Submitted:
13 July 2023
Posted:
14 July 2023
You are already at the latest version
Abstract
Keywords:
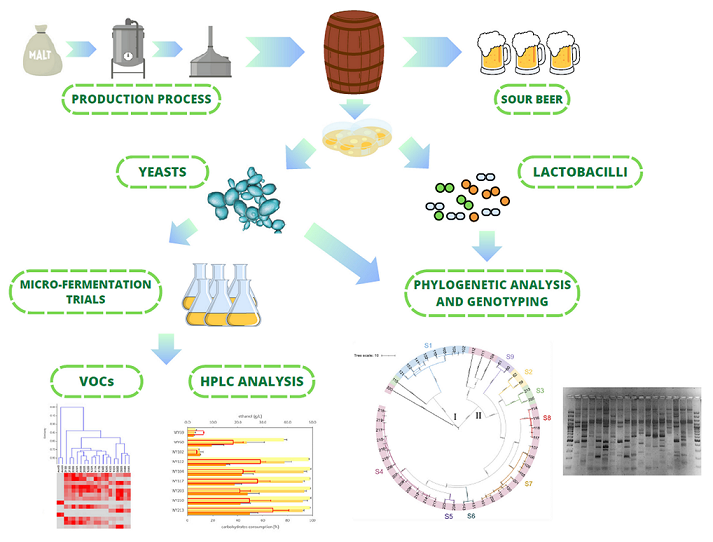
1. Introduction
2. Materials and Methods
2.1. Chemicals, Media, and Reference Strains
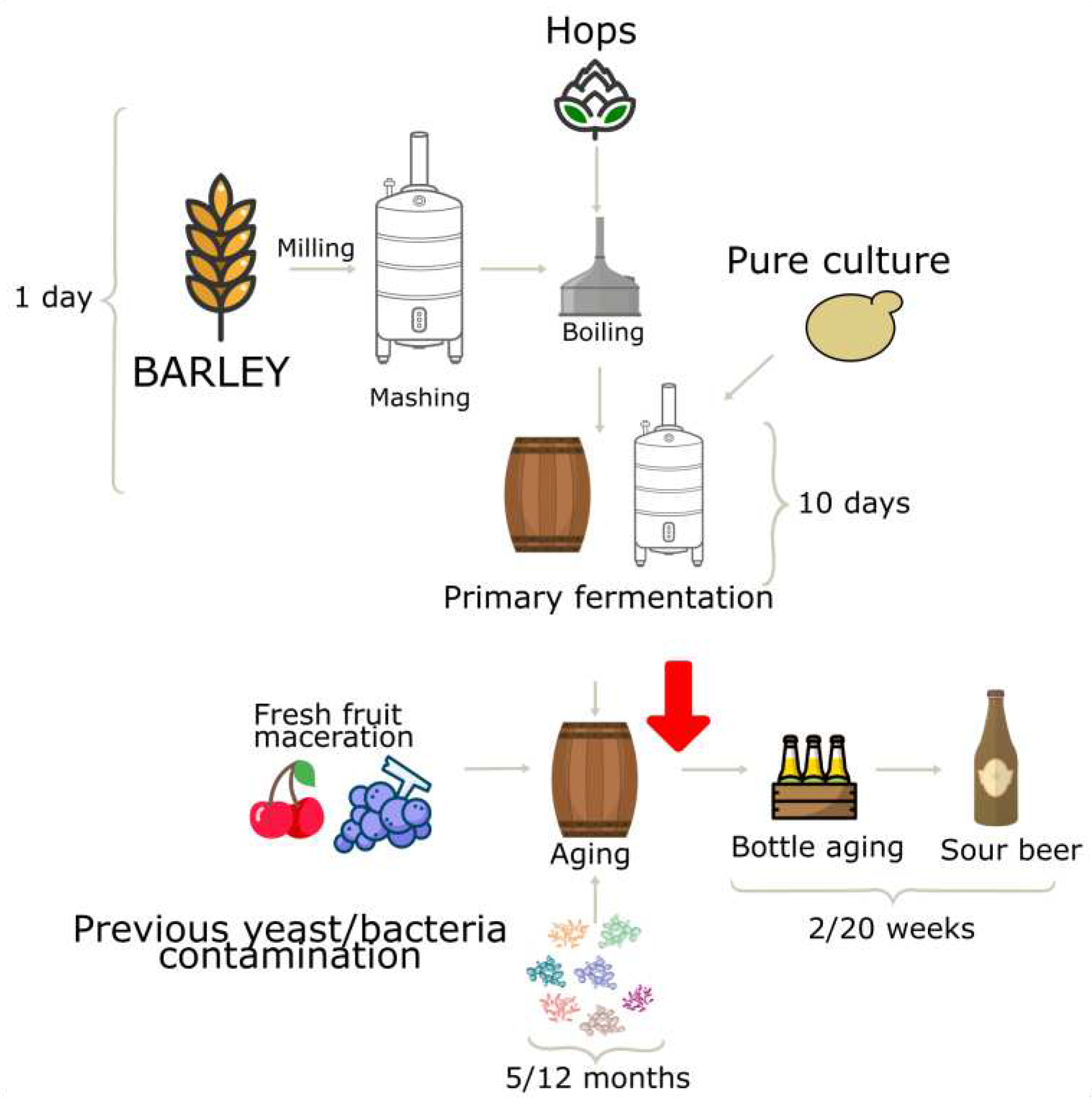
2.2. Sour Beer Production and Sampling
2.3. Microbial Counts, Bacterial and Yeast Isolation
2.4. Yeast and LAB Identification
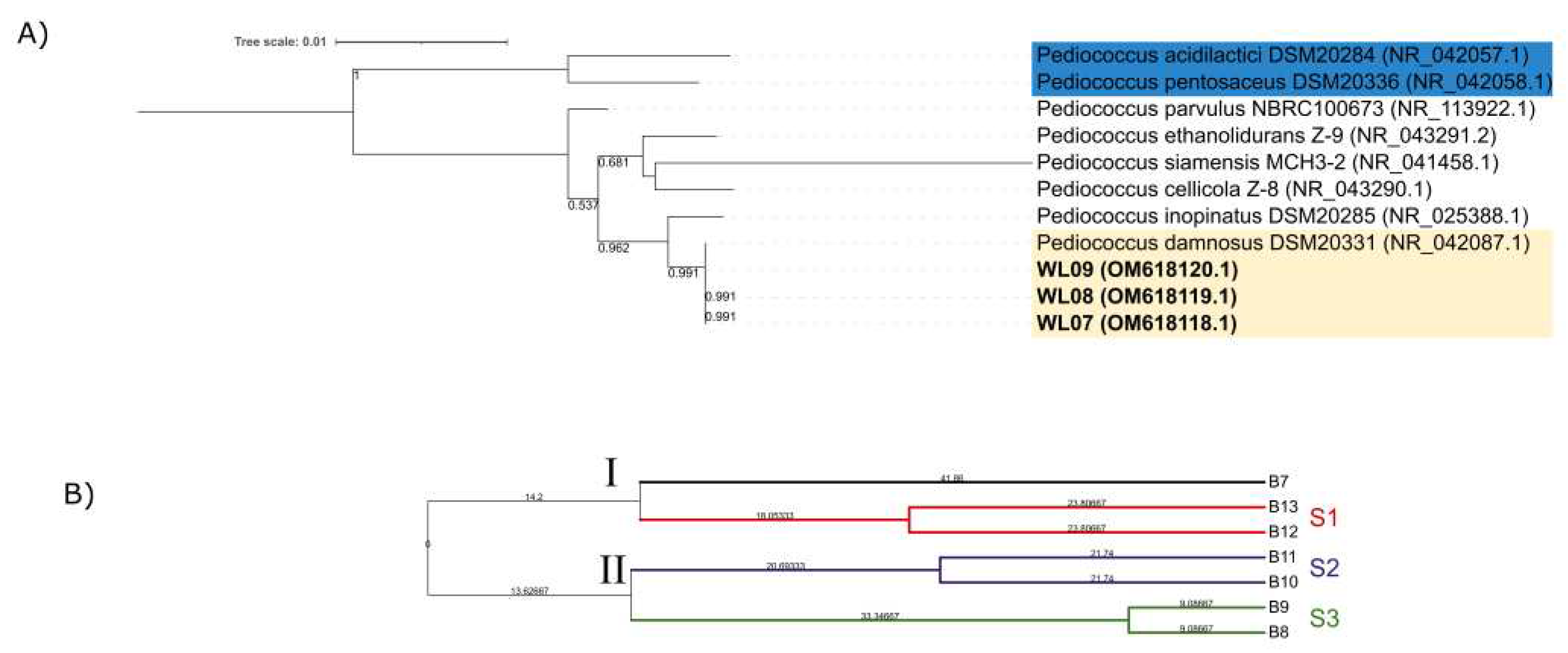
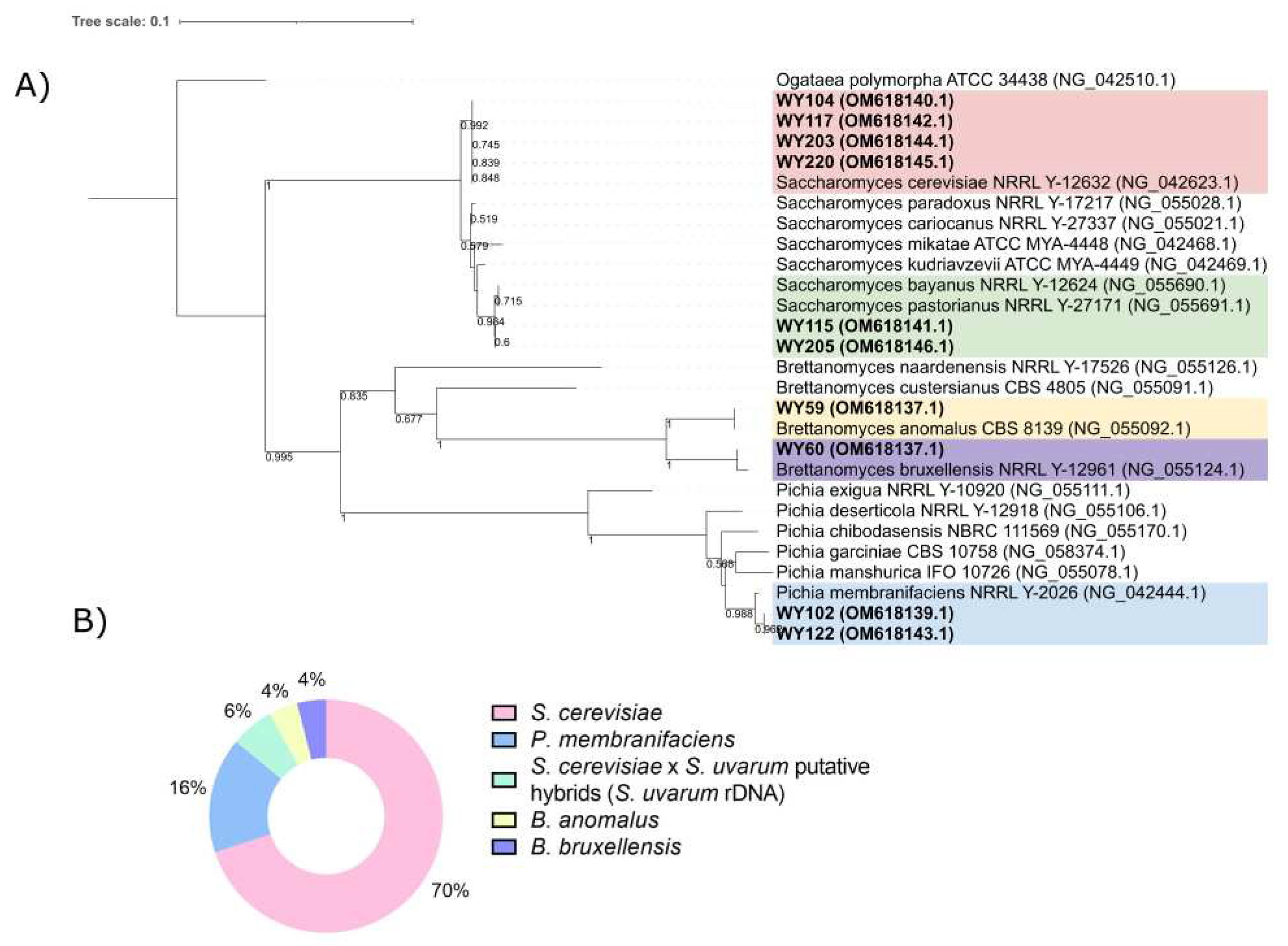
2.5. Phylogenetic Analysis
2.6. Genotyping Analysis
2.7. Flocculation, Sporulation Assays and MAT Locus Genotyping
2.8. Maltose Fermentation Tests
2.9. Wort Preparation and Micro-Scale Fermentation Trials
2.10. Physicochemical Analysis
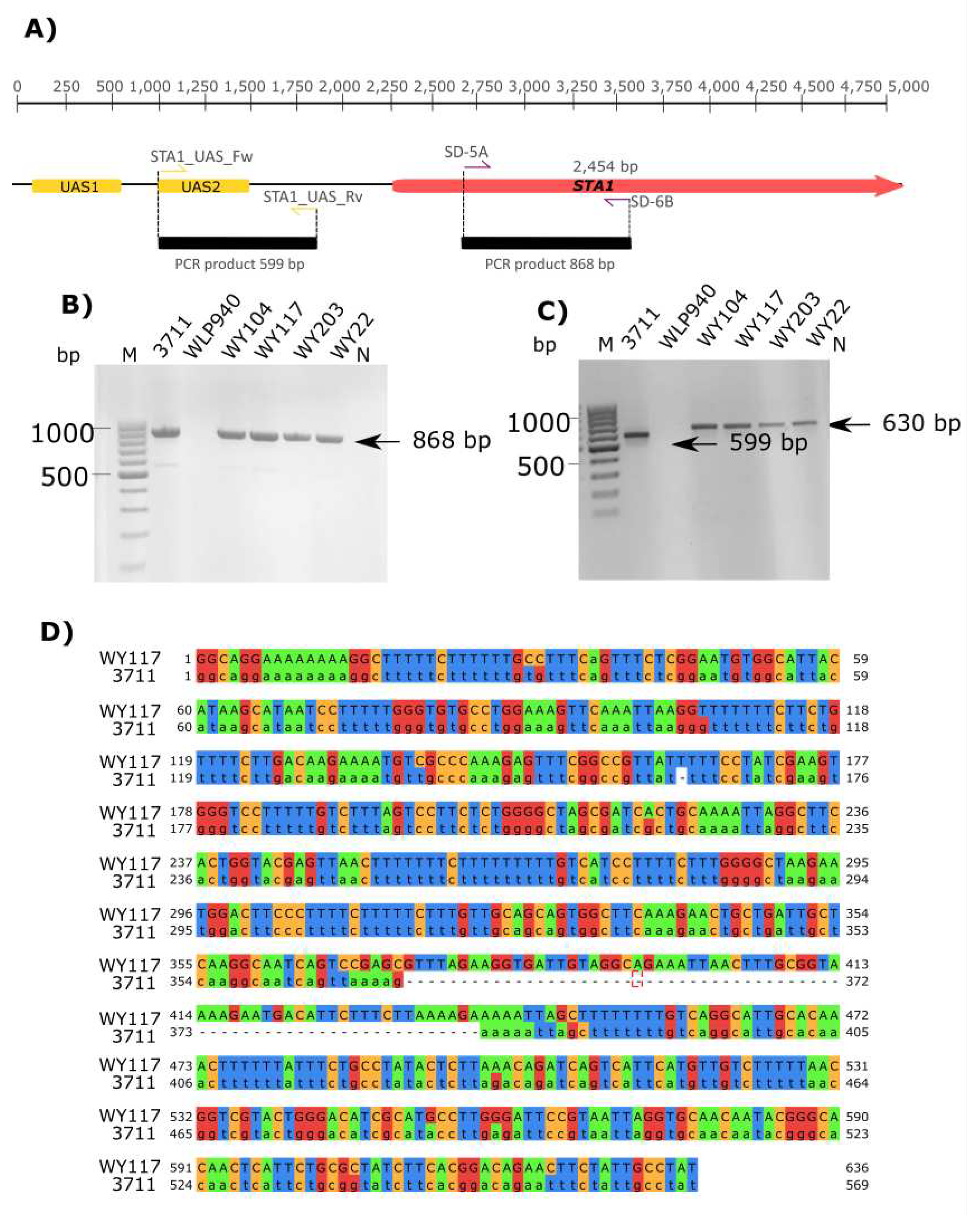
2.11. Screening of STA1 Gene
2.12. Statistical Analysis
3. Results
3.1. Sour Beer Physicochemical Caracterization and Microbiological Analysis
3.2. LAB Identification and Genotyping
3.3. Yeast Identification
3.4. Yeast Genotyping
3.5. Maltose Consumption and Flocculation
3.6. Sporulation and MAT Genotyping
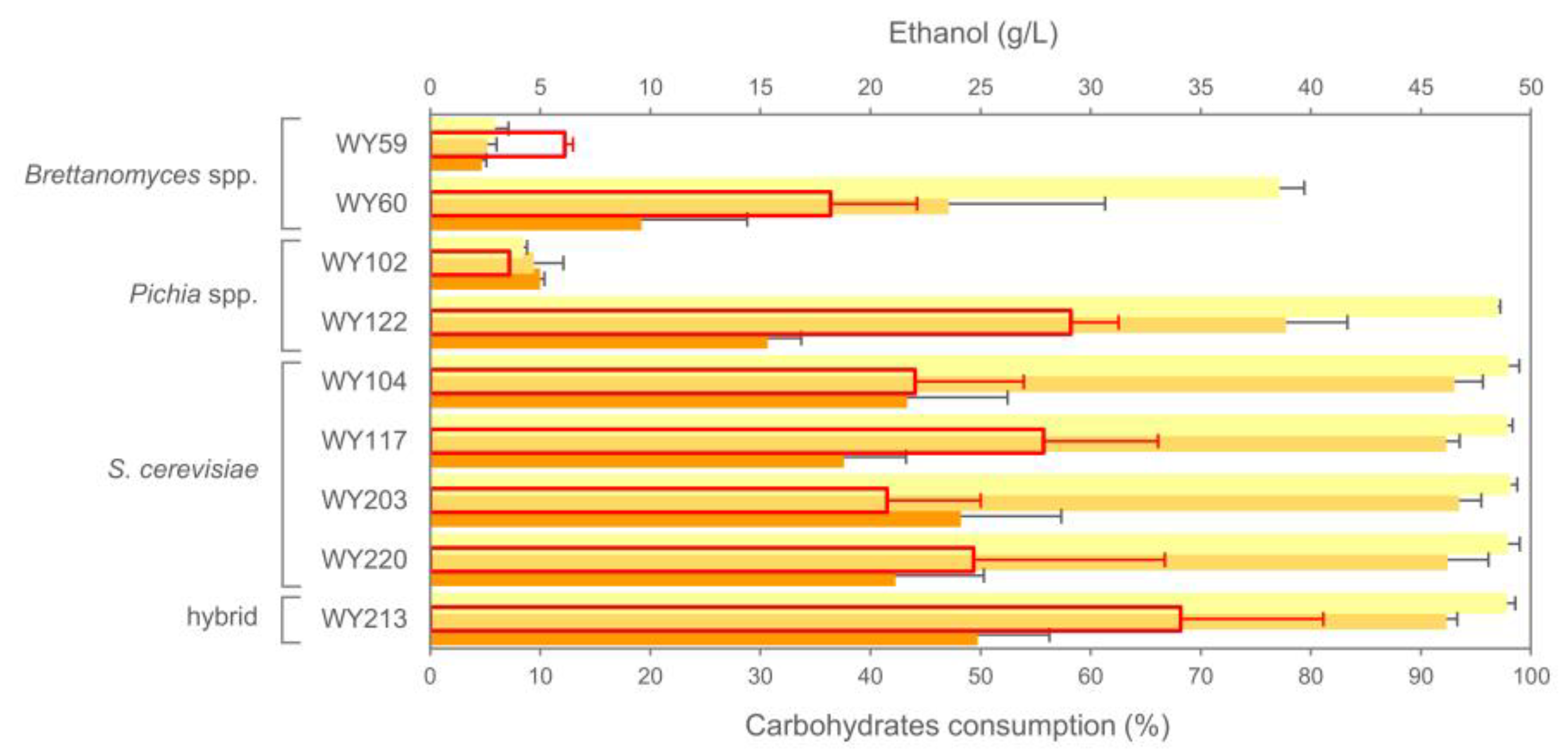
3.7. Micro-Scale Wort Fermentation
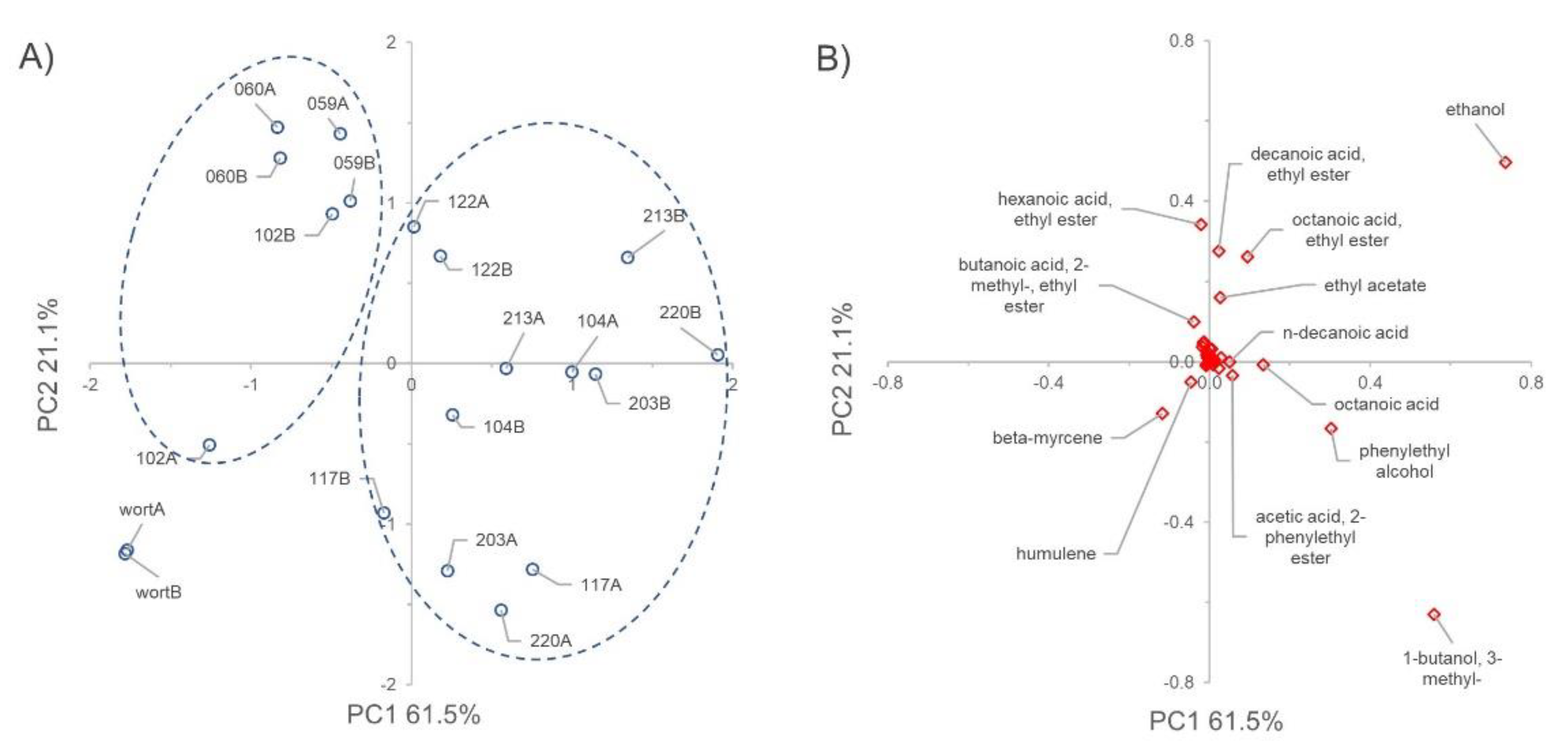
3.8. VOCs Determination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Carbone, A.; Quici, L. Craft beer mon amour: an exploration of Italian craft consumers. Br. Food J. 2020, 122, 2671–2687. [Google Scholar] [CrossRef]
- Baiano, A. Craft beer: An overview. Compr. Rev. Food Sci. Food Saf. 2021, 20, 1829–1856. [Google Scholar] [CrossRef] [PubMed]
- Aquilani, B.; Laureti, T.; Poponi, S.; Secondi, L. Beer choice and consumption determinants when craft beers are tasted: An exploratory study of consumer preferences. Food Qual. Pref. 2015, 41, 214–224. [Google Scholar] [CrossRef]
- Viejo, C.G.; Fuentes, S. Beer aroma and quality traits assessment using artificial intelligence. Fermentation 2020, 6, 56. [Google Scholar] [CrossRef]
- Pires, E.J.; Teixeira, J.A.; Brányik, T.; Vicente, A.A. Yeast: the soul of beer’s aroma—a review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl. Microbiol Biotechnol. 2014, 98, 1937–1949. [Google Scholar] [CrossRef] [PubMed]
- Svedlund, N.; Evering, S.; Gibson, B.; Krogerus, K. Fruits of their labour: biotransformation reactions of yeasts during brewery fermentation. Appl Microbiol Biotechnol. 2022, 106, 4929–4944. [Google Scholar]
- Holt, S.; Mukherjee, V.; Lievens, B. Verstrepen, K.J.; Thevelein, J.M. Bioflavoring by non-conventional yeasts in sequential beer fermentations. Food Microbiol. 2018, 72, 55–66. [Google Scholar] [CrossRef]
- Borneman, A.R.; Forgan, A.H.; Kolouchova, R.; Fraser, J.A.; Schmidt, S.A. Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3: Genes, Genomes, Genetics 2016, 6, 957–971. [Google Scholar] [CrossRef]
