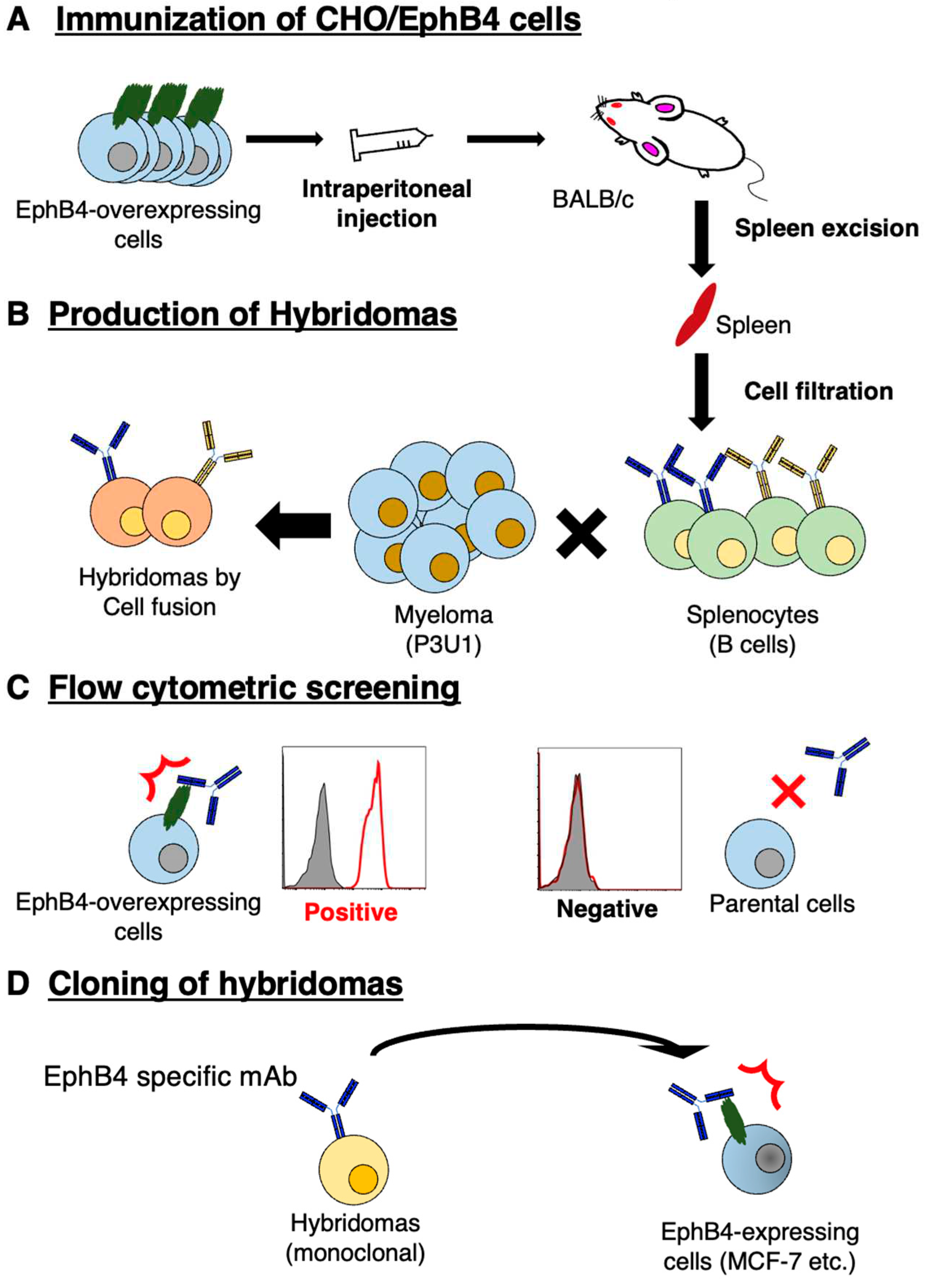
2.3. Hybridoma production
2.4. Flow cytometry
2.5. Determination of dissociation constant (KD) by flow cytometry
2.6. Western blot analysis
2.7. Immunohistochemical analysis
3. Results
3.1. Establishment of anti-EphB4 mAbs
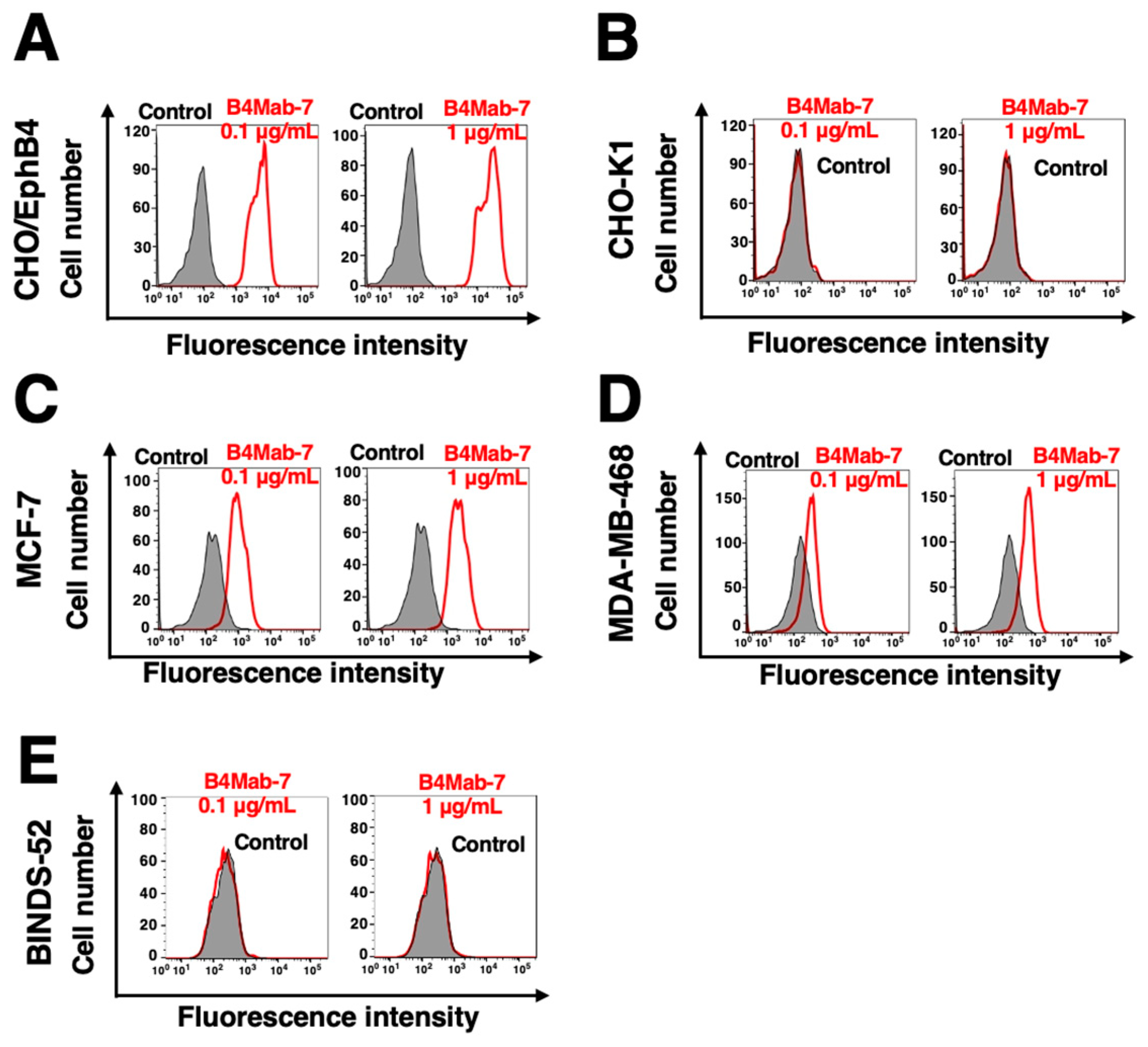
3.2. Flow cytometric analyses
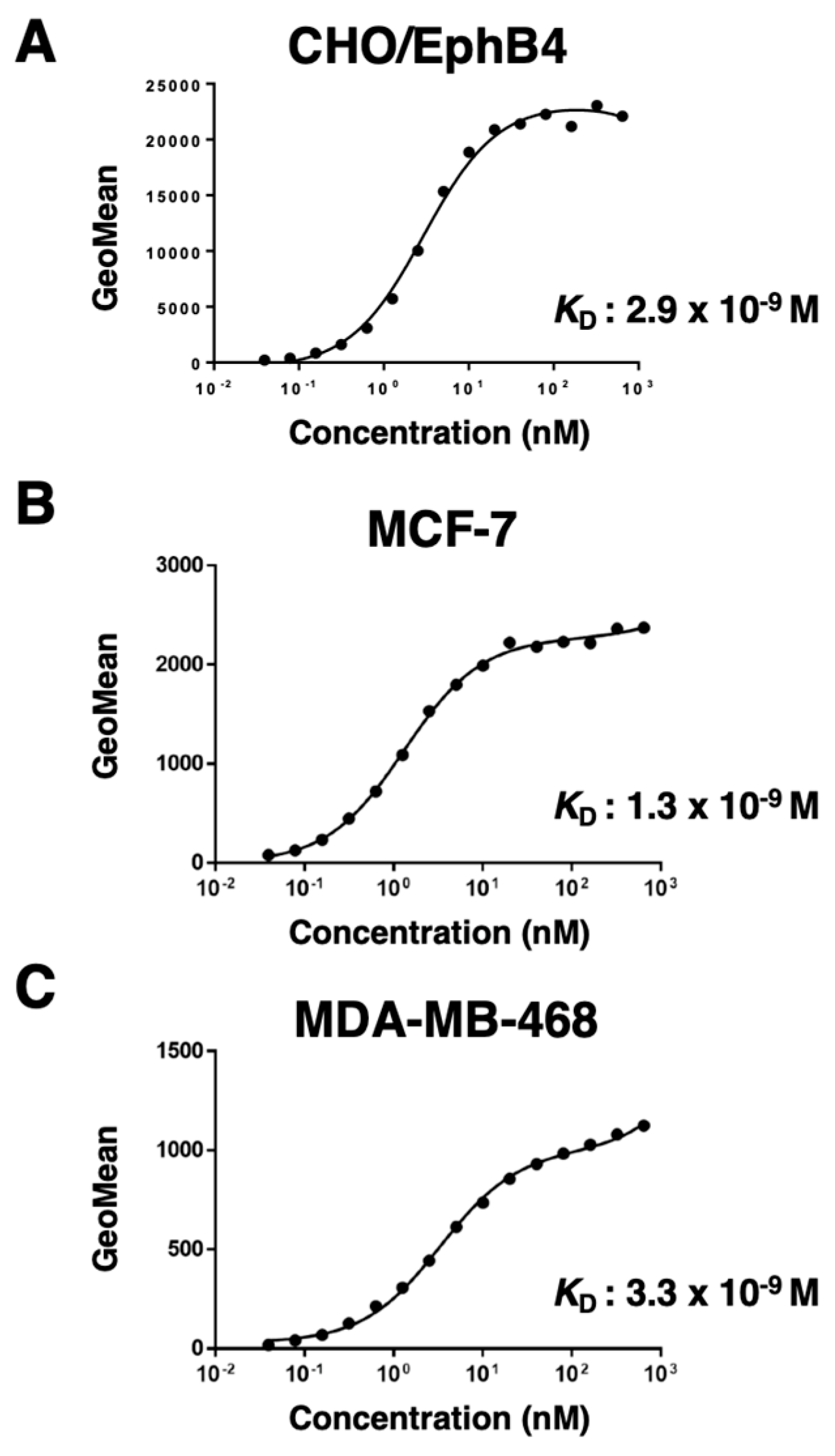
3.3. Determination of the binding affinity of B4Mab-7
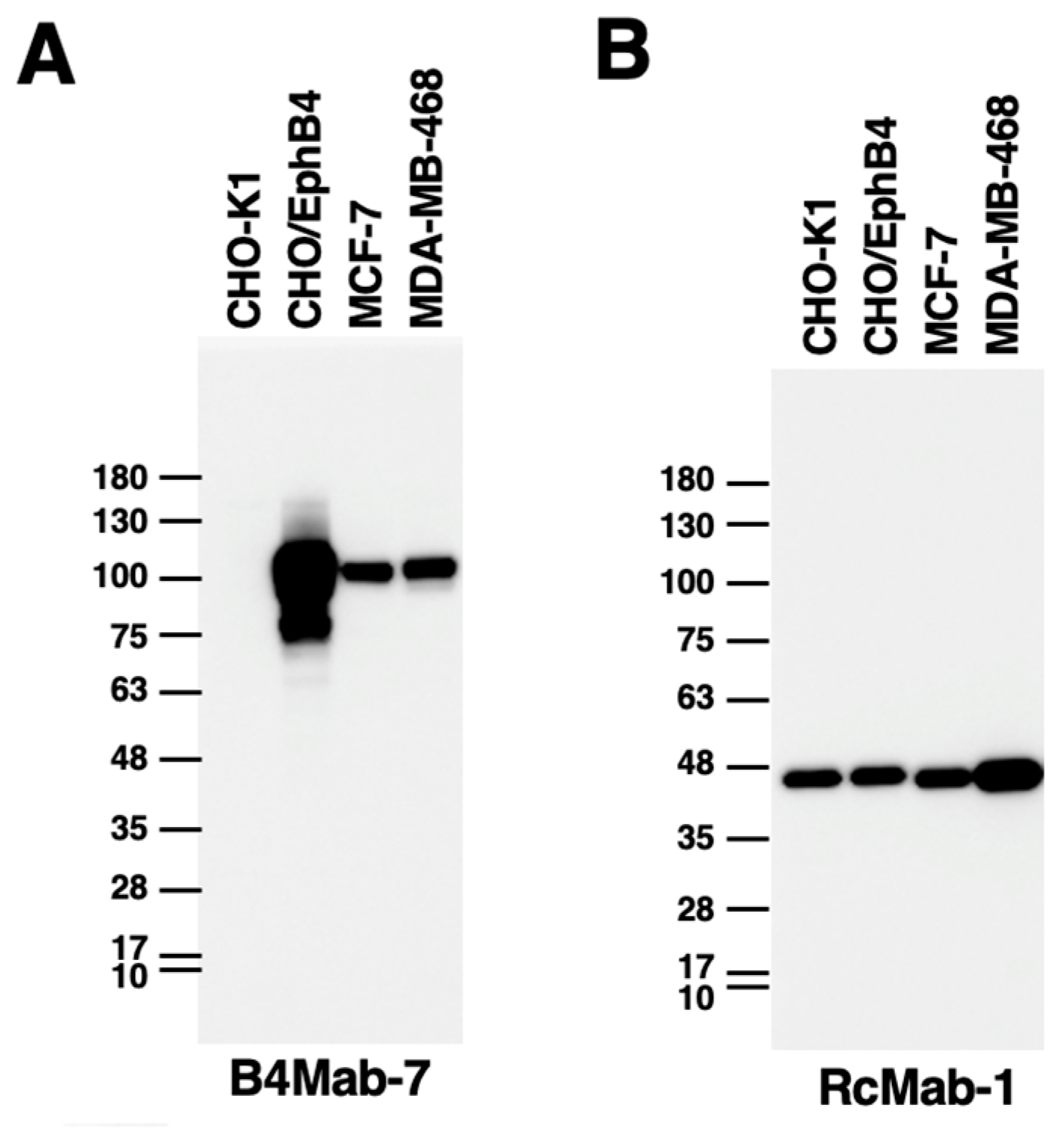
3.4. Western blot analysis
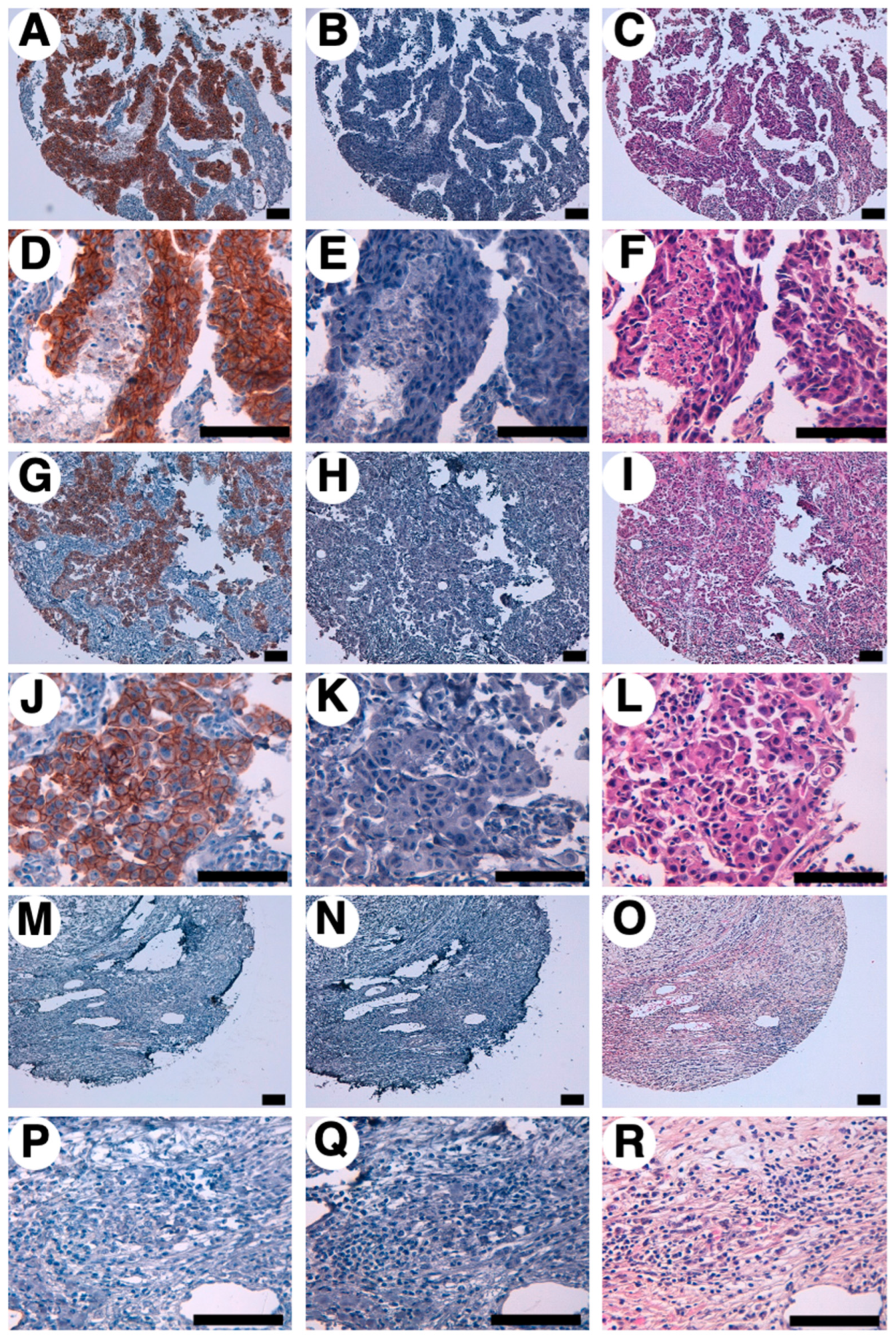
3.5. Immunohistochemical analysis against breast cancers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Weir, H.K.; Carreira, H.; Harewood, R.; Spika, D.; Wang, X.S.; Bannon, F.; Ahn, J.V.; Johnson, C.J.; Bonaventure, A.; et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015, 385, 977–1010. [Google Scholar] [CrossRef] [PubMed]
- Massat, N.J.; Dibden, A.; Parmar, D.; Cuzick, J.; Sasieni, P.D.; Duffy, S.W. Impact of Screening on Breast Cancer Mortality: The UK Program 20 Years On. Cancer Epidemiol Biomarkers Prev 2016, 25, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Bleyer, A.; Welch, H.G. Effect of three decades of screening mammography on breast-cancer incidence. N Engl J Med 2012, 367, 1998–2005. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- McDonald, E.S.; Clark, A.S.; Tchou, J.; Zhang, P.; Freedman, G.M. Clinical Diagnosis and Management of Breast Cancer. J Nucl Med 2016, 57 Suppl 1, 9S–16S. [Google Scholar] [CrossRef]
- Fisusi, F.A.; Akala, E.O. Drug Combinations in Breast Cancer Therapy. Pharm Nanotechnol 2019, 7, 3–23. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thurlimann, B.; Senn, H.J.; Panel, m. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galvan, P.; Fernandez, A.; Gaba, L.; Diez, M.; Viladot, M.; Arance, A.; Munoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24 Suppl 2, S26–35. [Google Scholar] [CrossRef]
- Mohammed, A.A. The clinical behavior of different molecular subtypes of breast cancer. Cancer Treat Res Commun 2021, 29, 100469. [Google Scholar] [CrossRef]
- Tuzi, N.L.; Gullick, W.J. eph, the largest known family of putative growth factor receptors. Br J Cancer 1994, 69, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Lisabeth, E.M.; Falivelli, G.; Pasquale, E.B. Eph receptor signaling and ephrins. Cold Spring Harb Perspect Biol 2013, 5. [Google Scholar] [CrossRef]
- Hirai, H.; Maru, Y.; Hagiwara, K.; Nishida, J.; Takaku, F. A novel putative tyrosine kinase receptor encoded by the eph gene. Science 1987, 238, 1717–1720. [Google Scholar] [CrossRef] [PubMed]
- Darling, T.K.; Lamb, T.J. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front Immunol 2019, 10, 1473. [Google Scholar] [CrossRef] [PubMed]
- Himanen, J.P.; Rajashankar, K.R.; Lackmann, M.; Cowan, C.A.; Henkemeyer, M.; Nikolov, D.B. Crystal structure of an Eph receptor-ephrin complex. Nature 2001, 414, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Kania, A.; Klein, R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol 2016, 17, 240–256. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph-ephrin promiscuity is now crystal clear. Nat Neurosci 2004, 7, 417–418. [Google Scholar] [CrossRef]
- Himanen, J.P.; Chumley, M.J.; Lackmann, M.; Li, C.; Barton, W.A.; Jeffrey, P.D.; Vearing, C.; Geleick, D.; Feldheim, D.A.; Boyd, A.W.; et al. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci 2004, 7, 501–509. [Google Scholar] [CrossRef]
- Forse, G.J.; Uson, M.L.; Nasertorabi, F.; Kolatkar, A.; Lamberto, I.; Pasquale, E.B.; Kuhn, P. Distinctive Structure of the EphA3/Ephrin-A5 Complex Reveals a Dual Mode of Eph Receptor Interaction for Ephrin-A5. PLoS One 2015, 10, e0127081. [Google Scholar] [CrossRef]
- North, H.A.; Zhao, X.; Kolk, S.M.; Clifford, M.A.; Ziskind, D.M.; Donoghue, M.J. Promotion of proliferation in the developing cerebral cortex by EphA4 forward signaling. Development 2009, 136, 2467–2476. [Google Scholar] [CrossRef]
- Murai, K.K.; Pasquale, E.B. 'Eph'ective signaling: forward, reverse and crosstalk. J Cell Sci 2003, 116, 2823–2832. [Google Scholar] [CrossRef]
