1. Introduction
Lung cancer is one of the most common and deadliest cancers worldwide. Most lung cancers are mainly classified into two categories: non-small cell lung cancer (NSCLC) or small cell lung cancer (SCLC), with NSCLC accounting for more than 80% of cases. Although recent declines in mortality have been reported, likely due to reductions in smoking and advances in early detection and major advances in treatment for NSCLC, the 5-year relative survival rate for lung cancer is still dismal at 22% overall [
1]. The diagnosis of lung cancer is highly dependent on imaging technology. Screening for lung cancer using low-dose computed tomography (LDCT) has already been shown to reduce lung cancer deaths [
2]. Therefore, LDCT is a promising method for lung cancer diagnosis. However, it has the major disadvantages of high false-positive rates and high costs [
2,
3]. Diagnosis at early stages (0-I) compared to metastatic stages (III-IV) has great potential to reduce mortality and increase the 5-year survival rate up to 60% in lung cancer patients [
2,
4]. However, only 24% of lung cancers are diagnosed at early stages because signs and respiratory symptoms do not usually appear until the cancer is advanced [
1]. Several candidates have been reported as cancer-associated biomarkers for detecting lung cancer with peripheral blood and targeting different sources, such as genetic, epigenetic, proteomic, and metabolic materials [
5,
6,
7,
8,
9]. Tumour markers such as cytokeratin 19 fragment (CYFRA 21-1), carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), squamous cell carcinoma antigen (SCC Ag), pro-gastrin-releasing peptide (proGRP) and cancer antigen 125 (CA 125) are currently used in clinical laboratories for the diagnosis and monitoring of lung cancer but are underutilized due to unsatisfactory diagnostic performance, in particular low sensitivity for lung cancer [
10].
Autoantibodies are produced and circulated early in the immune system of patients after exposure to cancer proteins and have been continuously attracting attention as useful markers for the early detection of cancer [
11,
12]. Moreover, even if the antigen exists at a low concentration, autoantibodies are produced at a relatively high concentration and have a continuous blood concentration and long half-life owing to limited proteolysis and removal [
13]. However, the detection of autoantibodies in the blood of patients with cancer has been technically challenging. As well as traditional biomarkers such as p53, CYFRA21-1, ProGRP, and NSE, several biomarkers associated with lung cancer, such as c-Myc, survivin, HER2, NY-ESO-1, SOX1 have been used in various diagnostic technologies based on the concept of autoantibodies [
4,
6,
14,
15,
16]. In addition, their diagnostic performance in lung cancer was improved by combining different biomarkers rather than using a single autoantibody [
17,
18,
19,
20]. However, the different methods for detecting these materials have insufficient sensitivity and specificity for use as diagnostic sensors in clinical laboratories [
16,
17,
18,
19,
20,
21]. Recently, a new method using a 9G DNA chip to measure the antigen-autoantibody immune complex (AIC) and its antigen based on the ratio of AIC to its free antigen was verified for its efficiency in detecting lung cancer [
22,
23,
24]. The clinical usefulness of AIC and its free antigen against CYFRA21-1 has been reported in the screening of NSCLC [
23,
24]. The present study investigated possible candidate biomarkers for NSCLC by measuring the AIC and its free antigens for four proteins (CYFRA21-1, ProGRP, neutrophil gelatinase associated lipocalin (NGAL), and NSE) using a 9G DNA chip to detect stage 0 (carcinoma in situ (CIS))–IV NSCLC.
2. Materials and Methods
2.1. Study participants
Clinical samples from patients with lung cancer (n = 85) and healthy individuals (n = 120) were collected and tested in 2019 at the Korea Cancer Central Hospital. We retrospectively reviewed the electronic medical records. All patients with lung cancer were screened using chest radiography and LDCT, followed by a biopsy for individuals with abnormal findings on LDCT. All healthy controls (HCs) were screened for lung cancer using chest radiography. Those with a history of cancer were excluded through a questionnaire in the yearly health check-up program. Patients with infectious diseases at the time of the evaluation were excluded. All blood samples from patients with lung cancer were collected before treatments, including surgery, chemotherapy, radiotherapy, etc. K2 ethylenediaminetetraacetic acid (EDTA)-anticoagulated remnant blood specimens were obtained from all subjects after a complete blood cell count test. All samples were stored at 4°C before centrifugation for 10 min at 2000 × g at 4°C. Plasma samples were then archived in the biobank at ˗70°C. The samples used in this study were obtained from the Korea Institute of Radiological and Medical Sciences (KIRAMS) Radiation Biobank (KRB-2019-I006). We chose eight candidate markers for four proteins (CYFRA21-1, ProGRP, NGAL, and NSE) that have been reported to be useful in the detection of lung cancer in previous studies [
25,
26,
27,
28,
29,
30]. Eight markers were measured in the plasma samples of all subjects: CYFRA21-1-anti-CYFRA21-1 AIC (CIC), CYFRA21-1 antigen, ProGRP-anti-ProGRP AIC (PrGIC), ProGRP antigen, NGAL-anti-NGAL AIC (NGIC), NGAL antigen, NSE-anti-NSE AIC (NSIC), and NSE. This study was approved by the institutional review board of the Korea Institute of Radiological and Medical Sciences (KIRAMS) (IRB#-2019-07-009-001).
2.2. Methods
According to the previous studies, bio-conjugates were prepared by six types of syntheses: a marker protein-capture antibody (cAb)-DNA, anti-mouse IgG-Cy5, anti-human IgG-Cy5, a marker protein-detection Ab (dAb)-fluorescent beads (FB), anti-human-IgG-FB, anti-human-IgG-FB, and Cy5-DNA for four protein markers including CYFRA21-1, ProGRP, NGAL, and NSE, respectively [
22,
24]. The lateral flow strip membranes (LFSM) were also prepared for the detection of the eight markers (CIC, CYFRA21-1, PrGIC, ProGRP, NGIC, NGAL, NSIC, and NSE). The LFSM was manufactured based on the 9G DNA membranes. The 9G DNA membranes were lining 18 pmol/L solutions of the oligonucleotide Probe 1-Probe 8 appended with 9 consecutive guanines (9G) corresponding to the test line and hybridization control line and allowed to immobilize. After the immobilization step, the membranes were soaked in a blocking solution and were dried.