- Krogerus, K.; Magalhães, F.; Vidgren, V.; Gibson, B. Novel brewing yeast hybrids: creation and application. Appl. Microbiol. Biotechnol. 2017, 10, 65–78. [Google Scholar]
- Cubillos, F.A.; Gibson, B.; Grijalva-Vallejos, N.; Krogerus, K.; Nikulin, J. Bioprospecting for brewers: exploiting natural diversity for naturally diverse beers. Yeast. 2019, 36, 383–3980. [Google Scholar] [CrossRef]
- Iattici, F.; Catallo, M.; Solieri, L. Designing new yeasts for craft brewing: when natural biodiversity meets biotechnology. Beverages 2020, 6, 3. [Google Scholar] [CrossRef]
- Denby, C.M.; Li RA, Vu VT. ; Costello, Z.; Lin, W.; Chan, L.J.G; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; Scheller, H.V.; Martin, H.G.; Keasling, J.D. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 965. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, J.C.; Aisala, H.; Jokinen, I.; Krogerus, K.; Rischer, H.; Toivari, M. Production and sensory analysis of grape flavoured beer by co-fermentation of an industrial and a genetically modified laboratory yeast strain. Eur. Food Res. Technol. 2023, 30, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Astola, A.; Durán-Guerrero, E. , Díaz, A.B.; Lasanta, C.; Castro, R. Impact of the genetic improvement of fermenting yeasts on the organoleptic properties of beer. Eur. Food Res. Technol. 2023, 249, 1677–1687. [Google Scholar] [CrossRef]
- Alperstein, L.; Gardner, J.M.; Sundstrom, J.F.; Sumby, K.M.; Jiranek, V. Yeast bioprospecting versus synthetic biology—which is better for innovative beverage fermentation? Appl. Microbiol. Biotechnol. 2020, 104, 1939–1953. [Google Scholar]
- Dysvik, A.; Leanti La Rosa, S.; De Rouck, G.; Rukke, E-O. ; Westereng, B.; Wicklund, T. Microbial dynamics in traditional and modern sour beer production. Appl. Environ. Microbiol. 2020, 86, e00566–20. [Google Scholar] [CrossRef] [PubMed]
- Spitaels, F.; Wieme, A.D.; Janssens, M.; Aerts, M. Van Landschoot, A.; De Vuyst, L.; Vandamme, P. The microbial diversity of an industrially produced lambic beer shares members of a traditionally produced one and reveals a core microbiota for lambic beer fermentation. Food Microbiol. 2015, 49, 23–32. [Google Scholar] [CrossRef]
- De Roos, J.; Vandamme, P.; De Vuyst, L. Wort substrate consumption and metabolite production during lambic beer fermentation and maturation explain the successive growth of specific bacterial and yeast species. Front. Microbiol. 2018, 9, 2763. [Google Scholar] [CrossRef]
- De Roos, J.; Verce, M.; Weckx, S.; De Vuyst, L. Temporal shotgun metagenomics revealed the potential metabolic capabilities of specific microorganisms during lambic beer production. Front Microbiol. 2020, 11, 1692. [Google Scholar] [CrossRef]