- Depaepe, V.; Suarez-Gonzalez, N.; Dufour, A.; Passante, L.; Gorski, J.A.; Jones, K.R.; Ledent, C.; Vanderhaeghen, P. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature 2005, 435, 1244–1250. [Google Scholar] [CrossRef]
- Mignon, L.; Vourc'h, P.; Romero-Ramos, M.; Osztermann, P.; Young, H.E.; Lucas, P.A.; Chesselet, M.F. Transplantation of multipotent cells extracted from adult skeletal muscles into the subventricular zone of adult rats. J Comp Neurol 2005, 491, 96–108. [Google Scholar] [CrossRef]
- Jensen, P.L. Eph receptors and ephrins. Stem Cells 2000, 18, 63–64. [Google Scholar] [CrossRef]
- Salvucci, O.; Tosato, G. Essential roles of EphB receptors and EphrinB ligands in endothelial cell function and angiogenesis. Adv Cancer Res 2012, 114, 21–57. [Google Scholar] [CrossRef]
- Cheng, N.; Brantley, D.M.; Chen, J. The ephrins and Eph receptors in angiogenesis. Cytokine Growth Factor Rev 2002, 13, 75–85. [Google Scholar] [CrossRef]
- Makinen, T.; Adams, R.H.; Bailey, J.; Lu, Q.; Ziemiecki, A.; Alitalo, K.; Klein, R.; Wilkinson, G.A. PDZ interaction site in ephrinB2 is required for the remodeling of lymphatic vasculature. Genes Dev 2005, 19, 397–410. [Google Scholar] [CrossRef]
- Zhang, G.; Brady, J.; Liang, W.C.; Wu, Y.; Henkemeyer, M.; Yan, M. EphB4 forward signalling regulates lymphatic valve development. Nat Commun 2015, 6, 6625. [Google Scholar] [CrossRef]
- Gerety, S.S.; Wang, H.U.; Chen, Z.F.; Anderson, D.J. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell 1999, 4, 403–414. [Google Scholar] [CrossRef]
- Adams, R.H.; Wilkinson, G.A.; Weiss, C.; Diella, F.; Gale, N.W.; Deutsch, U.; Risau, W.; Klein, R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev 1999, 13, 295–306. [Google Scholar] [CrossRef]
- Wang, H.U.; Chen, Z.F.; Anderson, D.J. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 1998, 93, 741–753. [Google Scholar] [CrossRef]
- Wang, Z.; Cohen, K.; Shao, Y.; Mole, P.; Dombkowski, D.; Scadden, D.T. Ephrin receptor, EphB4, regulates ES cell differentiation of primitive mammalian hemangioblasts, blood, cardiomyocytes, and blood vessels. Blood 2004, 103, 100–109. [Google Scholar] [CrossRef]
- de Muijnck, C.; van Gorkom, Y.; van Duijvenvoorde, M.; Eghtesadi, M.; Dekker-Ensink, G.; Bhairosingh, S.S.; Affinito, A.; Kuppen, P.J.K.; Vahrmeijer, A.L.; Sier, C.F.M. Evaluation of EphB4 as Target for Image-Guided Surgery of Breast Cancer. Pharmaceuticals (Basel) 2020, 13. [Google Scholar] [CrossRef]
- Kumar, S.R.; Singh, J.; Xia, G.; Krasnoperov, V.; Hassanieh, L.; Ley, E.J.; Scehnet, J.; Kumar, N.G.; Hawes, D.; Press, M.F.; et al. Receptor tyrosine kinase EphB4 is a survival factor in breast cancer. Am J Pathol 2006, 169, 279–293. [Google Scholar] [CrossRef]
- Brantley-Sieders, D.M.; Jiang, A.; Sarma, K.; Badu-Nkansah, A.; Walter, D.L.; Shyr, Y.; Chen, J. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PLoS One 2011, 6, e24426. [Google Scholar] [CrossRef]
- Zheng, M.F.; Ji, Y.; Wu, X.B.; Ye, S.G.; Chen, J.Y. EphB4 gene polymorphism and protein expression in non-small-cell lung cancer. Mol Med Rep 2012, 6, 405–408. [Google Scholar] [CrossRef]
- Ferguson, B.D.; Liu, R.; Rolle, C.E.; Tan, Y.H.; Krasnoperov, V.; Kanteti, R.; Tretiakova, M.S.; Cervantes, G.M.; Hasina, R.; Hseu, R.D.; et al. The EphB4 receptor tyrosine kinase promotes lung cancer growth: a potential novel therapeutic target. PLoS One 2013, 8, e67668. [Google Scholar] [CrossRef]
- Nanamiya, R.; Saito-Koyama, R.; Miki, Y.; Inoue, C.; Asavasupreechar, T.; Abe, J.; Sato, I.; Sasano, H. EphB4 as a Novel Target for the EGFR-Independent Suppressive Effects of Osimertinib on Cell Cycle Progression in Non-Small Cell Lung Cancer. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Yin, J.; Cui, Y.; Li, L.; Ji, J.; Jiang, W.G. Overexpression of EPHB4 Is Associated with Poor Survival of Patients with Gastric Cancer. Anticancer Res 2017, 37, 4489–4497. [Google Scholar] [CrossRef]
- Stammes, M.A.; Prevoo, H.A.; Ter Horst, M.C.; Groot, S.A.; Van de Velde, C.J.; Chan, A.B.; de Geus-Oei, L.F.; Kuppen, P.J.; Vahrmeijer, A.L.; Pasquale, E.B.; et al. Evaluation of EphA2 and EphB4 as Targets for Image-Guided Colorectal Cancer Surgery. Int J Mol Sci 2017, 18. [Google Scholar] [CrossRef]