Measurements of CIC, CYFRA21-1, PrGIC, ProGRP, NGIC, NGAL, NSIC, and NSE in plasma samples of healthy individuals and patients with lung cancer were performed using the sandwich immunoassay of DNA-guided method based on 9G DNA chip (Biometrix Technology Inc., Chuncheon, South Korea). The 9G DNA chip quantified plasma levels of all markers in 30 min at room temperature: for free antigen (or AIC) detection, a 20 μL plasma sample was incubated with the 100 μL of the solution containing a marker protein-cAb-DNA conjugate (10 fmol/mL), a marker protein-dAb-FB conjugate (or anti-hum-IgG-FB conjugate for AIC detection)(0.07 fmol/mL), and FB-DNA (10 fmol/mL) in an e-tube and incubated at 25°C for 10 min in a thermo-controller. After the incubation step, 60 μL of reaction buffer containing FB-DNA complementary to the DNA immobilized on the hybridization control line was added to the reaction tube. The reaction mixture was then loaded on the LFSM and hybridized for 10 min at 25°C. The highly specific DNA-DNA hybridization allowed the capture of a marker protein-dAb-FB-a marker protein-a marker protein-cAb-DNA for free antigen detection or the anti-human-IgG-FB-AIC-a marker protein-cAb-DNA for AIC detection, bimolecular complexes and the Cy5-dNA on the test line and hybridization control line, respectively. The unbound biomolecular complexes were then washed away at 25°C in a washing step that took 10 minutes by adding 170 mL of washing solution (0.1% SDS in 4 × SSC, pH = 7.4). After the washing step, the bound materials of each marker are then detected and quantified by scanning the LFSM in the BMT Membrane Reader
TM (Biometrix Technology Inc., South Korea). Fluorescence intensity was expressed in arbitrary units by reference to validated standard curves [
22,
24].
The levels of CIC, CYFRA21-1, PrGIC, ProGRP, NGIC, NGAL, NSIC, and NSE in plasma samples of HCs and patients with NSCLC were recorded. These levels were calculated using the ratios of CIC to CYFRA21-1, PrGIC to ProGRP, NGIC to NGAL, and NSIC to NSE. In particular, CYFRA21-1 was required in the combination equations because it has been shown to represent the leading marker in the diagnosis of NSCLC with the highest sensitivity, either by the same or in different methods [
22,
31]. Finally, combination ratios for two to four markers, including CIC/CYFRA 21-1, were determined according to equations (1)–(7) and used to discriminate between a healthy population and patients with lung cancer.
(1) C2-1 = [(CIC/CYFRA21-1) × (PrGIC/ProGRP)]
(2) C2-2 = [(CIC/CYFRA21-1) × (NGIC/NGAL)]
(3) C2-3 = [(CIC/CYFRA21-1) × (NSIC/NSE)]
(4) C3-1 = [(CIC/CYFRA21-1) × (PrGIC/ProGRP) × (NGIC/NGAL)]
(5) C3-2 = [(CIC/CYFRA21-1) × (PrGIC/ProGRP) × (NGIC/NGAL)]
(6) C3-3 = [(CIC/CYFRA21-1) × (NGIC/NGAL) × (NSIC/NSE)]
(7) C4-1 = [(CIC/CYFRA21-1) × (PrGIC/ProGRP) × (NGIC/NGAL) × (NSIC/NSE)]
Sensitivity and specificity were determined using the cutoff levels to distinguish between the healthy population and patients with lung cancer. Ultimately, the cutoff levels were aimed first at the area under the curve (AUC) and then at the minimal difference between sensitivity and specificity [
32].
2.3. Statistical Analysis
The data were not normally distributed. Continuous data are expressed as medians with interquartile ranges (IQR). Categorical data are presented as counts and percentages. The difference between healthy individuals and patients with lung cancer was calculated using the Mann–Whitney U test. For all analyses, two-sided p values <0.05 were considered statistically significant. The receiver operating characteristics (ROC) curve was applied to assess the overall diagnostic performance, and Delong’s method was used to compare the ROC curves. The optimal cut-off point in the coordinates of the ROC curve was determined using the closest top-left method. The sensitivity, specificity, accuracy, positive predictive value (PPV), and negative predictive value (NPV) were calculated at a 95% confidence interval (CI). Statistical analyses were performed using SPSS software version 22 (IBM Corp., Armonk, NY, USA) and Rex-Pro version 3.6.1.0 (RexSoft Inc. Seoul, Korea).