- Bossaert, S.; Winne, V.; Van Opstaele, F.; Buyse, J.; Verreth, C.; Herrera-Malaver, B.; Van Geel, M.; Verstrepen, K.J.; Crauwels, S.; De Rouck, G.; Lievens, B. Description of the temporal dynamics in microbial community composition and beer chemistry in sour beer production via barrel ageing of finished beers. Int. J. Food Microbiol. 2021, 339, 109030. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Bamforth, C.W.; Mills, D.A. Brewhouse-resident microbiota are responsible for multi-stage fermentation of American coolship ale. PloS one 2012, 7, e35507. [Google Scholar] [CrossRef] [PubMed]
- Bongaerts, D.; De Roos, J.; De Vuyst, L. Technological and environmental features determine the uniqueness of the lambic beer microbiota and production process. Appl. Environment. Microbiol. 2021, 87, e00612–21. [Google Scholar] [CrossRef] [PubMed]
- Bossaert, S.; Crauwels, S.; Lievens, B.; De Rouck, G. The power of sour—A review: Old traditions, new opportunities. BrSc. 2019, 72, 78–88. [Google Scholar]
- Hoffman, C.S. , Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformaion of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Gala, E.; Landi, S.; Solieri, L.; Nocetti, M.; Pulvirenti, A.; Giudici, P. Diversity of lactic acid bacteria population in ripened Parmigiano Reggiano cheese. Int. J. Food Microbiol. 2008, 125, 347–351. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8 S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Evol. Microbiol. 1999, 49, 329–337. [Google Scholar] [CrossRef]
- Muir, A.; Harrison, E.; Wheals, A. A multiplex set of species-specific primers for rapid identification of members of the genus Saccharomyces. FEMS Yeast Res. 2011, 11, 552–563. [Google Scholar] [CrossRef]
- Pengelly, R.J.; Wheals, A.E. Rapid identification of Saccharomyces eubayanus and its hybrids. " FEMS Yeast Res. 2013, 13, 156–161. [Google Scholar] [CrossRef]
- Martini, S.; Bonazzi, S.; Malorgio, I.; Pizzamiglio, V.; Tagliazucchi, D.; Solieri, L. Characterization of yeasts isolated from Parmigiano Reggiano cheese natural whey starter: from spoilage agents to potential cell factories for whey valorization. Microorganisms 2021, 9, 2288. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 4, D733–D745. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids. Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N; Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 1987, 4, 406–425. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 2, 256–259. [Google Scholar] [CrossRef]
- Versalovic, J.; Schneider, M.; De Bruijn, F.J.; Lupski, J.R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Meth. Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Solieri, L.; Bianchi, A.; Giudici, P. Inventory of non starter lactic acid bacteria from ripened Parmigiano Reggiano cheese as assessed by a culture dependent multiphasic approach. Syst. Appl. Microbiol. 2012, 35, 270–277. [Google Scholar] [CrossRef]