- Lv, J.; Xia, Q.; Wang, J.; Shen, Q.; Zhang, J.; Zhou, X. EphB4 promotes the proliferation, invasion, and angiogenesis of human colorectal cancer. Exp Mol Pathol 2016, 100, 402–408. [Google Scholar] [CrossRef]
- Merchant, A.A.; Jorapur, A.; McManus, A.; Liu, R.; Krasnoperov, V.; Chaudhry, P.; Singh, M.; Harton, L.; Agajanian, M.; Kim, M.; et al. EPHB4 is a therapeutic target in AML and promotes leukemia cell survival via AKT. Blood Adv 2017, 1, 1635–1644. [Google Scholar] [CrossRef]
- Alam, S.M.; Fujimoto, J.; Jahan, I.; Sato, E.; Tamaya, T. Coexpression of EphB4 and ephrinB2 in tumor advancement of uterine cervical cancers. Gynecol Oncol 2009, 114, 84–88. [Google Scholar] [CrossRef]
- Tu, Y.; He, S.; Fu, J.; Li, G.; Xu, R.; Lu, H.; Deng, J. Expression of EphrinB2 and EphB4 in glioma tissues correlated to the progression of glioma and the prognosis of glioblastoma patients. Clin Transl Oncol 2012, 14, 214–220. [Google Scholar] [CrossRef]
- Kumar, S.R.; Masood, R.; Spannuth, W.A.; Singh, J.; Scehnet, J.; Kleiber, G.; Jennings, N.; Deavers, M.; Krasnoperov, V.; Dubeau, L.; et al. The receptor tyrosine kinase EphB4 is overexpressed in ovarian cancer, provides survival signals and predicts poor outcome. Br J Cancer 2007, 96, 1083–1091. [Google Scholar] [CrossRef]
- Alam, S.M.; Fujimoto, J.; Jahan, I.; Sato, E.; Tamaya, T. Coexpression of EphB4 and ephrinB2 in tumour advancement of ovarian cancers. Br J Cancer 2008, 98, 845–851. [Google Scholar] [CrossRef]
- Sagar, V.; Vatapalli, R.; Lysy, B.; Pamarthy, S.; Anker, J.F.; Rodriguez, Y.; Han, H.; Unno, K.; Stadler, W.M.; Catalona, W.J.; et al. EPHB4 inhibition activates ER stress to promote immunogenic cell death of prostate cancer cells. Cell Death Dis 2019, 10, 801. [Google Scholar] [CrossRef]
- Xia, G.; Kumar, S.R.; Masood, R.; Zhu, S.; Reddy, R.; Krasnoperov, V.; Quinn, D.I.; Henshall, S.M.; Sutherland, R.L.; Pinski, J.K.; et al. EphB4 expression and biological significance in prostate cancer. Cancer Res 2005, 65, 4623–4632. [Google Scholar] [CrossRef]
- Xuqing, W.; Lei, C.; Zhengfa, M.; Shengchun, D.; Xin, F.; Jianguo, Q.; Jianxin, Z. EphB4 is overexpressed in papillary thyroid carcinoma and promotes the migration of papillary thyroid cancer cells. Tumour Biol 2012, 33, 1419–1427. [Google Scholar] [CrossRef]
- Sharma, G.K.; Dhillon, V.K.; Masood, R.; Maceri, D.R. Overexpression of EphB4, EphrinB2, and epidermal growth factor receptor in papillary thyroid carcinoma: A pilot study. Head Neck 2015, 37, 964–969. [Google Scholar] [CrossRef]
- Xia, G.; Kumar, S.R.; Stein, J.P.; Singh, J.; Krasnoperov, V.; Zhu, S.; Hassanieh, L.; Smith, D.L.; Buscarini, M.; Broek, D.; et al. EphB4 receptor tyrosine kinase is expressed in bladder cancer and provides signals for cell survival. Oncogene 2006, 25, 769–780. [Google Scholar] [CrossRef]
- Li, X.; Choi, W.W.; Yan, R.; Yu, H.; Krasnoperov, V.; Kumar, S.R.; Schuckman, A.; Klumpp, D.J.; Pan, C.X.; Quinn, D.; et al. The differential expression of EphB2 and EphB4 receptor kinases in normal bladder and in transitional cell carcinoma of the bladder. PLoS One 2014, 9, e105326. [Google Scholar] [CrossRef]
- Ferguson, B.D.; Carol Tan, Y.H.; Kanteti, R.S.; Liu, R.; Gayed, M.J.; Vokes, E.E.; Ferguson, M.K.; John Iafrate, A.; Gill, P.S.; Salgia, R. Novel EPHB4 Receptor Tyrosine Kinase Mutations and Kinomic Pathway Analysis in Lung Cancer. Sci Rep 2015, 5, 10641. [Google Scholar] [CrossRef]
- Masood, R.; Kumar, S.R.; Sinha, U.K.; Crowe, D.L.; Krasnoperov, V.; Reddy, R.K.; Zozulya, S.; Singh, J.; Xia, G.; Broek, D.; et al. EphB4 provides survival advantage to squamous cell carcinoma of the head and neck. Int J Cancer 2006, 119, 1236–1248. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, H.; Zhang, Y. Targeting receptor tyrosine kinase EphB4 in cancer therapy. Semin Cancer Biol 2019, 56, 37–46. [Google Scholar] [CrossRef]
- Noren, N.K.; Lu, M.; Freeman, A.L.; Koolpe, M.; Pasquale, E.B. Interplay between EphB4 on tumor cells and vascular ephrin-B2 regulates tumor growth. Proc Natl Acad Sci U S A 2004, 101, 5583–5588. [Google Scholar] [CrossRef]