3. Results
3.1. The levels of AICs and their antigens for CYFRA21-1, ProGRP, NGAL, and NSE
The characteristics of the participants in the NSCLC and HC groups are shown in
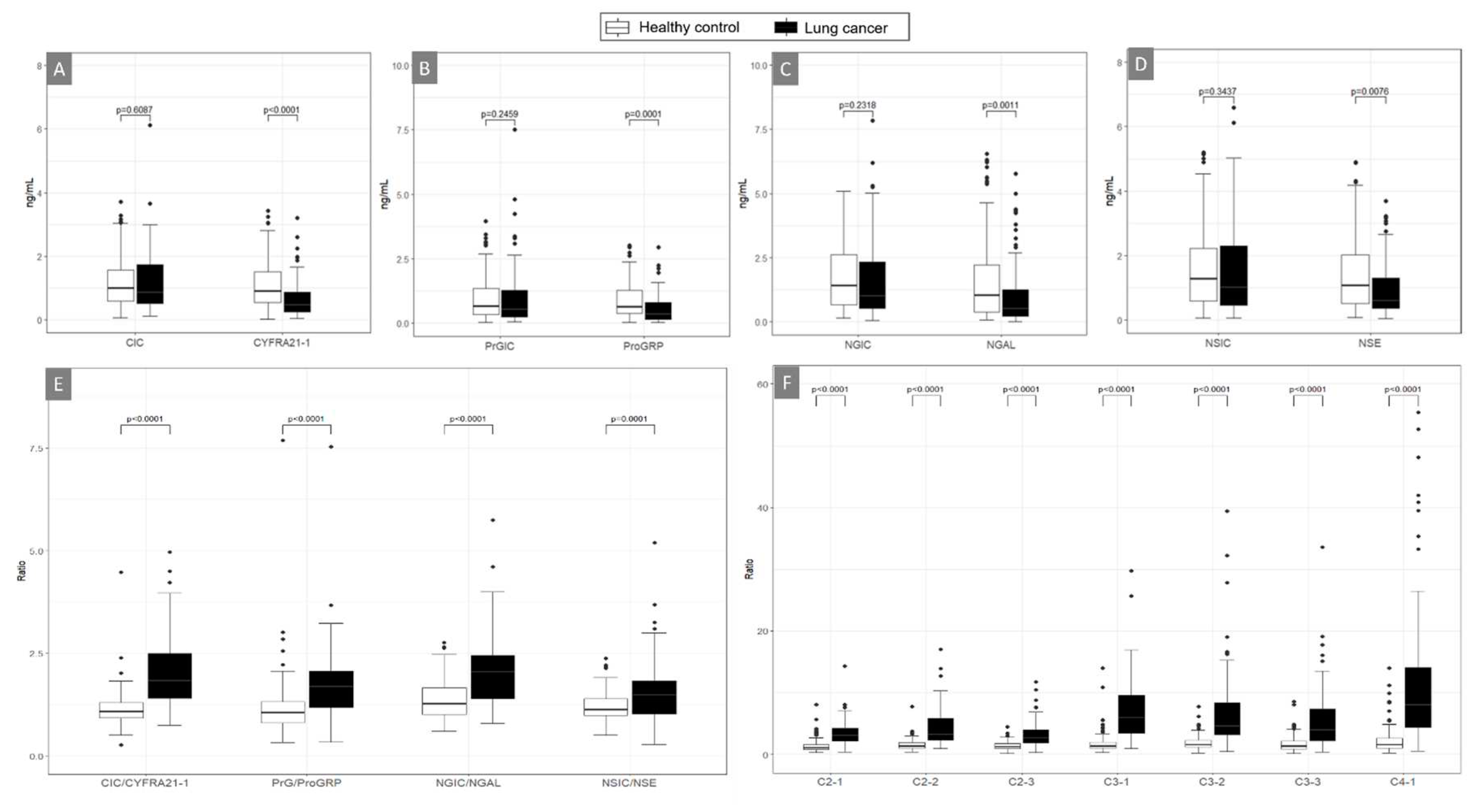
Table 1. The levels of AICs (CIC, PrGIC, NGIC, and NSIC) were significantly higher than those of their free antigens (CYFRA21-1, ProGRP, NGAL, and NSE) in all participants (all p < 0.0001), healthy controls (p < 0.0001, p = 0.0027, p = 0.0198, and p < 0.0001, respectively), and patients with NSCLC (all p < 0.0001) (Fig. S1).
3.2. Diagnostic performance
The levels of each free antigen distinguished patients with NSCLC from HCs better than AIC, and their levels in NSCLC were significantly lower than those in HC (
Figure 1A–1D). The ratios of CIC to CYFRA21-1, PrGIC to ProGRP, NGIC to NGAL, and NSIC to NSE were significantly higher in patients with lung cancer than in HC (
Figure 1E). Additionally, combination ratios (C2-1, C2-2, C2-3, C3-1, C3-2, C3-3, and C4-1) highly discriminated between patients with NSCLC and HCs (all, p < 0.0001) (
Figure 1F and
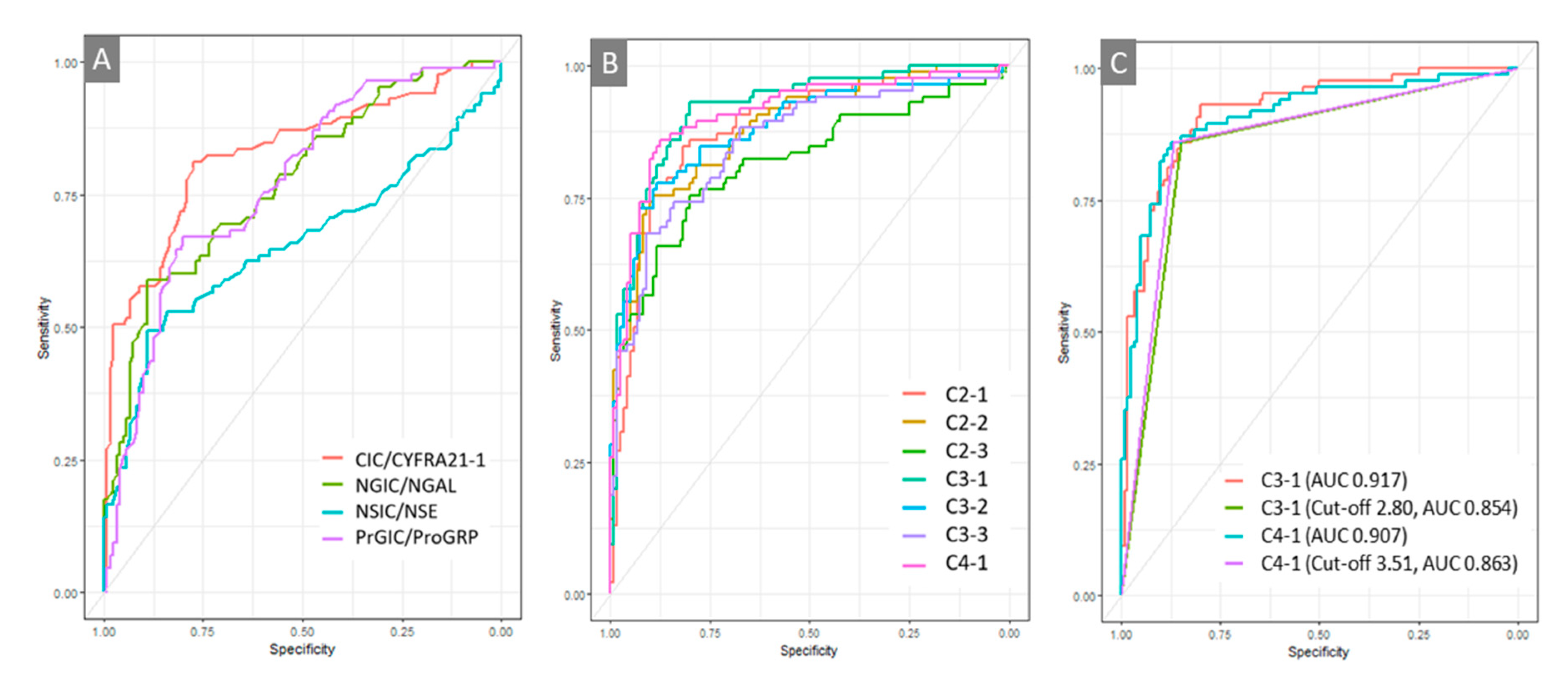
Table S1). The diagnostic performance of the eight single markers (CIC, CYFRA21-1, PrGIC, ProGRP, NGIC, NGAL, NSIC, and NSE); the ratios of CIC/CYFRA21-1, PrGIC/ProGRP, NGIC/NGAL, and NSIC/NSE; and the seven combinations were compared using ROC analyses (
Figure 2 and
Table S2). Good diagnostic performance was confirmed with CIC/CYFRA21-1 among all ratios from the other ratios for a protein, and better diagnostic performance was achieved with combinations of C3-1, C4-1, C2-1, C3-2, C2-2, C3-3, and C2-3, in the order of large area under the curve (AUC) of ROC (
Table S2). Excellent diagnostic performance after applying optimal cutoff point was obtained using the combinations of C4-1 and C3-1 at the diagnostic cut-off of 3.51 and 2.80, respectively, which had 85.9% and 85.9% sensitivity and 86.7% and 85% specificity with 0.863 and 0.854 AUC, respectively, in discriminating patients with NSCLC from HCs. However, there was no difference between the two ROC curves (p = 0.4063) (
Figure 2C). The sensitivity and specificity of the C4-1 combination were analysed according to the subgroup stages and pathological diagnosis. Higher sensitivity was observed in the very early stages (0-I), the localized tumours, than in the stages II to IV, with 86.7% specificity for C4-1 (
Table 3). Pathological diagnoses were better distinguished in patients with NSCLCs, with 92.1% for adenocarcinoma and 81.6% for squamous carcinoma at the 86.7% specificity for C4-1 (
Table 4).
Abbreviations: CIC, CYFRA21-1-Anti-CYFRA21-1 autoantibody immune complex; PrGIC, ProGRP-Anti-ProGRP autoantibody immune complex; NGIC, NGAL-Anti-NGAL autoantibody immune complex; NSIC, NSE-Anti-NSE autoantibody immune complex.
4. Discussion
Early screening benefits by reducing about 80% of deaths from lung cancer [
