Submitted:
13 July 2023
Posted:
14 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
Visualising CWDM in culture
| Basic Fuchsin | 24 gm (CARL ROTH GmbH &Co (C.I 42510) |
|---|---|
| Ethanol 95% | 120 ml |
| Distilled Water | 800 ml |
| Phenol | 48 ml |
2. Materials and Methods
2.1. ZIEHL-NEELSEN STAIN FOR CWDM
2.2. Method for Staining
2.3. Microscope Slides
2.4. Sample collection
2.5. Detection of CWDM
- 1)
- To a MGIT tube supplemented with OADC and PANTA, add enough sterile filtered sucrose solution in water to arrive at a final concentration in the MGIT of 1% sucrose. If MAC is suspected, add mycobactin J. (Allied Monitor Inc., Fayette, MO) according to manufacturer’s instructions.
- 2)
- The buffy coat from a human subject is harvested after centrifugation of the sodium citrated blood sample for 10 minutes at 3000 RPM. Some consideration should be given to separate culture of the erythrocyte component.[34]
- 3)
- Inoculate the MGIT tube with buffy coat and incubate for 30 days at 370c, with a primary reading of the culture deposit at 8 days, using the above ZN stain method.
3. Results

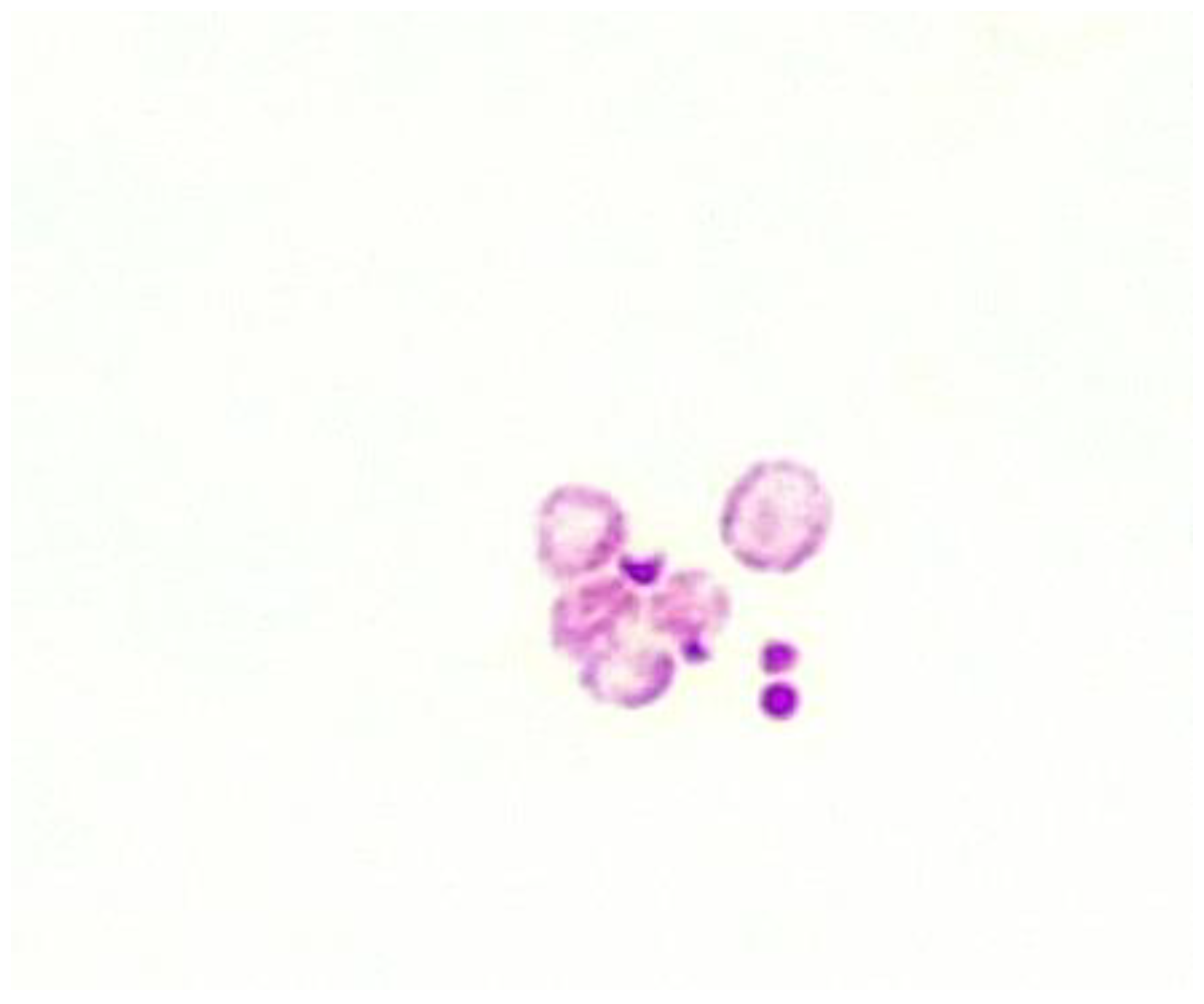
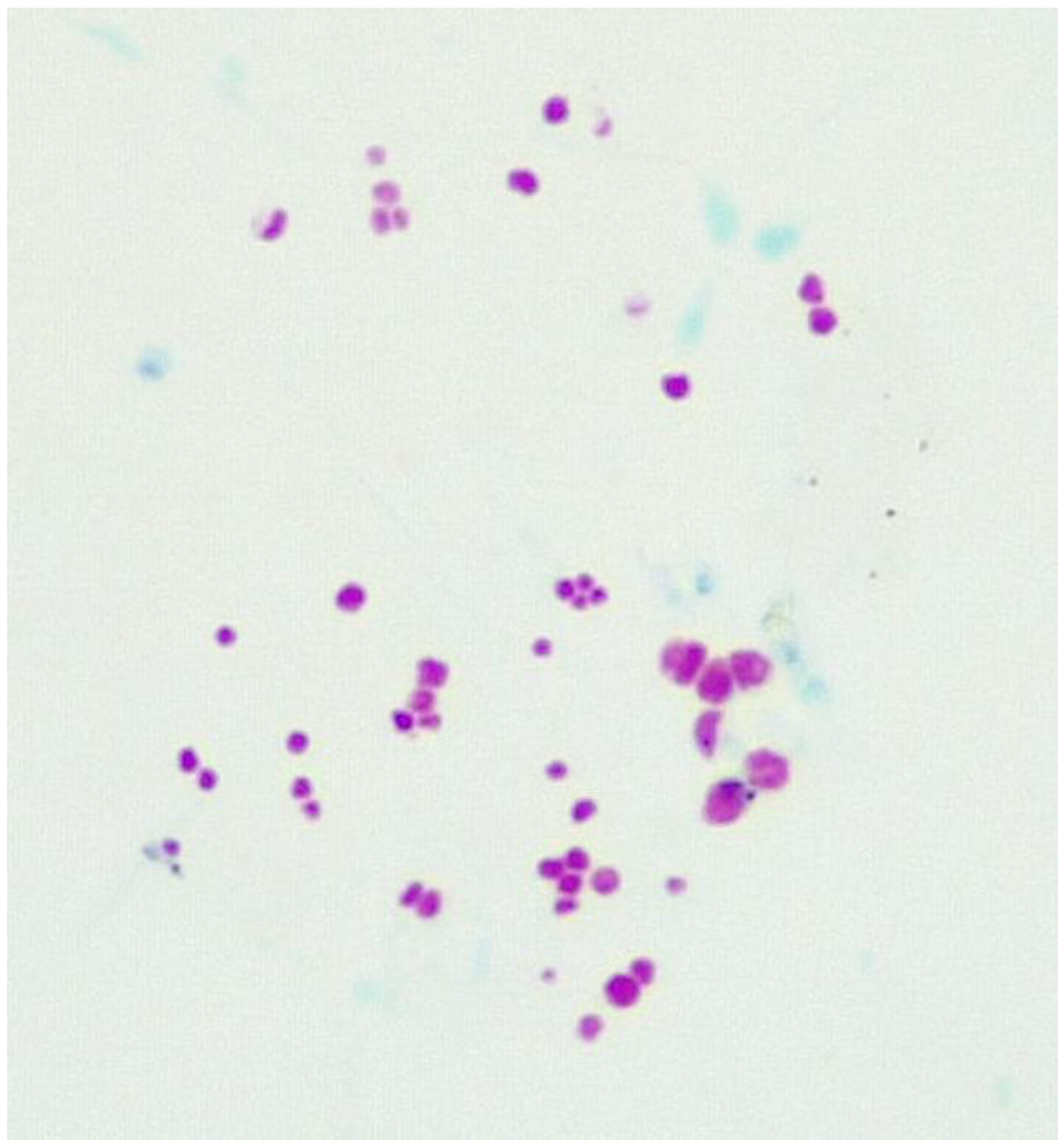
3.1. Fluid culture media for CWDM.
3.2. Solid culture media for CWDM
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- P. T. Elkington, “TUBERCULOSIS: TIME FOR A NEW PERSPECTIVE?,” J Infect, vol. 66, no. 4, pp. 299–302, Apr. 2013. [CrossRef]
- J. D. Ernst, “The immunological life cycle of tuberculosis,” Nat Rev Immunol, vol. 12, no. 8, Art. no. 8, Aug. 2012. [CrossRef]