- Piffko, A.; Broggini, T.; Harms, C.; Adams, R.H.; Vajkoczy, P.; Czabanka, M. Ligand-Dependent and Ligand-Independent Effects of Ephrin-B2-EphB4 Signaling in Melanoma Metastatic Spine Disease. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Aslam, M.I.; Abraham, J.; Mansoor, A.; Druker, B.J.; Tyner, J.W.; Keller, C. PDGFRbeta reverses EphB4 signaling in alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A 2014, 111, 6383–6388. [Google Scholar] [CrossRef]
- Noren, N.K.; Foos, G.; Hauser, C.A.; Pasquale, E.B. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nat Cell Biol 2006, 8, 815–825. [Google Scholar] [CrossRef]
- Wu, Q.; Suo, Z.; Risberg, B.; Karlsson, M.G.; Villman, K.; Nesland, J.M. Expression of Ephb2 and Ephb4 in breast carcinoma. Pathol Oncol Res 2004, 10, 26–33. [Google Scholar] [CrossRef]
- Ding, J.; Yao, Y.; Huang, G.; Wang, X.; Yi, J.; Zhang, N.; Liu, C.; Wang, K.; Zhang, Y.; Wang, M.; et al. Targeting the EphB4 receptor tyrosine kinase sensitizes HER2-positive breast cancer cells to Lapatinib. Cancer Lett 2020, 475, 53–64. [Google Scholar] [CrossRef]
- Zhu, M.; Tang, X.; Gong, Z.; Tang, W.; Zhang, Y. TAD1822-7 induces ROS-mediated apoptosis of HER2 positive breast cancer by decreasing E-cadherin in an EphB4 dependent manner. Life Sci 2021, 285, 119954. [Google Scholar] [CrossRef]
- Yamada, S.; Itai, S.; Nakamura, T.; Yanaka, M.; Kaneko, M.K.; Kato, Y. Detection of high CD44 expression in oral cancers using the novel monoclonal antibody, C(44)Mab-5. Biochem Biophys Rep 2018, 14, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Kaneko, M.K.; Kato, Y. MAP Tag: A Novel Tagging System for Protein Purification and Detection. Monoclon Antib Immunodiagn Immunother 2016, 35, 293–299. [Google Scholar] [CrossRef]
- Fujii, Y.; Kaneko, M.; Neyazaki, M.; Nogi, T.; Kato, Y.; Takagi, J. PA tag: a versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Protein Expr Purif 2014, 95, 240–247. [Google Scholar] [CrossRef]
- Nanamiya, R.; Takei, J.; Asano, T.; Tanaka, T.; Sano, M.; Nakamura, T.; Yanaka, M.; Hosono, H.; Kaneko, M.K.; Kato, Y. Development of Anti-Human CC Chemokine Receptor 9 Monoclonal Antibodies for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2021, 40, 101–106. [Google Scholar] [CrossRef]
- Tanaka, T.; Asano, T.; Sano, M.; Takei, J.; Hosono, H.; Nanamiya, R.; Nakamura, T.; Yanaka, M.; Harada, H.; Fukui, M.; et al. Development of Monoclonal Antibody PMab-269 Against California Sea Lion Podoplanin. Monoclon Antib Immunodiagn Immunother 2021, 40, 124–133. [Google Scholar] [CrossRef]
- Takei, J.; Asano, T.; Nanamiya, R.; Nakamura, T.; Yanaka, M.; Hosono, H.; Tanaka, T.; Sano, M.; Kaneko, M.K.; Harada, H.; et al. Development of Anti-human T Cell Immunoreceptor with Ig and ITIM Domains (TIGIT) Monoclonal Antibodies for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2021, 40, 71–75. [Google Scholar] [CrossRef]
- Sayama, Y.; Kaneko, M.K.; Takei, J.; Hosono, H.; Sano, M.; Asano, T.; Kato, Y. Establishment of a novel anti-TROP2 monoclonal antibody TrMab-29 for immunohistochemical analysis. Biochem Biophys Rep 2021, 25, 100902. [Google Scholar] [CrossRef]
- Sayama, Y.; Kaneko, M.K.; Kato, Y. Development and characterization of TrMab-6, a novel anti-TROP2 monoclonal antibody for antigen detection in breast cancer. Mol Med Rep 2021, 23. [Google Scholar] [CrossRef]
- Asano, T.; Nanamiya, R.; Takei, J.; Nakamura, T.; Yanaka, M.; Hosono, H.; Tanaka, T.; Sano, M.; Kaneko, M.K.; Kato, Y. Development of Anti-Mouse CC Chemokine Receptor 3 Monoclonal Antibodies for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2021, 40, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.K.; Sano, M.; Takei, J.; Asano, T.; Sayama, Y.; Hosono, H.; Kobayashi, A.; Konnai, S.; Kato, Y. Development and Characterization of Anti-Sheep Podoplanin Monoclonal Antibodies PMab-253 and PMab-260. Monoclon Antib Immunodiagn Immunother 2020, 39, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Kaneko, M.K.; Kato, Y. Establishment of C(20)Mab-11, a novel anti-CD20 monoclonal antibody, for the detection of B cells. Oncol Lett 2020, 20, 1961–1967. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Kaneko, M.K.; Kato, Y. Establishment of an Anti-CD20 Monoclonal Antibody (C(20)Mab-60) for Immunohistochemical Analyses. Monoclon Antib Immunodiagn Immunother 2020, 39, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Furusawa, Y.; Yamada, S.; Itai, S.; Takei, J.; Sano, M.; Kaneko, M.K. Establishment of a monoclonal antibody PMab-225 against alpaca podoplanin for immunohistochemical analyses. Biochem Biophys Rep 2019, 18, 100633. [Google Scholar] [CrossRef]