4]. Several issues related to cost-effectiveness and overdiagnosis due to high false-positive rates have been raised regarding LDCT-dependent diagnostic tools for screening lung cancer. After identifying that autoantibodies are produced during the early stages of tumourigenesis, the possibility of their use as diagnostic markers was suggested [
33,
34]. Studies on the biomarkers of autoantibodies against tumour-associated antigens have been continuously conducted using various methods. However, methods using blood samples have not been widely adopted in clinical practice because of their low sensitivity and specificity. Moreover, technical issues in measuring autoantibodies can be challenging.
The recent method using a 9G DNA chip for high affinity to AIC and its free antigen revealed good results in the method and clinical applicability [
22,
23,
24]. This study aimed to explore biomarkers for the diagnosis of NSCLC using a combination of the ratios of AIC to its free antigen for four well-known lung cancer-associated protein markers, including CYFRA21-1, which is one of the best discriminators of NSCLC [
25,
26,
27]. Levels of CYFRA21-1 above 10 ng/mL are thought to indicate malignant lung tumours or primary lung cancer, but CYFRA21-1 is released by all lung cancers, regardless of histological type [
31]. NGAL is a secreted protein that has a role in controlling cell proliferation and survival, and its alteration is associated with various malignant tumours, including lung cancer [
29,
35]. Overexpression of NGAL has been identified in lung adenocarcinoma progression [
35]. ProGRP and NSE are diagnostic biomarkers of SCLC with various sensitivities from 47% to 80% [
29,
30,
31,
36]. They are mainly released by lung cancers, while CYFRA21-1 and NGAL are released by various solid cancers [
31]. Much lower levels of free antigens than AICs for the four protein biomarkers were identified in patients with NSCLC, which is consistent with a previous study [
37]. However, no significant difference in the levels of AICs was observed between the patients with NSCLC and HCs. This is inconsistent with previous studies with different methods that found few tumour-associated autoantibodies in healthy individuals, even those at high risk of lung cancer [
10,
38]. However, our results are consistent with previous studies with the same method, the ratios of antigen-autoantibody immune complexes to their free antigens were more useful than every single marker (AICs and free antigens) in patients with NSCLC [
23,
24]. Although relatively low sensitivity of AIC/free antigen for ProGRP (67.1%) and NSE (63.5%) was observed, higher sensitivity was observed for the combination of the four fractions (C4-1) for CYFRA21-1, ProGRP, NGAL, and NSE. Although all patients were diagnosed with NSCLC, it excellently detected stages 0/I–IV with 85.9% sensitivity and 86.7% specificity compared to other combinations of ratios or a single ratio (
Table 3). The diagnostic performance was more pronounced in early lung cancer (stage 0/I) or localized tumours, with over 90% sensitivity and 86.7% specificity, reflecting the characteristics of tumour-associated autoantibodies.