- Dakal, T.C.; Solieri, L.; Giudici, P. Evaluation of fingerprinting techniques to assess genotype variation among Zygosaccharomyces strains. Food Microbiol. 2018, 72, 135–145. [Google Scholar] [CrossRef]
- Legras, J.-L.; Karst, F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol. Lett. 2003, 221, 2249–2255. [Google Scholar] [CrossRef]
- Corte, L.; Lattanzi, M.; Buzzini, P.; Bolano, A.; Fatichenti, F.; Cardinali, G. Use of RAPD and killer toxin sensitivity in Saccharomyces cerevisiae strain typing. J. Appl. Microbiol. 2005, 9, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Vigentini, I.; Barrera Cardenas, S.; Valdetara, F.; Faccincani, M.; Panont, C.A.; Picozzi, C.; Foschino, R. Use of native yeast strains for in-bottle fermentation to face the uniformity in sparkling wine production. Front. Microbiol. 2017, 8, 1225. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P., Fell; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In The yeasts, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, 2011; pp. 87–110. [Google Scholar]
- Catallo, M.; Iattici, F.; Randazzo, C.; Caggia, C.; Krogerus, K.; Magalhães, F.; Gibson, B.; Solieri, L. Hybridization of Saccharomyces cerevisiae Sourdough Strains with Cryotolerant Saccharomyces bayanus NBRC1948 as a Strategy to Increase Diversity of Strains Available for Lager Beer Fermentation. Microorganisms 2021, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Marongiu, A.; Zara, G.; Legras, J.-L.; Del Caro, A.; Mascia, I.; Fadda, C.; Budroni, M. Novel Starters for Old Processes: Use of Saccharomyces cerevisiae Strains Isolated from Artisanal Sourdough for Craft Beer Production at a Brewery Scale. J. Ind. Microbiol. Biotechnol. 2015, 42, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Kahm, M.; Hasenbrink, G.; Lichtenberg-Frate, H.; Ludwig, J.; Kschischo, M. Grofit: fitting biological growth curves with R. J. Stat. Softw. 2010, 33, 1–21. [Google Scholar] [CrossRef]
- Yamauchi, H.; Yamamoto, H.; Shibano, Y.; Amaya, N.; Saeki, T. Rapid methods for detecting Saccharomyces diastaticus, a beer spoilage yeast, using the polymerase chain reaction. J. Am. Soc. Brew. Chem. 1998, 56, 58–63. [Google Scholar]
- Krogerus, K.; Magalhães, F.; Kuivanen, J. A deletion in the STA1 promoter determines maltotriose and starch utilization in STA1+ Saccharomyces cerevisiae strains. Appl Microbiol Biotechnol. 2019, 103, 7597–7615. [Google Scholar] [CrossRef]
- Hammer, Øyvind, and David AT Harper. "Past: paleontological statistics software package for educaton and data anlysis." Palaeontologia electronica 4.1. 2001, 1.
- Bokulich, N.A.; Bamforth,C.W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. 2013, 77, 157–172. [CrossRef]
- De Roos, J.; Van der Veken, D.; De Vuyst, L. The interior surfaces of wooden barrels are an additional microbial inoculation source for lambic beer production. Appl. Environ. Microbiol. 2019, 85, e02226–18. [Google Scholar] [CrossRef]