- Ikota, H.; Nobusawa, S.; Arai, H.; Kato, Y.; Ishizawa, K.; Hirose, T.; Yokoo, H. Evaluation of IDH1 status in diffusely infiltrating gliomas by immunohistochemistry using anti-mutant and wild type IDH1 antibodies. Brain Tumor Pathol 2015, 32, 237–244. [Google Scholar] [CrossRef]
- Carles-Kinch, K.; Kilpatrick, K.E.; Stewart, J.C.; Kinch, M.S. Antibody targeting of the EphA2 tyrosine kinase inhibits malignant cell behavior. Cancer Res 2002, 62, 2840–2847. [Google Scholar]
- Coffman, K.T.; Hu, M.; Carles-Kinch, K.; Tice, D.; Donacki, N.; Munyon, K.; Kifle, G.; Woods, R.; Langermann, S.; Kiener, P.A.; et al. Differential EphA2 epitope display on normal versus malignant cells. Cancer Res 2003, 63, 7907–7912. [Google Scholar]
- Landen, C.N., Jr.; Lu, C.; Han, L.Y.; Coffman, K.T.; Bruckheimer, E.; Halder, J.; Mangala, L.S.; Merritt, W.M.; Lin, Y.G.; Gao, C.; et al. Efficacy and antivascular effects of EphA2 reduction with an agonistic antibody in ovarian cancer. J Natl Cancer Inst 2006, 98, 1558–1570. [Google Scholar] [CrossRef]
- Merritt, W.M.; Kamat, A.A.; Hwang, J.Y.; Bottsford-Miller, J.; Lu, C.; Lin, Y.G.; Coffey, D.; Spannuth, W.A.; Nugent, E.; Han, L.Y.; et al. Clinical and biological impact of EphA2 overexpression and angiogenesis in endometrial cancer. Cancer Biol Ther 2010, 10, 1306–1314. [Google Scholar] [CrossRef]
- Gokmen-Polar, Y.; Toroni, R.A.; Hocevar, B.A.; Badve, S.; Zhao, Q.; Shen, C.; Bruckheimer, E.; Kinch, M.S.; Miller, K.D. Dual targeting of EphA2 and ER restores tamoxifen sensitivity in ER/EphA2-positive breast cancer. Breast Cancer Res Treat 2011, 127, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Ansuini, H.; Meola, A.; Gunes, Z.; Paradisi, V.; Pezzanera, M.; Acali, S.; Santini, C.; Luzzago, A.; Mori, F.; Lazzaro, D.; et al. Anti-EphA2 Antibodies with Distinct In Vitro Properties Have Equal In Vivo Efficacy in Pancreatic Cancer. J Oncol 2009, 2009, 951917. [Google Scholar] [CrossRef] [PubMed]
- Wesa, A.K.; Herrem, C.J.; Mandic, M.; Taylor, J.L.; Vasquez, C.; Kawabe, M.; Tatsumi, T.; Leibowitz, M.S.; Finke, J.H.; Bukowski, R.M.; et al. Enhancement in specific CD8+ T cell recognition of EphA2+ tumors in vitro and in vivo after treatment with ligand agonists. J Immunol 2008, 181, 7721–7727. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, J.; Sue, M.; Yamato, M.; Ichikawa, J.; Ishida, S.; Shibutani, T.; Kitamura, M.; Wada, T.; Agatsuma, T. Novel anti-EPHA2 antibody, DS-8895a for cancer treatment. Cancer Biol Ther 2016, 17, 1158–1167. [Google Scholar] [CrossRef]
- Bruckheimer, E.M.; Fazenbaker, C.A.; Gallagher, S.; Mulgrew, K.; Fuhrmann, S.; Coffman, K.T.; Walsh, W.; Ready, S.; Cook, K.; Damschroder, M.; et al. Antibody-dependent cell-mediated cytotoxicity effector-enhanced EphA2 agonist monoclonal antibody demonstrates potent activity against human tumors. Neoplasia 2009, 11, 509–517, 502 p following 517. [Google Scholar] [CrossRef]
- Charmsaz, S.; Beckett, K.; Smith, F.M.; Bruedigam, C.; Moore, A.S.; Al-Ejeh, F.; Lane, S.W.; Boyd, A.W. EphA2 Is a Therapy Target in EphA2-Positive Leukemias but Is Not Essential for Normal Hematopoiesis or Leukemia. PLoS One 2015, 10, e0130692. [Google Scholar] [CrossRef]
- Jackson, D.; Gooya, J.; Mao, S.; Kinneer, K.; Xu, L.; Camara, M.; Fazenbaker, C.; Fleming, R.; Swamynathan, S.; Meyer, D.; et al. A human antibody-drug conjugate targeting EphA2 inhibits tumor growth in vivo. Cancer Res 2008, 68, 9367–9374. [Google Scholar] [CrossRef]
- Lee, J.W.; Stone, R.L.; Lee, S.J.; Nam, E.J.; Roh, J.W.; Nick, A.M.; Han, H.D.; Shahzad, M.M.; Kim, H.S.; Mangala, L.S.; et al. EphA2 targeted chemotherapy using an antibody drug conjugate in endometrial carcinoma. Clin Cancer Res 2010, 16, 2562–2570. [Google Scholar] [CrossRef]