This study had several limitations owing to the small number of retrospective cohort studies. The levels of AIC, free antigens, and their ratios did not allow us to assess differences for sex and age. However, no differences in autoantibodies by age, gender or race have been shown in a previous study with large cohorts [
39]. A small number of participants, especially those with stage 0, II, or IV NSCLC, were available. Although a significant difference after applying C4-1 was observed between the patients with stage II NSCLC and HCs, a lower sensitivity in stage II was presented than in the other stages (
Table 3). Additionally, a lower sensitivity was observed in the regional tumour state, comprising stages IIb and III. Furthermore, 12 of the 16 stage II patients did not have adenocarcinoma (squamous carcinoma, 10; other NSCLCs, 2), and showed lower sensitivity than adenocarcinoma (
Table 4). Pathologically, the diagnostic efficiency showed higher sensitivity in adenocarcinoma than in squamous cell carcinoma or other NSCLCs. These results can be thought of as a decrease in the ratio at some intermediate stage (II–III), due to a simultaneous increase in the number of autoantibodies and an increase in tumour antigens. Alternatively, this could be related to the increasing inaccuracy of staging in higher stages, which is expected to have lower sensitivity than that in the early stages [
40]. Second, clinical information, such as smoking history or occupations related to carcinogenic exposure of participants, which are known as potential risk factors for lung cancer, was not provided [
1]. This information may help validate the currently recommended sensitivity for identified lung cancer markers. Lastly, this study did not evaluate samples with SCLC or benign states, such as lung nodules or infective lung diseases, which could be difficult to diagnose using LDCT. Further research with wider cohorts including would help prove their clinical efficiency in discriminating against lung cancer. It would also be worth investigating its utility as a biomarker for post-treatment monitoring in lung cancer patients.
5. Conclusions
In our study, the ratio of AICs to tumour-associated free antigens was much more sensitive in distinguishing patients with NSCLC than measuring individual proteins (AICs and free antigens) by the 9G DNA chip method, which has a high affinity for autoantibodies and antigens present at very low concentrations. Moreover, a combination of all four ratios of AICs to their free antigens for CYFRA21-1, ProGRP, NGAL, and NSE discriminated between patients with NSCLC and HCs with excellent diagnostic performance. Complementing the imaging-based diagnosis of lung cancer, this test would help to minimize the risk of lung cancer by screening at-risk populations and diagnosing NSCLC at an early stage. This also has great potential for the diagnosis of NSCLC with clinical efficiency in terms of accessibility and cost-effectiveness with liquid biopsy.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: The levels of AICs and their antigens for CYFRA21-1, ProGRP, NGAL, and NSE in all participants, in healthy controls, and in patients with lung cancers. Table S1: The levels of antigen-autoantibody immune complex, and free antigen, their ratios, and combinations from two to four ratios for four proteins; Table S2: The area under the curve (AUC) for each marker.
Author Contributions
Conceptualization, Young Jun Hong and Heyjin Kim; Formal analysis, Ae-Chin Oh and Hye-Ryoun Kim; Investigation, Heyjin Kim, Ae-Chin Oh and Hye-Ryoun Kim; Project administration, Heyjin Kim; Supervision, Young Jun Hong; Writing – original draft, Heyjin Kim; Writing – review & editing, Jin Kyung Lee.
Funding
This work was supported by the Technology Innovation Program (or Industrial Strategic Technology Development Program) (20003670, Development and Production of Diagnostic Technology for Early Screening and Curing Process Monitoring of Stage 1 Lung Cancer), funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of the Korea Institute of Radiological and Medical Sciences (KIRAMS) (IRB#-2019-07-009-001).
Informed Consent Statement
Patient consent was waived due to the retrospective study using archived samples from the patient’s remnant samples from a biobank with anonymous clinical data.
Acknowledgments
We thank the Korea Institute of Radiological and Medical Sciences (KIRAMS) Radiation Biobank (KRB) for providing samples. We would like to thank Editage (
www.editage.co.kr) for English language editing.
Conflicts of Interest
The authors declare no conflict of interest. Biometrix Technology Inc. does not have a conflict of interest.
References
- American Cancer Society (ACS). Cancer facts & figures 2022. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2022.html.
- Jonas, D.E.; Reuland, D.S.; Reddy, S.M.; Nagle, M.; Clark, S.D.; Weber, R.P.; Enyioha, C.; Malo, T.L.; Brenner, A.T.; Armstrong, C, Coker-Schwimmer, M. ; Middleton, J.C.; Voisin, C.; Harris, R.P. Screening for lung cancer with low-dose computed tomography: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA 2021, 325, 971–987. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.W.; Li, W.; Han, F.J.; Liu, Y.D. Screening for lung cancer using low-dose computed tomography: concerns about the application in low-risk individuals. Transl Lung Cancer Res. 2015, 4, 275–286. [Google Scholar] [PubMed]
- International Early Lung Cancer Action Program Investigators; Henschke, C. I.; Yankelevitz, D.F.; Libby, D.M.; Pasmantier, M.W.; Smith, J.P.; Miettinen, O.S. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med 2006, 355, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Goebel, C.; Louden, C.L.; Mckenna, R. Jr.; Onugha, O.; Wachtel, A.; Long, T. Blood test shows high accuracy in detecting stage I non-small cell lung cancer. BMC Cancer 2020, 20, 137. [Google Scholar] [CrossRef] [PubMed]
- Zamay, T.N.; Zamay, G.S.; Kolovskaya, O.S.; Zukov, R.A.; Petrova, M.M.; Gargaun, A.; Berezovski, M.V.; Kichkailo, A.S. Current and prospective protein biomarkers of lung cancer. Cancers (Basel) 2017, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Pennell, N.A.; Arcila, M.E.; Gandara, D.R.; West, H. Biomarker testing for patients with advanced non-small cell lung cancer: real-world issues and tough choices. Am Soc Clin Oncol Educ Book 2019, 39, 531–542. [Google Scholar] [CrossRef]
- Duruisseaux, M.; Esteller, M. Lung cancer epigenetics: from knowledge to applications. Semin Cancer Biol 2018, 51, 116–128. [Google Scholar] [CrossRef]
- Qi, S.A.; Wu, Q.; Chen, Z.; Zhang, W.; Zhou, Y.; Mao, K.; Li, J.; Li, Y.; Chen, J.; Huang, Y.; Huang, Y. High-resolution metabolomic biomarkers for lung cancer diagnosis and prognosis. Sci Rep 2021, 11, 11805. [Google Scholar] [CrossRef]