- Caldwell, S.L.; McMahon, D.J.; Oberg, C.J. , Broadbent, J.R. Development and characterization of lactose-positive Pediococcus species for milk fermentation. Appl. Environ. Microbiol. 1996, 62, 936–941. [Google Scholar] [CrossRef]
- Spitaels, F.; Wieme, A.D.; Janssens, M.; Aerts, M.; Daniel, H-M. ; Van Landschoot, A.; De Vuyst, L.; Vandamme, P. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS One, 2014, 9, e95384. [Google Scholar] [CrossRef] [PubMed]
- Rankine, B.C. Decomposition of L-malic acid by wine yeasts. J. Sci. Food Agric. 1966, 17, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Saez, J.; Lopes, C.A; Kirs, V.E.; Sangorrín, M. Production of volatile phenols by Pichia manshurica and Pichia membranifaciens isolated from spoiled wines and cellar environment in Patagonia. Food Microbiol. 2011, 28, 503–509. [Google Scholar] [CrossRef]
- Wang, X.; Ren, X.; Shao, Q.; Peng, X.; Zou, W.; Sun, Z.; Zhang, L.; Li, H. Transformation of Microbial Negative Correlations into Positive Correlations by Saccharomyces cerevisiae Inoculation during Pomegranate Wine Fermentation. Appl. Environ. Microbiol. 2020, 86, e01847–20. [Google Scholar] [CrossRef] [PubMed]
- Lachance, M.-A. Yeast Communities in a Natural Tequila Fermentation. Antonie Van Leeuwenhoek 1995, 68, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Vianna, C.R.; Cadete. R.M.; Santos, R.O.; Gomes, F.C.; Oliveira, E.S.; Rosa, C.A. Selection, growth, and chemo-sensory evaluation of flocculent starter culture strains of Saccharomyces cerevisiae in the large-scale production of traditional Brazilian cachaça. Int J Food Microbiol. 2009, 131, 203–210.
- Bossaert, S.; Kocijan, T.; Winne, V.; Schlich, J.; Herrera-Malaver, B.; Verstrepen, K.J.; Van Opstaele, F.; De Rouck, G.; Crauwels, S.; Lievens, B. Beer ethanol and iso-α-acid level affect microbial community establishment and beer chemistry throughout wood maturation of beer. Int J Food Microbiol. 2022, 374, 109724. [Google Scholar] [CrossRef]
- Tomičić, R.; Tomičić, Z.; Thaler, N.; Humar, M.; Raspor, P. Factors Influencing Adhesion of Bacteria Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus and Yeast Pichia membranifaciens to Wooden Surfaces. Wood Sci. Technol. 2020, 54, 1663–1676. [Google Scholar] [CrossRef]
- Ponzoni, C.; Gasparetti, C.; Goretti, M.; Turchetti, B.; Pagnoni, U.M.; Cramarossa. M.R.; Forti, L.; Buzzini, P. Biotransformation of acyclic monoterpenoids by Debaryomyces sp., Kluyveromyces sp., and Pichia sp. strains of environmental origin. Chem Biodivers. 2008, 5, 471–83. [CrossRef]
- Vaughan-Martini, A.; Martini, A. Saccharomyces Meyen ex Reess (1870). In The yeasts 5th ed; Kurtzman, C.P.; Fell, J.W.; Boekhout, T., Eds.; Elsevier, Amsterdam, 2011, pp. 733-746.