- Shitara, K.; Satoh, T.; Iwasa, S.; Yamaguchi, K.; Muro, K.; Komatsu, Y.; Nishina, T.; Esaki, T.; Hasegawa, J.; Kakurai, Y.; et al. Safety, tolerability, pharmacokinetics, and pharmacodynamics of the afucosylated, humanized anti-EPHA2 antibody DS-8895a: a first-in-human phase I dose escalation and dose expansion study in patients with advanced solid tumors. J Immunother Cancer 2019, 7, 219. [Google Scholar] [CrossRef]
- Annunziata, C.M.; Kohn, E.C.; LoRusso, P.; Houston, N.D.; Coleman, R.L.; Buzoianu, M.; Robbie, G.; Lechleider, R. Phase 1, open-label study of MEDI-547 in patients with relapsed or refractory solid tumors. Invest New Drugs 2013, 31, 77–84. [Google Scholar] [CrossRef]
- Sakamoto, A.; Kato, K.; Hasegawa, T.; Ikeda, S. An Agonistic Antibody to EPHA2 Exhibits Antitumor Effects on Human Melanoma Cells. Anticancer Res 2018, 38, 3273–3282. [Google Scholar] [CrossRef] [PubMed]
- Vearing, C.; Lee, F.T.; Wimmer-Kleikamp, S.; Spirkoska, V.; To, C.; Stylianou, C.; Spanevello, M.; Brechbiel, M.; Boyd, A.W.; Scott, A.M.; et al. Concurrent binding of anti-EphA3 antibody and ephrin-A5 amplifies EphA3 signaling and downstream responses: potential as EphA3-specific tumor-targeting reagents. Cancer Res 2005, 65, 6745–6754. [Google Scholar] [CrossRef] [PubMed]
- Charmsaz, S.; Al-Ejeh, F.; Yeadon, T.M.; Miller, K.J.; Smith, F.M.; Stringer, B.W.; Moore, A.S.; Lee, F.T.; Cooper, L.T.; Stylianou, C.; et al. EphA3 as a target for antibody immunotherapy in acute lymphoblastic leukemia. Leukemia 2017, 31, 1779–1787. [Google Scholar] [CrossRef] [PubMed]
- Vail, M.E.; Murone, C.; Tan, A.; Hii, L.; Abebe, D.; Janes, P.W.; Lee, F.T.; Baer, M.; Palath, V.; Bebbington, C.; et al. Targeting EphA3 inhibits cancer growth by disrupting the tumor stromal microenvironment. Cancer Res 2014, 74, 4470–4481. [Google Scholar] [CrossRef] [PubMed]
- Swords, R.T.; Greenberg, P.L.; Wei, A.H.; Durrant, S.; Advani, A.S.; Hertzberg, M.S.; Jonas, B.A.; Lewis, I.D.; Rivera, G.; Gratzinger, D.; et al. KB004, a first in class monoclonal antibody targeting the receptor tyrosine kinase EphA3, in patients with advanced hematologic malignancies: Results from a phase 1 study. Leuk Res 2016, 50, 123–131. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, F.; Airoldi, I.; Di Carlo, E.; Marotta, P.; Falco, G.; Simeon, V.; Laurenzana, I.; Trino, S.; De Luca, L.; Todoerti, K.; et al. EphA3 targeting reduces in vitro adhesion and invasion and in vivo growth and angiogenesis of multiple myeloma cells. Cell Oncol (Dordr) 2017, 40, 483–496. [Google Scholar] [CrossRef]
- Day, B.W.; Stringer, B.W.; Al-Ejeh, F.; Ting, M.J.; Wilson, J.; Ensbey, K.S.; Jamieson, P.R.; Bruce, Z.C.; Lim, Y.C.; Offenhauser, C.; et al. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell 2013, 23, 238–248. [Google Scholar] [CrossRef]
- Chu, L.; Wang, A.; Ni, L.; Yan, X.; Song, Y.; Zhao, M.; Sun, K.; Mu, H.; Liu, S.; Wu, Z.; et al. Nose-to-brain delivery of temozolomide-loaded PLGA nanoparticles functionalized with anti-EPHA3 for glioblastoma targeting. Drug Deliv 2018, 25, 1634–1641. [Google Scholar] [CrossRef]
- Offenhauser, C.; Al-Ejeh, F.; Puttick, S.; Ensbey, K.S.; Bruce, Z.C.; Jamieson, P.R.; Smith, F.M.; Stringer, B.W.; Carrington, B.; Fuchs, A.V.; et al. EphA3 Pay-Loaded Antibody Therapeutics for the Treatment of Glioblastoma. Cancers (Basel) 2018, 10. [Google Scholar] [CrossRef]
- Taki, S.; Kamada, H.; Inoue, M.; Nagano, K.; Mukai, Y.; Higashisaka, K.; Yoshioka, Y.; Tsutsumi, Y.; Tsunoda, S. A Novel Bispecific Antibody against Human CD3 and Ephrin Receptor A10 for Breast Cancer Therapy. Plos One 2015, 10. [Google Scholar] [CrossRef]
- Mao, W.; Luis, E.; Ross, S.; Silva, J.; Tan, C.; Crowley, C.; Chui, C.; Franz, G.; Senter, P.; Koeppen, H.; et al. EphB2 as a therapeutic antibody drug target for the treatment of colorectal cancer. Cancer Res 2004, 64, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Krasnoperov, V.; Kumar, S.R.; Ley, E.; Li, X.; Scehnet, J.; Liu, R.; Zozulya, S.; Gill, P.S. Novel EphB4 monoclonal antibodies modulate angiogenesis and inhibit tumor growth. Am J Pathol 2010, 176, 2029–2038. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, S.A.; Douglas, E.L.; Mertens-Walker, I.; Lisle, J.E.; Maharaj, M.S.; Herington, A.C. Anti-tumour effects of antibodies targeting the extracellular cysteine-rich region of the receptor tyrosine kinase EphB4. Oncotarget 2015, 6, 7554–7569. [Google Scholar] [CrossRef] [PubMed]