- Qin, J.; Zeng, N.; Yang, T.; Wan, C.; Chen, L.; Shen, Y.; Wen, F. Diagnostic value of autoantibodies in lung cancer: a systematic review and meta-analysis. Cell Physiol Biochem 2018, 51, 2631–2646. [Google Scholar] [CrossRef]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Solassol, J.; Maudelonde, T.; Mange, A.; Pujol, J.L. Clinical relevance of autoantibody detection in lung cancer. J Thorac Oncol 2011, 6, 955–962. [Google Scholar] [CrossRef]
- Pedersen, J.W.; Wandall, H.H. Autoantibodies as biomarkers in cancer. Lab Med 2011, 42, 623–628. [Google Scholar] [CrossRef]
- Chapman, C.J.; Murray, A.; McElveen, J.E.; Sahin, U.; Luxemburger, U.; Türeci, O.; Wiewrodt, R.; Barnes, A.C.; Robertson, J.F. Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax 2008, 63, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Rohayem, J.; Diestelkoetter, P.; Weigle, B.; Oehmichen, A.; Schmitz, M.; Mehlhorn, J.; Conrad, K.; Rieber, E.P. Antibody to the tumor-associated inhibitor of apoptosis protein survivin in cancer patients. Cancer Res 2000, 60, 1815–1817. [Google Scholar] [PubMed]
- Yang, B.; Li, X.; Ren, T.; Yin, Y. Autoantibodies as diagnostic biomarkers for lung cancer: A systematic review. Cell Death Discov 2019, 5, 126. [Google Scholar] [CrossRef] [PubMed]
- Boyle, P.; Chapman, C.J.; Holdenrieder, S.; Murray, A.; Robertson, C.; Wood, W.C.; Maddison, P.; Healey, G.; Fairley, G.H.; Barnes, A.C.; Robertson, JF. Clinical validation of an autoantibody test for lung cancer. Ann Oncol 2011, 22, 383–389. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Casiano, C.A.; Peng, X.X.; Koziol, J.A.; Chan, E.K.; Tan, E.M. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol Biomarkers Prev 2003, 12, 136–143. [Google Scholar]
- Zhong, L.; Coe, S.P.; Stromberg, A.J.; Khattar, N.H.; Jett, J.R.; Hirschowitz, E.A. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J Thorac Oncol 2006, 1, 513–519. [Google Scholar] [CrossRef]
- Broodman, I.; Lindemans, J.; van Sten, J.; Bischoff, R.; Luider, T. Serum protein markers for the early detection of lung cancer: A focus on autoantibodies. J Proteome Res 2017, 16, 3–13. [Google Scholar] [CrossRef]
- Dai, N.; Cao, X.J.; Li, M.X.; Qing, Y.; Liao, L.; Lu, X.F.; Zhang, S.H.; Li, Z.; Yang, Y.X.; Wang, D. Serum APE1 autoantibodies: a novel potential tumor marker and predictor of chemotherapeutic efficacy in non-small cell lung cancer. PLOS ONE 2013, 8, e58001. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Nimse, S.B.; Warkad, S.D.; Oh, A.C.; Kim, T.; Hong, Y.J. Quantification of CYFRA 21–1 and a CYFRA 21–1–anti-CYFRA 21–1 autoantibody immune complex for detection of early stage lung cancer. Chem Commun 2019, 55, 10060–10063. [Google Scholar] [CrossRef]
- Choe, W.; Chae, J.D.; Lee, B.H.; Kim, S.H.; Park, S.Y.; Nimse, S.B.; Kim, J.; Warkad, S.D.; Song, K.S.; Oh, A. C, Hong, Y.J.; Kim, T. 9G TestTM cancer/lung: A desirable companion to LDCT for lung cancer screening. Cancers (Basel) 2020, 12, 3192. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Nimse, S.B.; Warkad, S.D.; Kim, J.H.; Kim, H.J.; Kim, T. Detection and quantification of Tp53 and p53-anti-p53 autoantibody immune complex: promising biomarkers in early stage lung cancer diagnosis. Biosensors (Basel) 2022, 12, 127. [Google Scholar] [CrossRef]
- Pujol, J.L.; Molinier, O.; Ebert, W.; Daurès, J.P.; Barlesi, F.; Buccheri, G.; Paesmans, M.; Quoix, E.; Moro-Sibilot, D.; Szturmowicz, M.; Bréchot, J.M.; Muley, T.; Grenier, J. CYFRA 21–1 is a prognostic determinant in non-small-cell lung cancer: results of a meta-analysis in 2063 patients. Br J Cancer 2004, 90, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Karnak, D.; Ulubay, G.; Kayacan, O.; Beder, S.; Ibis, E.; Oflaz, G. Evaluation of Cyfra 21–1: a potential tumor marker for non-small cell lung carcinomas. Lung 2001, 179, 57–65. [Google Scholar] [CrossRef]
- Wieskopf, B.; Demangeat, C.; Purohit, A.; Stenger, R.; Gries, P.; Kreisman, H.; Quoix, E. Cyfra 21–1 as a biologic marker of non-small cell lung cancer. Evaluation of sensitivity, specificity, and prognostic role. Chest 1995, 108, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Crescenzi, E.; Leonardi, A.; Pacifico, F. NGAL as a potential target in tumor microenvironment. Int J Mol Sci 2021, 22, 12333. [Google Scholar] [CrossRef] [PubMed]
- Shibayama, T.; Ueoka, H.; Nishii, K.; Kiura, K.; Tabata, M.; Miyatake, K.; Kitajima, T.; Harada, M. Complementary roles of pro-gastrin-releasing peptide (ProGRP) and neuron specific enolase (NSE) in diagnosis and prognosis of small-cell lung cancer (SCLC). Lung Cancer 2001, 32, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.; Auge, J.M.; Filella, X.; Viñolas, N.; Alicarte, J.; Domingo, J.M.; Ballesta, A.M. Pro-gastrin-releasing peptide (proGRP) in patients with benign and malignant diseases: comparison with CEA, SCC, CYFRA 21–1 and NSE in patients with lung cancer. Anticancer Res 2005, 25, 1773–1778. [Google Scholar]
- Molina, R.; Holdenrieder, S.; Auge, J.M.; Schalhorn, A.; Hatz, R.; Stieber, P. Diagnostic relevance of circulating biomarkers in patients with lung cancer. Cancer Biomark 2010, 6, 163–178. [Google Scholar] [CrossRef]
- Unal, I. Defining an optimal cut-point value in ROC analysis: an alternative approach. Comput Math Methods Med 2017, 2017, 3762651. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.M.; Zhang, J. Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol Rev 2008, 222, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.S.; LaBaer, J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res 2005, 4, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Mongre, R.K.; Sodhi, S.S.; Sharma, N.; Ghosh, M.; Kim, J.H.; Kim, N.; Park, Y.H.; Shin, Y.G.; Kim, S.J.; Jiao, Z.J.; Huynh, do L. ; Jeong, D.K. Epigenetic induction of epithelial to mesenchymal transition by LCN2 mediates metastasis and tumorigenesis, which is abrogated by NF-κB inhibitor BRM270 in a xenograft model of lung adenocarcinoma. Int J Oncol 2016, 48, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, E.; Kulpa, J.K. Pro-gastrin-releasing peptide (ProGRP) as a biomarker in small-cell lung cancer diagnosis, monitoring and evaluation of treatment response. Lung Cancer (Auckl) 2017, 8, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Broodman, I.; Lindemans, J.; van Sten, J.; Bischoff, R.; Luider, T. Serum protein markers for the early detection of lung cancer: a focus on autoantibodies. J Proteome Res 2017, 16, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Trivers, G. E.; De Benedetti, V. M.; Cawley, H. L.; Caron, G.; Harrington, A. M.; Bennett, W. P.; Jett, J. R.; Colby, T. V.; Tazelaar, H.; Pairolero, P.; Miller, R. D.; Harris, C. C. Anti-p53 antibodies in sera from patients with chronic obstructive pulmonary disease can predate a diagnosis of cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 1996, 2, 1767–1775. [Google Scholar]
- Mathew J.; Healey G.; Jewell W.; Murray A.; Chapman C.; Peek L.; Barnes A.; Wood W.; Robertson JF.; Boyle P. Demographics of populations at high risk of lung cancer and results of the Early CDT-Lung test. Journal of Clinical Oncology 2010; 28, 15_suppl. 7033. 7033.