- Peŕez-Través, L.; Lopes, C.A.; Querol, A.; Barrio, E. On the complexity of the Saccharomyces bayanus taxon: Hybridization and potential hybrid speciation. PLoS ONE 2014, 9, e93729. [Google Scholar] [CrossRef]
- Libkind, D.; Hittinger, C.T.; Valério, E.; Gonçalves, C.; Dover, J.; Johnston, M.; Gonçalves, P.; Sampaio, J.P. Microbe Domestication and the Identification of the Wild Genetic Stock of Lager-Brewing Yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.I.; James, S.A.; Naumova, E.S.; Louis, E.J.; Roberts, I.N. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Sys. Evol. Microbiol. 2000, 39050, 1931–1942. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Taxonomic and physiological diversity of Saccharomyces bayanus. Biodivers. Biotechnol. Wine Yeasts 2002, 53–69. [Google Scholar]
- Demuyter, C.; Lollier, M.; Legras, J.-L.; Le Jeune, C. Predominance of Saccharomyces uvarum during spontaneous alcoholic fermentation, for three consecutive years, in an Alsatian winery. J. Appl. Microbiol. 2004, 97, 1140–1148. [Google Scholar] [CrossRef]
- Coton, E.; Coton, M.; Levert, D.; Casaregola, S.; Sohier, D. Yeast Ecology in French Cider and Black Olive Natural Fermentations. Int. J. Food. Microbiol. 2006, 108, 130–135. [Google Scholar] [CrossRef]
- Valles, B.S.; Bedriñana, R.P.; Tascón, N.F.; Simón, A.Q.; Madrera, R.R. Yeast species associated with the spontaneous fermentation of cider. Food Microbiol. 2007, 24, 25–31. [Google Scholar] [CrossRef]
- Rodríguez ME, Perez-Traves L, Sangorrín MP, Barrio E, Querol A, Lopes CA. Saccharomyces uvarum is responsible for the traditional fermentation of apple chicha in Patagonia. FEMS Yeast Res. 2017, 17, fow109.
- Rainieri, S.; Kodama, Y.; Kaneko, Y.; Mikata, K.; Nakao, Y.; Ashikari, T. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 2006, 72, 3968–3974. [Google Scholar] [CrossRef]
- Gibson, B.; Dahabieh, M.; Krogerus, K.; Jouhten, P.; Magalhães, F.; Pereira, R.; Siewers, V.; Vidgren, V. Adaptive Laboratory Evolution of Ale and Lager Yeasts for Improved Brewing Efficiency and Beer Quality. Annu. Rev. Food Sci. Technol. 2020, 11, 23–44. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Pichia EC Hansen (1904). In The yeasts, 5th ed; Kurtzman, C.P.; Fell, J.W.; Boekhout, T., Eds.; Elsevier, Amsterdam, 2011, pp. 273-352.
- Gallone, B.; Steensels, J.; Prahl, T.; Soriaga, L.; Saels, V.; Herrera-Malaver, B.; Merlevede, A.; Roncoroni, M.; Voordeckers, K.; Miraglia, L.; et al. Domestication and Divergence of Saccharomyces cerevisiae Beer Yeasts. Cell 2016, 166, 1397–1410. [Google Scholar] [CrossRef]
- Steensels, J.; Daenen, L.; Malcorps, P.; Derdelinckx, G.; Verachtert, H.; Verstrepen, K.J. Brettanomyces Yeasts—From Spoilage Organisms to Valuable Contributors to Industrial Fermentations. Int. J. Food Microbiol. 2015, 206, 24–38. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Derdelinckx, G.; Dufour, J.-P.; Winderickx, J.; Thevelein, J.M.; Pretorius, I.S.; Delvaux, F.R. Flavor-Active Esters: Adding Fruitiness to Beer. J. Biosci. Bioeng. 2003, 96, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Saerens, S.; Delvaux, F.; Verstrepen, K.; Van Dijck, P.; Thevelein, J.; Delvaux, F. Parameters Affecting Ethyl Ester Production by Saccharomyces cerevisiae during Fermentation. Appl. Environ. Microbiol. 2008, 74, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.; Nikulin, J.; Juvonen, R.; Krogerus, K.; Magalhães, F.; Mikkelson, A.; Nuppunen-Puputti, M.; Sohlberg, E.; de Francesco, G.; Perretti, G.; et al. Sourdough Cultures as Reservoirs of Maltose-Negative Yeasts for Low-Alcohol Beer Brewing. Food Microbiol. 2021, 94, 103629. [Google Scholar] [CrossRef]
- Krogerus, K.; Eerikainen, R.; Aisala, H.; Gibson, B. Repurposing brewery contaminant yeast as production strains for low-alcohol beer fermentation. Yeast 2022, 39, 156–169. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, J. The Mystery of Lambic Beer. Sci. Am. 1996, 275, 74–80. [Google Scholar] [CrossRef]
- Piskur, J.; Rozpedowska, E.; Polakova, S.; Merico, A.; Compagno, C. How Did Saccharomyces Evolve to Become a Good Brewer? Trends Genet. 2006, 22, 183–186. [Google Scholar] [CrossRef]
- Shayevitz, A.; Harrison, K.; Curtin, C.D. Barrel-Induced Variation in the Microbiome and Mycobiome of Aged Sour Ale and Imperial Porter Beer. J. Am. Soc. Brew. Chem. 2021, 79, 33–40. [Google Scholar] [CrossRef]
- Sampaio, J.P.; Gonçalves, P. Natural Populations of Saccharomyces kudriavzevii in Portugal Are Associated with Oak Bark and Are Sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 2008, 74, 2144–2152. [Google Scholar] [CrossRef]
- Paraíso, F.; Pontes, A.; Neves, J.; Lebani, K.; Hutzler, M.; Zhou, N.; Sampaio, J.P. Do microbes evade domestication?-Evaluating potential ferality among diastatic Saccharomyces cerevisiae. Food Microbiol. 2023, 7, 104320. [Google Scholar] [CrossRef]
- Coelho, E.; Azevedo, M.; Teixeira, J.A.; Tavares, T.; Oliveira, J.M.; Domingues, L. Evaluation of multi-starter S. cerevisiae/D. bruxellensis cultures for mimicking and accelerating transformations occurring during barrel ageing of beer. Food Chem. 2020, 323, 126826. [Google Scholar] [CrossRef] [PubMed]
- Rozpędowska, E.; Hellborg, L.; Ishchuk, O.P.; Orhan, F.; Galafassi, S.; Merico, A.; Woolfit, M.; Compagno, C.; Piskur, J. Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts. Nat. Commun. 2011, 2, 302. [Google Scholar] [CrossRef] [PubMed]
- Meier-Dörnberg T, Kory OI, Jacob F, Michel M, Hutzler M. Saccharomyces cerevisiae variety diastaticus friend or foe?-spoilage potential and brewing ability of different Saccharomyces cerevisiae variety diastaticus yeast isolates by genetic, phenotypic and physiological characterization. FEMS Yeast Res. 2018, 18, foy023.