- Janes, P.W.; Vail, M.E.; Gan, H.K.; Scott, A.M. Antibody Targeting of Eph Receptors in Cancer. Pharmaceuticals (Basel) 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.; Cher, L.; Inglis, P.; Lwin, Z.; Lau, E.; Ackermann, U.; Coombs, N.; Remen, K.; Guo, N.; Lee, S.T.; et al. Preliminary Findings of a Phase I Safety and Bioimaging Trial of Kb004 (Ifabotuzumab) in Patients with Glioblastoma. Neuro-Oncology 2019, 21, 6–6. [Google Scholar] [CrossRef]
- Boyd, A.W.; Ward, L.D.; Wicks, I.P.; Simpson, R.J.; Salvaris, E.; Wilks, A.; Welch, K.; Loudovaris, M.; Rockman, S.; Busmanis, I. Isolation and characterization of a novel receptor-type protein tyrosine kinase (hek) from a human pre-B cell line. J Biol Chem 1992, 267, 3262–3267. [Google Scholar] [CrossRef]
- Lin, Q.; Ba, T.; Ho, J.; Chen, D.; Cheng, Y.; Wang, L.; Xu, G.; Xu, L.; Zhou, Y.; Wei, Y.; et al. First-in-Human Trial of EphA2-Redirected CAR T-Cells in Patients With Recurrent Glioblastoma: A Preliminary Report of Three Cases at the Starting Dose. Front Oncol 2021, 11, 694941. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Wang, Y.N.; Chu, Y.Y.; Wang, C.H.; Lee, H.H.; Xia, W.; Shyu, W.C.; Liu, S.P.; Yao, J.; et al. Ephrin receptor A10 monoclonal antibodies and the derived chimeric antigen receptor T cells exert an antitumor response in mouse models of triple-negative breast cancer. J Biol Chem 2022, 298, 101817. [Google Scholar] [CrossRef]
- Sharma, P.; Roberts, C.; Herpai, D.; Fokt, I.D.; Priebe, W.; Debinski, W. Drug Conjugates for Targeting Eph Receptors in Glioblastoma. Pharmaceuticals (Basel) 2020, 13. [Google Scholar] [CrossRef]
- Hammond, S.A.; Lutterbuese, R.; Roff, S.; Lutterbuese, P.; Schlereth, B.; Bruckheimer, E.; Kinch, M.S.; Coats, S.; Baeuerle, P.A.; Kufer, P.; et al. Selective targeting and potent control of tumor growth using an EphA2/CD3-Bispecific single-chain antibody construct. Cancer Res 2007, 67, 3927–3935. [Google Scholar] [CrossRef]
- Janowicz, P.W.; Houston, Z.H.; Bunt, J.; Fletcher, N.L.; Bell, C.A.; Cowin, G.; Howard, C.B.; Taslima, D.; Westra van Holthe, N.; Prior, A.; et al. Understanding nanomedicine treatment in an aggressive spontaneous brain cancer model at the stage of early blood brain barrier disruption. Biomaterials 2022, 283, 121416. [Google Scholar] [CrossRef] [PubMed]
- Qazi, M.A.; Vora, P.; Venugopal, C.; Adams, J.; Singh, M.; Hu, A.; Gorelik, M.; Subapanditha, M.K.; Savage, N.; Yang, J.; et al. Cotargeting Ephrin Receptor Tyrosine Kinases A2 and A3 in Cancer Stem Cells Reduces Growth of Recurrent Glioblastoma. Cancer Res 2018, 78, 5023–5037. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, W.; Swindell, E.; Pien, C.; Luus, L.; Cain, J.; Pham, M.; Kandela, I.; Huang, Z.R.; Tipparaju, S.K.; Koshkaryev, A.; et al. Targeting EphA2 in Bladder Cancer Using a Novel Antibody-Directed Nanotherapeutic. Pharmaceutics 2020, 12. [Google Scholar] [CrossRef] [PubMed]
| No. | Sex | Age | Organ | Pathology diagnosis | Differentiation | TNM | B4Mab-7 |
| 1 | F | 44 | Breast | Invasive ductal carcinoma | Moderately | T2N2M1 | 2+ |
| 2 | F | 58 | Breast | Medullary carcinoma | Moderately | T2N2M1 | 3+ |
| 3 | F | 40 | Breast | Invasive ductal carcinoma | Moderately | T2N1M0 | 1+ |
| 4 | F | 52 | Breast | Invasive ductal carcinoma | Moderately | T2N2M1 | 0 |
| 5 | F | 60 | Breast | Invasive ductal carcinoma | Moderately | T2N1M1 | 1+ |
| 6 | F | 57 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 3+ |
| 7 | F | 48 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 2+ |
| 8 | F | 66 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 0 |
| 9 | F | 58 | Breast | Adenocarcinoma | Moderately | T2N2M1 | 0 |
| 10 | F | 63 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 1+ |
| 11 | F | 32 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 1+ |
| 12 | F | 59 | Breast | Invasive lobular carcinoma | Well | T2N2M0 | 0 |
| 13 | F | 44 | Breast | Invasive lobular carcinoma | Well | T2N2M0 | 0 |
| 14 | F | 60 | Breast | Invasive lobular carcinoma | Moderately | T2N1M0 | 1+ |