- S. W. Park et al., “Growth of Mycobacteria on Carbon Monoxide and Methanol,” J Bacteriol, vol. 185, no. 1, pp. 142–147, Jan. 2003. [CrossRef]
- S. Ratnam and S. Chandrasekhar, “The pathogenicity of spheroplasts of Mycobacterium tuberculosis,” American Review of Respiratory Disease, vol. 114, no. 3, pp. 549–554, 1976.
- Velayati et al., “Populations of latent Mycobacterium tuberculosis lack a cell wall: Isolation, visualization, and whole-genome characterization,” International Journal of Mycobacteriology, vol. 5, no. 1, pp. 66–73, Mar. 2016. [CrossRef]
- W. R. Burnham, J. E. Lennard-Jones, J. L. Stanford, and R. G. Bird, “MYCOBACTERIA AS A POSSIBLE CAUSE OF INFLAMMATORY BOWEL DISEASE,” The Lancet, vol. 312, no. 8092, pp. 693–696, Sep. 1978. [CrossRef]
- G. Gitnick et al., “Preliminary report on isolation of mycobacteria from patients with Crohn’s disease,” Digest Dis Sci, vol. 34, no. 6, pp. 925–932, Jun. 1989. [CrossRef]
- D. Raoult et al., “Cultivation of the Bacillus of Whipple’s Disease,” New England Journal of Medicine, vol. 342, no. 9, pp. 620–625, Mar. 2000. [CrossRef]
- J.-C. Lagier, S. Edouard, I. Pagnier, O. Mediannikov, M. Drancourt, and D. Raoult, “Current and past strategies for bacterial culture in clinical microbiology,” Clin Microbiol Rev, vol. 28, no. 1, pp. 208–236, Jan. 2015. [CrossRef]
- J.-C. Lagier, P. Hugon, S. Khelaifia, P.-E. Fournier, B. La Scola, and D. Raoult, “The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota,” Clin Microbiol Rev, vol. 28, no. 1, pp. 237–264, Jan. 2015. [CrossRef]