- Peris, D.; Pérez-Torrado, R.; Hittinger, C.T.; Barrio, E.; Querol, A. On the origins and industrial applications of Saccharomyces cerevisiae × Saccharomyces kudriavzevii hybrids. Yeast 2018, 35, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Preiss, R.; Tyrawa, C.; Krogerus, K.; Garshol, L.M.; van der Merwe, G. Traditional Norwegian kveik are a genetically distinct group of domesticated Saccharomyces cerevisiae brewing yeasts. Front. Microbiol. 2018, 9, 2137. [Google Scholar] [CrossRef] [PubMed]
- Krogerus, K.; Preiss, R.; Gibson, B. A Unique Saccharomyces cerevisiae × Saccharomyces uvarum Hybrid Isolated From Norwegian Farmhouse Beer: Characterization and Reconstruction. Front Microbiol. 2018, 9, 2253. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Sadiq, F.A.; Yang, H.; Gu, J.; Yuan, L.; Lee, Y.K.; He, G. Predominant yeasts in Chinese traditional sourdough and their influence on aroma formation in Chinese steamed bread. Food Chem. 2018, 242, 404–411. [Google Scholar] [CrossRef]
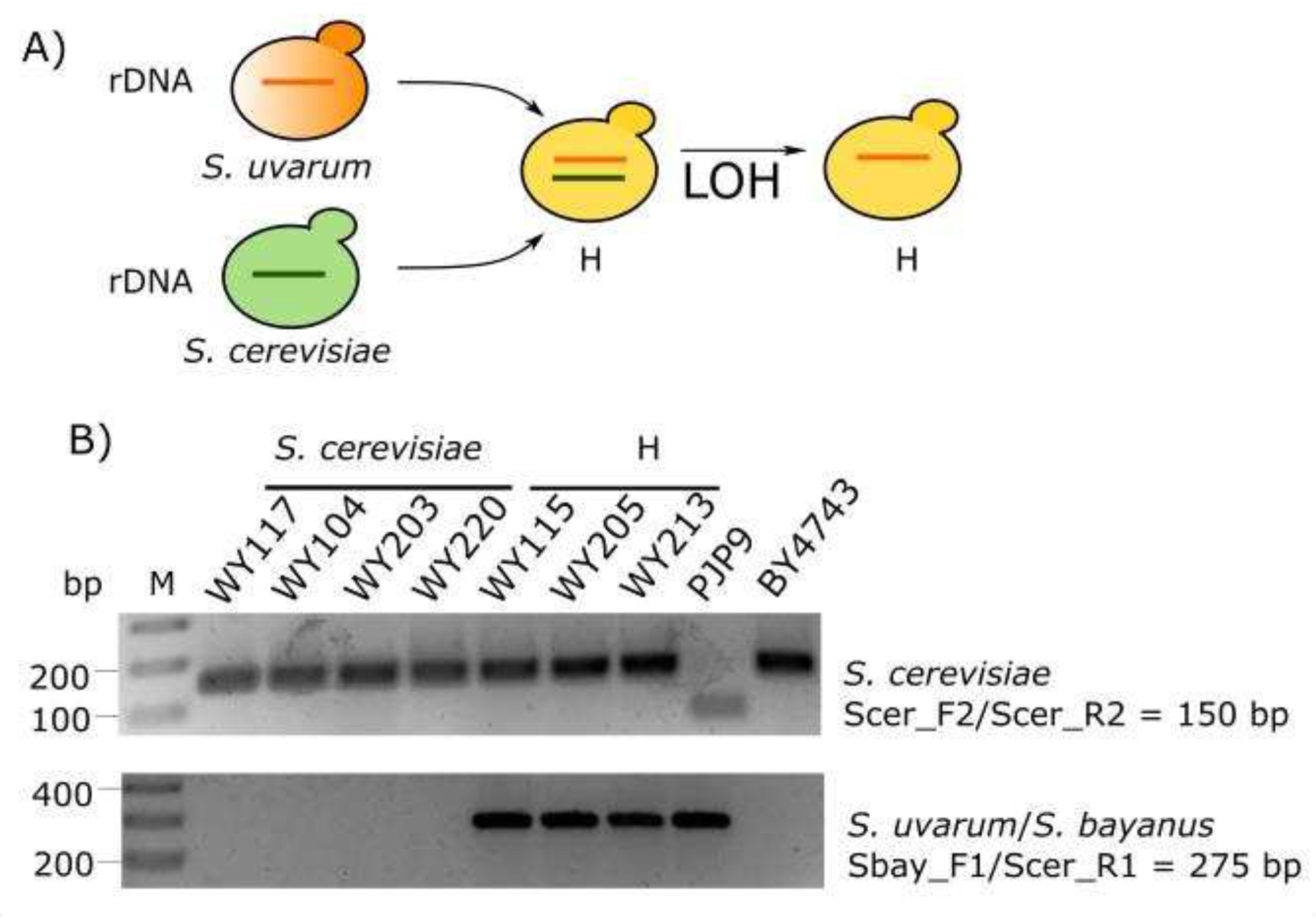
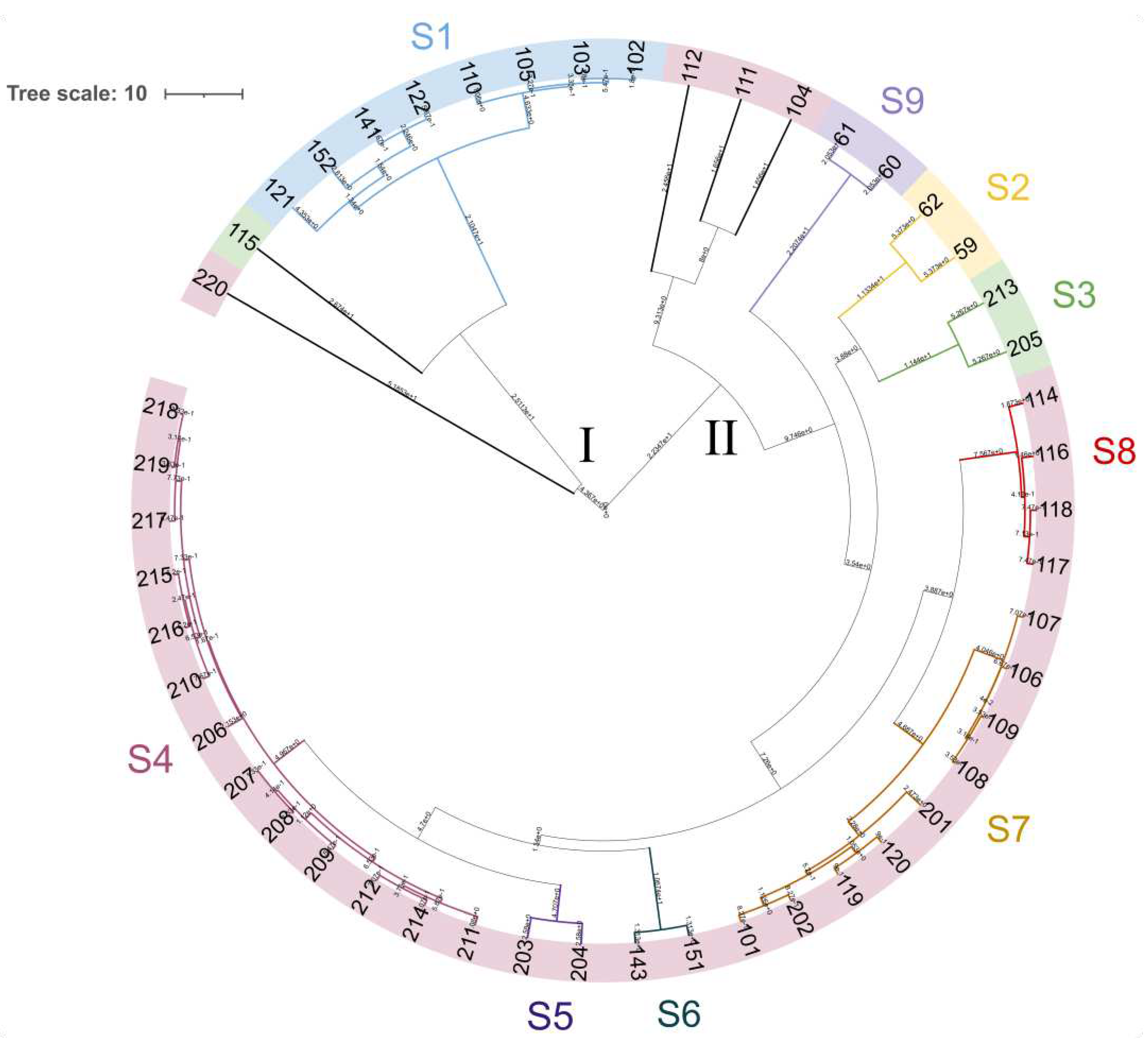
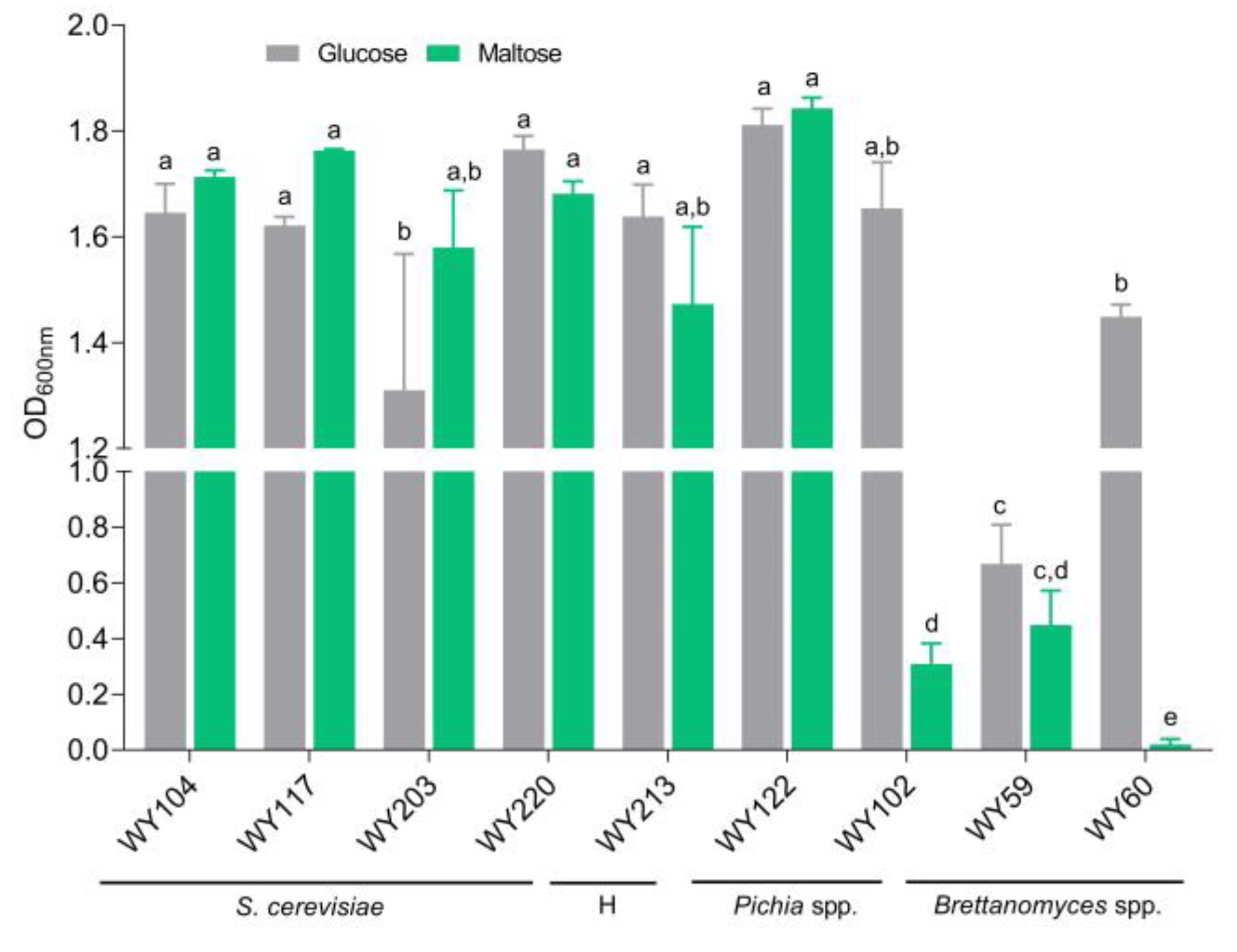
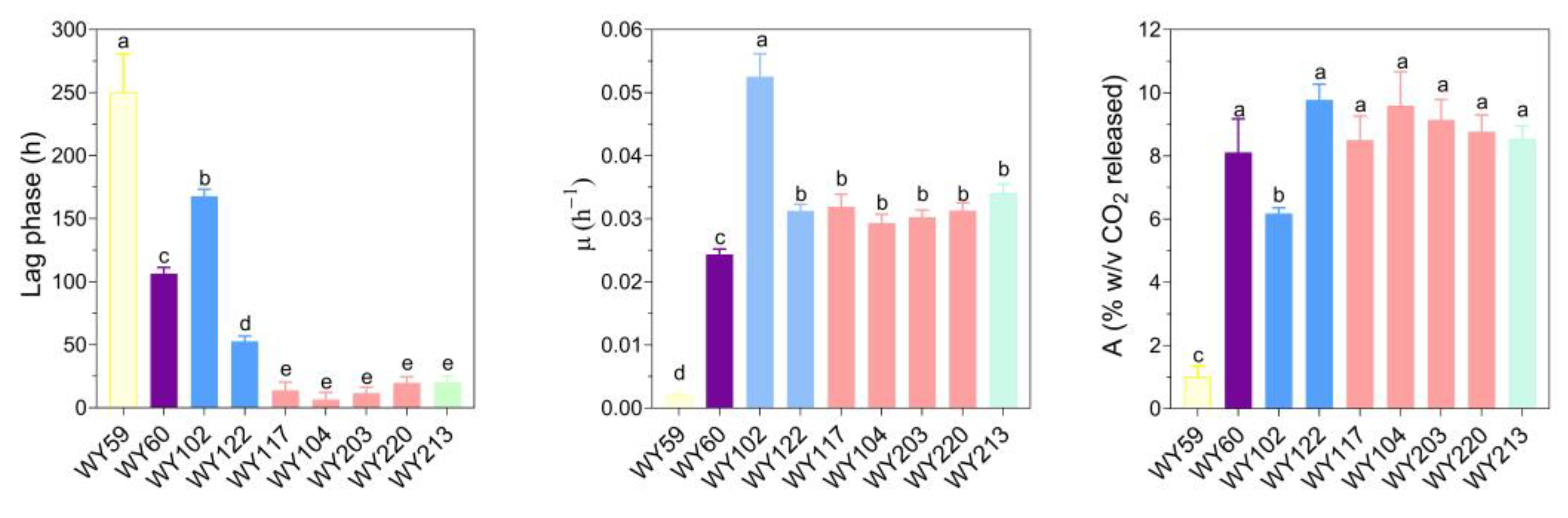