- Heineman, D.J.; Daniels, J.M.; Schreurs, W.H. Clinical staging of NSCLC: current evidence and implications for adjuvant chemotherapy. Ther Adv Med Oncol 2017, 9, 599–609. [Google Scholar] [CrossRef]
Figure 1.
Boxplots for distinguishing patients with lung cancer (non-small cell lung cancer) and healthy controls using four protein markers, including antigen-autoantibody immune complexes and their free antigen (A–D), their ratios (E) and combination ratios of two to four markers, including CIC/CYFRA21-1: A) CIC and CYFRA21-1, B) ProGIC and ProGRP, C) NGIC and NGAL, D) NSIC and NSE; combinations of the ratios E) CIC/CYFRA21-1, PrGIC/ProGRP, NGIC/NGAL, and NSIC/NSE, and F) C2-1 (CIC/CYFRA21-1 × PrGIC/ProGRP), C2-2 (CIC/CYFRA 21-1 × NGIC/NGAL), C2-3 (CIC/CYFRA21-1 × NSIC/NSE), C3-1 (CIC/CYFRA21-1 × PrGIC/ProGRP × NGIC/NGAL), C3-2(CIC/CYFRA21-1 × NGIC/NGAL × NSIC/NSE), C3-3 (CIC/CYFRA21-1 × PrGIC/ProGRP × NSIC/NSE), C4-1 (CIC/CYFRA21-1 × PrGIC/ProGRP × NGIC/NGAL × NSIC/NSE) (Patients with non-small cell lung cancer, n = 85; Healthy controls, n = 120).
Figure 1.
Boxplots for distinguishing patients with lung cancer (non-small cell lung cancer) and healthy controls using four protein markers, including antigen-autoantibody immune complexes and their free antigen (A–D), their ratios (E) and combination ratios of two to four markers, including CIC/CYFRA21-1: A) CIC and CYFRA21-1, B) ProGIC and ProGRP, C) NGIC and NGAL, D) NSIC and NSE; combinations of the ratios E) CIC/CYFRA21-1, PrGIC/ProGRP, NGIC/NGAL, and NSIC/NSE, and F) C2-1 (CIC/CYFRA21-1 × PrGIC/ProGRP), C2-2 (CIC/CYFRA 21-1 × NGIC/NGAL), C2-3 (CIC/CYFRA21-1 × NSIC/NSE), C3-1 (CIC/CYFRA21-1 × PrGIC/ProGRP × NGIC/NGAL), C3-2(CIC/CYFRA21-1 × NGIC/NGAL × NSIC/NSE), C3-3 (CIC/CYFRA21-1 × PrGIC/ProGRP × NSIC/NSE), C4-1 (CIC/CYFRA21-1 × PrGIC/ProGRP × NGIC/NGAL × NSIC/NSE) (Patients with non-small cell lung cancer, n = 85; Healthy controls, n = 120).
Figure 2.
Figure 2. Receiver operating characteristic (ROC) curves (A) ROC curve analysis with the ratios of autoantibody immune complexes to their free antigens (CIC/CYFRA 21-1, PrGIC/ProGRP, NGIC, NGAL, NSIC, and NSE). (B) ROC curve analysis comparing seven combinations comprising two to four markers, including CIC/CYFRA21-1 (C2-1, C2-2, C2-3, C3-1, C3-2, C3-3, C4-1). (C) Comparison of ROC curves for C3-1 and C4-1.
Figure 2.
Figure 2. Receiver operating characteristic (ROC) curves (A) ROC curve analysis with the ratios of autoantibody immune complexes to their free antigens (CIC/CYFRA 21-1, PrGIC/ProGRP, NGIC, NGAL, NSIC, and NSE). (B) ROC curve analysis comparing seven combinations comprising two to four markers, including CIC/CYFRA21-1 (C2-1, C2-2, C2-3, C3-1, C3-2, C3-3, C4-1). (C) Comparison of ROC curves for C3-1 and C4-1.
Table 1.
Characteristics of the study participants (n = 205).
Table 1.