- R. B. Gearry, J. M. Aitken, R. L. Roberts, S. Ismail, J. Keenan, and M. L. Barclay, “Images of interest. Gastrointestinal: Mycobacterium avium paratuberculosis and Crohn’s disease,” J Gastroenterol Hepatol, vol. 20, no. 12, p. 1943, Dec. 2005. [CrossRef]
- J. M. Aitken, T. J. Borody, and G. Agrawal, “A revaluation of the use of conventional Ziehl-Neelsen stain for detection of non-tuberculous mycobacteria,” New Zealand Journal of Medical Laboratory Science, vol. 73, no. 3, Art. no. 3, 2019.
- J. T. Kuenstner et al., “Presence of Infection by Mycobacterium avium subsp. paratuberculosis in the Blood of Patients with Crohn’s Disease and Control Subjects Shown by Multiple Laboratory Culture and Antibody Methods,” Microorganisms, vol. 8, no. 12, Art. no. 12, Dec. 2020. [CrossRef]
- H. M. Fidler, W. Thurrell, N. M. Johnson, G. A. Rook, and J. J. McFadden, “Specific detection of Mycobacterium paratuberculosis DNA associated with granulomatous tissue in Crohn’s disease,” Gut, vol. 35, no. 4, pp. 506–510, Apr. 1994. [CrossRef]
- S. A. Naser, S. R. Sagramsingh, A. S. Naser, and S. Thanigachalam, “Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients,” World J Gastroenterol, vol. 20, no. 23, pp. 7403–7415, Jun. 2014. [CrossRef]
- R. J. Chiodini, H. J. Van Kruiningen, W. R. Thayer, and J. A. Coutu, “Spheroplastic phase of mycobacteria isolated from patients with Crohn’s disease.,” Journal of Clinical Microbiology, vol. 24, no. 3, pp. 357–363, 1986.