| 15 | F | 44 | Breast | Invasive ductal carcinoma | Moderately | T2N2M0 | 3+ |
| 16 | F | 82 | Breast | Invasive ductal carcinoma | Moderately | T2N1M1 | 1+ |
| 17 | F | 58 | Breast | Adenocarcinoma | Moderately | T2N1M1 | 2+ |
| 18 | F | 57 | Breast | Invasive ductal carcinoma | Poorly | T3N3M0 | 1+ |
| 19 | F | 41 | Breast | Invasive ductal carcinoma | Moderately | T2N1M0 | 3+ |
| 20 | F | 44 | Breast | Invasive ductal carcinoma | Moderately | T2N2M0 | 0 |
| 21 | F | 78 | Breast | Invasive ductal carcinoma | Moderately | T2N1M0 | 1+ |
| 22 | F | 60 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 1+ |
| 23 | F | / | Breast | Invasive ductal carcinoma | Moderately | T2N1M1 | 1+ |
| 24 | F | 46 | Breast | Invasive ductal carcinoma | Moderately | T2N3M1 | 0 |
| 25 | F | 41 | Breast | Invasive ductal carcinoma | Moderately | T2N2M0 | 0 |
| 26 | F | 59 | Breast | Invasive ductal carcinoma | Poorly | T2N0M0 | 1+ |
| 27 | F | 45 | Breast | Invasive ductal carcinoma | Poorly | T2N0M0 | 0 |
| 28 | F | 43 | Breast | Invasive ductal carcinoma | N/A | T2N1M1 | 0 |
| 29 | F | 26 | Breast | Fibroadenoma | N/A | T1N0M0 | 2+ |
| 30 | F | 40 | Breast | Invasive ductal carcinoma | N/A | T1N0M0 | 0 |
| 31 | F | 38 | Breast | Fibroadenoma | N/A | T2N0M0 | 1+ |
| 32 | F | 51 | Breast | Invasive ductal carcinoma | Moderately | T2N2M0 | 0 |
| 33 | F | 45 | Breast | Invasive ductal carcinoma | Poorly | T2N0M0 | 1+ |
| 34 | F | 45 | Breast | Invasive ductal carcinoma | Poorly | T2N1M0 | 2+ |
| 35 | F | 47 | Breast | Invasive ductal carcinoma | Moderately | T2N1M0 | 0 |
| 36 | F | 55 | Breast | Invasive ductal carcinoma | Moderately | T2N3M1 | 2+ |
| 37 | F | 58 | Breast | Invasive ductal carcinoma | Moderately | T3N3M0 | 1+ |
| 38 | F | 47 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 3+ |
| 39 | F | 38 | Breast | Invasive ductal carcinoma | Poorly | T2N0M0 | 1+ |
| 40 | F | 40 | Breast | Invasive ductal carcinoma | Poorly | T2N0M0 | 2+ |
| 41 | F | 57 | Breast | Invasive ductal carcinoma | Poorly | T2N0M0 | 0 |
| 42 | F | 42 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 2+ |
| 43 | F | 60 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 2+ |
| 44 | F | 58 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 0 |
| 45 | F | 41 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 0 |
| 46 | F | 50 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 1+ |
| 47 | F | 60 | Breast | Invasive ductal carcinoma | Moderately | T2N2M1 | 0 |
| 48 | F | 53 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 3+ |
| 49 | F | 65 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 0 |
| 50 | F | 43 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 2+ |
| 51 | F | 57 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 2+ |
| 52 | F | 37 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 1+ |
| 53 | F | 50 | Breast | Invasive ductal carcinoma | Moderately | T2N3M0 | 0 |
| 54 | F | 48 | Breast | Invasive ductal carcinoma | Poorly | T2N1M0 | 0 |
| 55 | F | 50 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 1+ |
| 56 | F | 53 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 3+ |
| 57 | F | 49 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 2+ |
| 58 | F | 65 | Breast | Invasive ductal carcinoma | Moderately | T2N1M0 | 0 |
| 59 | F | 43 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 2+ |
| 60 | F | 58 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 0 |
| 61 | F | 48 | Breast | Invasive ductal carcinoma | Moderately | T2N0M0 | 1+ |
| 62 | F | / | Breast | Invasive ductal carcinoma | Moderately | TxNxMx | 2+ |
| 63 | F | / | Breast | Invasive ductal carcinoma | Moderately | TxNxMx | 1+ |
| Pathology diagnosis | No. of cases | B4Mab-7 immunostaining | No. of positive cases | |||
| 3+ | 2+ | 1+ | 0 | |||
| Invasive ductal carcinoma | 55 | 6 | 12 | 18 | 19 | 36/55 (65.4%) |
| Invasive lobular carcinoma | 3 | 0 | 0 | 1 | 2 | 1/3 (33.3%) |
| Adenocarcinoma | 2 | 0 | 1 | 0 | 1 | 1/2 (50.0%) |
| Fibroadenoma | 2 | 0 | 1 | 1 | 0 | 2/2 (100%) |
| Medullary carcinoma | 1 | 1 | 0 | 0 | 0 | 1/1 (100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