Characteristics of the study participants (n = 205).
| Characteristic |
Patients with NSCLC (n = 85) |
Healthy Control
(n = 120) |
| Age, years (median, range) |
66 (39 – 82) |
42 (25 – 66) |
| Male gender, n (%) |
70 (82.35%) |
60 (50%) |
| |
|
|
| Stage |
|
- |
| - CIS (0) |
2 (2.35%) |
|
| - I |
39 (45.88%) |
|
| - II |
16 (18.82%) |
|
| - III |
24 (28.24%) |
|
| - IV |
4 (4.71%) |
|
| |
|
|
| Pathologic diagnosis |
|
|
| -Adenocarcinoma |
38 (44.70%) |
|
| -Squamous carcinoma |
37 (43.54%) |
|
| -Other types |
11 (12.79%) |
|
| Pleomorphic carcinoma |
4 (4.71%) |
|
| Bronchoalveolar carcinoma |
2 (2.35%) |
|
| Adenosquamous carcinoma |
1 (1.18%) |
|
| High-grade mucoepidermoid carcinoma |
1 (1.18%) |
|
| Mucinous carcinoma |
1 (1.18%) |
|
| - Non-small cell carcinoma |
1 (1.18%) |
|
Table 2.
Diagnostic efficiency in terms of sensitivity, specificity, accuracy, PPV, and NPV of the ratios comprising four protein biomarkers and their combinations from the ratios for discrimination of patients with NSCLC (n = 85) and healthy controls (n = 120)
Table 2.
Diagnostic efficiency in terms of sensitivity, specificity, accuracy, PPV, and NPV of the ratios comprising four protein biomarkers and their combinations from the ratios for discrimination of patients with NSCLC (n = 85) and healthy controls (n = 120)
| Variable |
Sensitivity
(95% CI) |
Specificity
(95% CI) |
Accuracy
(95% CI) |
PPV
(95% CI) |
NPV
(95% CI) |
| CIC/CYFRA 21-1 |
81.2
(71.8–88.8) |
77.5
(69.0–84.6) |
79.0
(72.8–84.4) |
71.9
(61.8–80.6) |
85.3
(77.3–91.4) |
| PrGIC/ProGRP |
67.1
(56.0–76.9) |
79.2
(70.8–86.0) |
74.2
(67.6–80.0) |
69.5
(58.4–68.8) |
77.2
(68.8–84.3) |
| NGIC/NGAL |
69.4
(58.5–79.0) |
70.8
(61.8–78.8) |
70.2
(63.5–76.4) |
62.8
(52.2–72.5) |
76.6
(67.6–84.1) |
| NSIC/NSE |
63.5
(52.4–73.7) |
60.8
(51.5–69.6) |
62.0
(54.9–68.6) |
53.5
(43.3–63.5) |
70.2
(60.4–78.8) |
| C2-1 |
85.9
(76.6–92.5) |
80.0
(71.7–86.8) |
82.4
(76.5–87.4) |
75.3
(65.5–83.5) |
88.9
(81.4–94.1) |
| C2-2 |
81.9
(71.2–88.8) |
78.3
(69.9–85.3) |
79.5
(73.3–84.8) |
72.6
(62.5–81.3) |
85.5
(77.5–91.5) |
| C2-3 |
76.5
(66.0–85.0) |
76.7
(68.1–83.9) |
76.6
(70.2–82.2) |
69.9
(59.5–79.0) |
82.1
(73.8–88.7) |
| C3-1 |
85.9
(76.6–92.5) |
85.0
(77.3–90.9) |
85.4
(79.8–89.9) |
80.2
(70.6–87.8) |
89.5
(82.3–94.4) |
| C3-2 |
84.7
(75.3–91.6) |
77.5
(69.0–84.6) |
80.5
(74.4–85.7) |
72.7
(62.9–81.2) |
87.7
(80.0–93.3) |
| C3-3 |
75.2
(64.8–84.0) |
76.7
(68.1–83.9) |
76.1
(69.7–81.8) |
69.6
(59.1–78.7) |
81.4
(73.0–88.1) |
| C4-1 |
85.9
(76.6–92.5) |
86.7
(79.3–92.2) |
86.3
(80.9–90.7) |
82.0
(72.5–89.4) |
89.7
(82.6–94.5) |
Table 3.
The diagnostic efficiency of the C4-1 according to stages in patients with NSCLC
Table 3.
The diagnostic efficiency of the C4-1 according to stages in patients with NSCLC
| Stage (number) |
Sensitivity
(95% CI) |
Specificity
(95% CI) |
PPV
(95% CI) |
NPV
(95% CI)
|
| CIS (n=2) |
100
(19.8–100) |
86.7
(79.3–92.2) |
11.1
(1.9–36.1) |
100
(95.6–100) |
| Stage I (n=39) |
89.7
(74.8–96.7) |
68.6
(54.0–80.5) |
96.3
(90.2–98.8) |
| Stage II (n=16) |
62.5
(35.9–83.7)
|
38.5
(20.9–59.3) |
94.5
(88.0–97.8) |
| Stage III (n=24) |
91.7
(71.5–98.5) |
57.9
(40.9–73.2) |
98.1
(92.7–99.7) |
| Stage IV (n=4) |
100
(39.6–100) |
20.0
(6.6–44.3) |
100
(55.7–93.4) |
| Very early-stage CIS (0)–I (n=41) |
90.2
(75.9–96.8) |
86.7
(79.3–92.2) |
69.8
(55.5–81.3) |
96.3
(90.2–98.8) |
| Stage II–IV (n=44) |
81.8
(64.2–89.7) |
69.2
(54.7–80.9) |
92.9
(86.0–96.6) |
Table 4.
The diagnostic efficiency of C4-1 according to pathologic diagnosis in patients with NSCLC
Table 4.
The diagnostic efficiency of C4-1 according to pathologic diagnosis in patients with NSCLC
Type
(number) |
Sensitivity
(95% CI) |
Specificity
(95% CI) |
PPV
(95% CI) |
NPV
(95% CI)
|
| NSCLC (n=85) |
85.9
(76.6–92.5) |
86.7
(79.3–92.2) |
82.0
(72.5–89.4) |
89.7
(82.6–94.5) |
Squamous carcinoma
(n=38, 14/10/12/2*) |
81.6
(65.1–91.7) |
66.0
(50.6–78.7) |
93.7
(87.0–97.2) |
Adenocarcinoma
(n=38, 24/4/9/1*) |
92.1
(77.5–97.9) |
68.6
(54.0–80.5) |
97.2
(91.4–99.3) |
Other NSCLCs
(n=9, 3/2/3/1*) |
77.8
(40.2–96.1) |
30.4
(14.1–53.0) |
98.1
(92.7–99.7) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).