- K. L. Josephson, C. P. Gerba, and I. L. Pepper, “Polymerase chain reaction detection of nonviable bacterial pathogens,” Appl Environ Microbiol, vol. 59, no. 10, pp. 3513–3515, Oct. 1993. [CrossRef]
- H. Kobayashi, M. Oethinger, M. J. Tuohy, G. W. Procop, G. S. Hall, and T. W. Bauer, “Limiting false-positive polymerase chain reaction results: detection of DNA and mRNA to differentiate viable from dead bacteria,” Diagnostic Microbiology and Infectious Disease, vol. 64, no. 4, pp. 445–447, Aug. 2009. [CrossRef]
- R. A. Juste et al., “Association between Mycobacterium avium subsp. paratuberculosis DNA in blood and cellular and humoral immune response in inflammatory bowel disease patients and controls,” International Journal of Infectious Diseases, vol. 13, no. 2, pp. 247–254, Mar. 2009. [CrossRef]
- P. L. Almenoff, A. Johnson, M. Lesser, and L. H. Mattman, “Growth of acid fast L forms from the blood of patients with sarcoidosis.,” Thorax, vol. 51, no. 5, pp. 530–533, 1996.
- S. T. Brown et al., “Recovery of cell wall-deficient organisms from blood does not distinguish between patients with sarcoidosis and control subjects,” Chest, vol. 123, no. 2, pp. 413–417, Feb. 2003. [CrossRef]
- T. Udou, M. Ogawa, and Y. Mizuguchi, “Spheroplast formation of Mycobacterium smegmatis and morphological aspects of their reversion to the bacillary form,” J Bacteriol, vol. 151, no. 2, pp. 1035–1039, Aug. 1982. [CrossRef]
- T. Udou, M. Ogawa, and Y. Mizuguchi, “An improved method for the preparation of mycobacterial spheroplasts and the mechanism involved in the reversion to bacillary form: electron microscopic and physiological study,” Can. J. Microbiol., vol. 29, no. 1, pp. 60–68, Jan. 1983. [CrossRef]
- L. H. Mattman, Cell wall deficient forms: stealth pathogens. Boca Raton: CRC Press, 2001.
- Wohlfarth et al., “L-form conversion in Gram-positive bacteria enables escape from phage infection,” Nat Microbiol, pp. 1–13, Jan. 2023. [CrossRef]
- S. Gagneux, “Host–pathogen coevolution in human tuberculosis,” Philos Trans R Soc Lond B Biol Sci, vol. 367, no. 1590, pp. 850–859, Mar. 2012. [CrossRef]
- J. W. Saelens, G. Viswanathan, and D. M. Tobin, “Mycobacterial Evolution Intersects With Host Tolerance,” Front Immunol, vol. 10, p. 528, Mar. 2019. [CrossRef]
- L. L. Lu, T. J. Suscovich, S. M. Fortune, and G. Alter, “Beyond binding: antibody effector functions in infectious diseases,” Nat Rev Immunol, vol. 18, no. 1, pp. 46–61, Jan. 2018. [CrossRef]
- G. G. Meynell and E. Meynell, “Theory and Practice in Experimental Bacteriology.,” Theory and Practice in Experimental Bacteriology., 1965. [Online]. Available: https://www.cabdirect.org/cabdirect/abstract/19652704624 (accessed Jun. 14 2023).
- G. S. Wilson and M. A. Topley, “Wilson’s Principles of bacteriology, virology and immunity,” London: Edward Arnold, pp. 1855–1863, 1975.
- L. Lande et al., “Mycobacterium avium in Community and Household Water, Suburban Philadelphia, Pennsylvania, USA, 2010–2012,” Emerg Infect Dis, vol. 25, no. 3, pp. 473–481, Mar. 2019. [CrossRef]
- J. Aitken, G. Agrawal, N. Markova, G. Slavchev, W. Chamberlin, and T. J. Borody, “Demonstration of Intracellular Mycobacterium Species in Crohn’s Disease Using Novel Technologies: 1990,” Official journal of the American College of Gastroenterology | ACG, vol. 110, p. S843, Oct. 2015.