| Strain | Species | Features | Source |
|---|---|---|---|
| BY4742 | S. cerevisiae | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 | Euroscarf |
| BY4743 | S. cerevisiae | MATa/α; his3Δ1/ his3Δ1; leu2Δ0/ leu2Δ0; met15Δ0/MET15; LYS2/lys2Δ0; ura3Δ0/ura3Δ0 | Euroscarf |
| 3711 | S. cerevisiae var. diastaticus | STA1 positive French Saison ale yeast | Wyeast |
| WLP940 | Lager | STA1 negative lager yeast | White Lab |
| Atecrem | S. cerevisiae | Atecrem Belgian Saison | Bioenologia 2.0 |
| WLP565 | S. cerevisiae | Belgian Saison I Ale Yeast | White Lab |
| 3724 | S. cerevisiae var. diastaticus | Belgian Saison Yeast | Wyeast |
| PJP9 | S. uvarum/S. bayanus | nd | Gift from P. Marullo |
| Species | Strains | Sporulation efficiency (%) | Spore viability (%) | MAT loci | |
|---|---|---|---|---|---|
| 3 d | 14 d | ||||
| S. cerevisiae | WY104 | 22.96 | 24.56 | 75 | MATa/MATα |
| WY117 | 23.84 | 32.72 | 75 | MATa/MATα | |
| WY203 | 22.70 | 27.85 | 81 | MATa/MATα | |
| WY220 | 22.78 | 32.13 | 56 | MATa/MATα | |
| hybrids | WY205 | 1.22 | 0.89 | nd | nd |
| WY213 | 2.48 | 0.41 | nd | nd | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