- S. Finnegan and S. L. Percival, “EDTA: An Antimicrobial and Antibiofilm Agent for Use in Wound Care,” Adv Wound Care (New Rochelle), vol. 4, no. 7, pp. 415–421, Jul. 2015. [CrossRef]
- Y. Nishiuchi et al., “Direct Attachment with Erythrocytes Augments Extracellular Growth of Pathogenic Mycobacteria,” Microbiol Spectr, vol. 10, no. 2, p. e0245421, Apr. 2022. [CrossRef]
- E. Mankiewicz and M. G. Tamari, “Lysogeny and Deoxyribonuclease Production in Mycobacteria,” Am Rev Respir Dis, vol. 106, no. 4, pp. 609–610, Oct. 1972. [CrossRef]
- Xinxin Zang et al., “Extracellular DNase MAP3916c attacks the neutrophil extracellular traps and is needed for Mycobacterium avium subsp. paratuberculosis virulence,” Veterinary microbiology, vol. 273, pp. 109529-, 2022. [CrossRef]
- T. Battaglioli, A. Soto, J. Agapito, V. Acurio, and P. Van der Stuyft, “Manual liquid culture on simple Middlebrook 7H9 or MGIT for the diagnosis of smear-negative pulmonary tuberculosis,” Trop Med Int Health, vol. 19, no. 12, pp. 1500–1503, Dec. 2014. [CrossRef]
- J. E. Meneguello et al., “Fast detection of Mycobacterium tuberculosis in culture-positive sputum samples by nitrate reductase activity,” Brazilian Journal of Pharmaceutical Sciences, vol. 54, no. 1, Art. no. 1, Jun. 2018. [CrossRef]
- M. BERNEY and L. BERNEY-MEYER, “Mycobacterium tuberculosis in the Face of Host-Imposed Nutrient Limitation,” Microbiol Spectr, vol. 5, no. 3, p. 10.1128/microbiolspec.TBTB2-0030–2016, Jun. 2017. [CrossRef]
- G. V. Mukamolova, O. Turapov, J. Malkin, G. Woltmann, and M. R. Barer, “Resuscitation-promoting factors reveal an occult population of tubercle Bacilli in Sputum,” Am J Respir Crit Care Med, vol. 181, no. 2, pp. 174–180, Jan. 2010. [CrossRef]
- M. L. Joloba et al., “What is the most reliable solid culture medium for tuberculosis treatment trials?,” Tuberculosis, vol. 94, no. 3, pp. 311–316, May 2014. [CrossRef]
- M. Drancourt, P. Carrieri, M.-J. Gévaudan, and D. Raoult, “Blood Agar and Mycobacterium tuberculosis: the End of a Dogma,” J Clin Microbiol, vol. 41, no. 4, pp. 1710–1711, Apr. 2003. [CrossRef]
- M. Drancourt and D. Raoult, “Cost-Effectiveness of Blood Agar for Isolation of Mycobacteria,” PLOS Neglected Tropical Diseases, vol. 1, no. 2, p. e83, Nov. 2007. [CrossRef]
- N. Markova, G. Slavchev, and L. Michailova, “Filterable forms and L-forms of Mycobacterium bovis BCG,” Human Vaccines & Immunotherapeutics, vol. 8, no. 6, pp. 759–764, Jun. 2012. [CrossRef]
- Petrovic Fabijan et al., “L-Form Switching in Escherichia coli as a Common β-Lactam Resistance Mechanism,” Microbiology Spectrum, vol. 10, no. 5, pp. e02419-22, Sep. 2022. [CrossRef]
- D. Wolf, P. Domínguez-Cuevas, R. A. Daniel, and T. Mascher, “Cell Envelope Stress Response in Cell Wall-Deficient L-Forms of Bacillus subtilis,” Antimicrob Agents Chemother, vol. 56, no. 11, pp. 5907–5915, Nov. 2012. [CrossRef]
- J. Errington, “Cell wall-deficient, L-form bacteria in the 21st century: a personal perspective,” Biochem Soc Trans, vol. 45, no. 2, pp. 287–295, Apr. 2017. [CrossRef]
- “One Health Zoonotic Disease Prioritization (OHZDP) | One Health | CDC,” Dec. 12, 2022. https://www.cdc.gov/onehealth/what-we-do/zoonotic-disease-prioritization/index.html (accessed Jun. 19, 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




