Submitted:
12 July 2023
Posted:
13 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates, Propagation And Species Determination
2.2. DsRNA Extraction, Virus-Like Particle Purification, Protein Analysis and Electron Microscopy
2.3. Virus Sequence Determination
2.4. Verification of Virus Presence by RT-PCR
3. Results
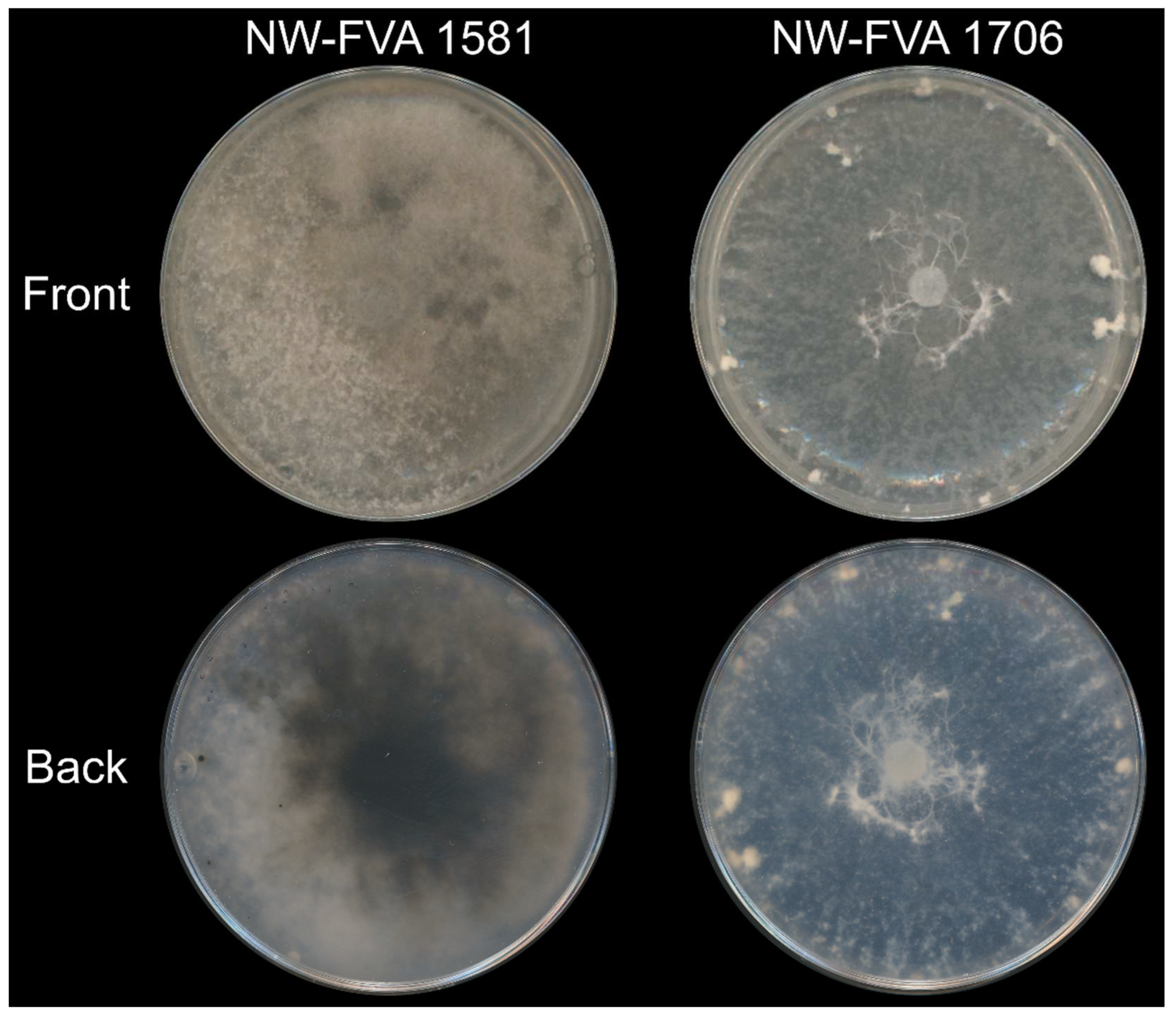
3.1. NW-FVA 1581 and NW-FVA 1706 were Determined as D. fraxini and Harbor dsRNAs
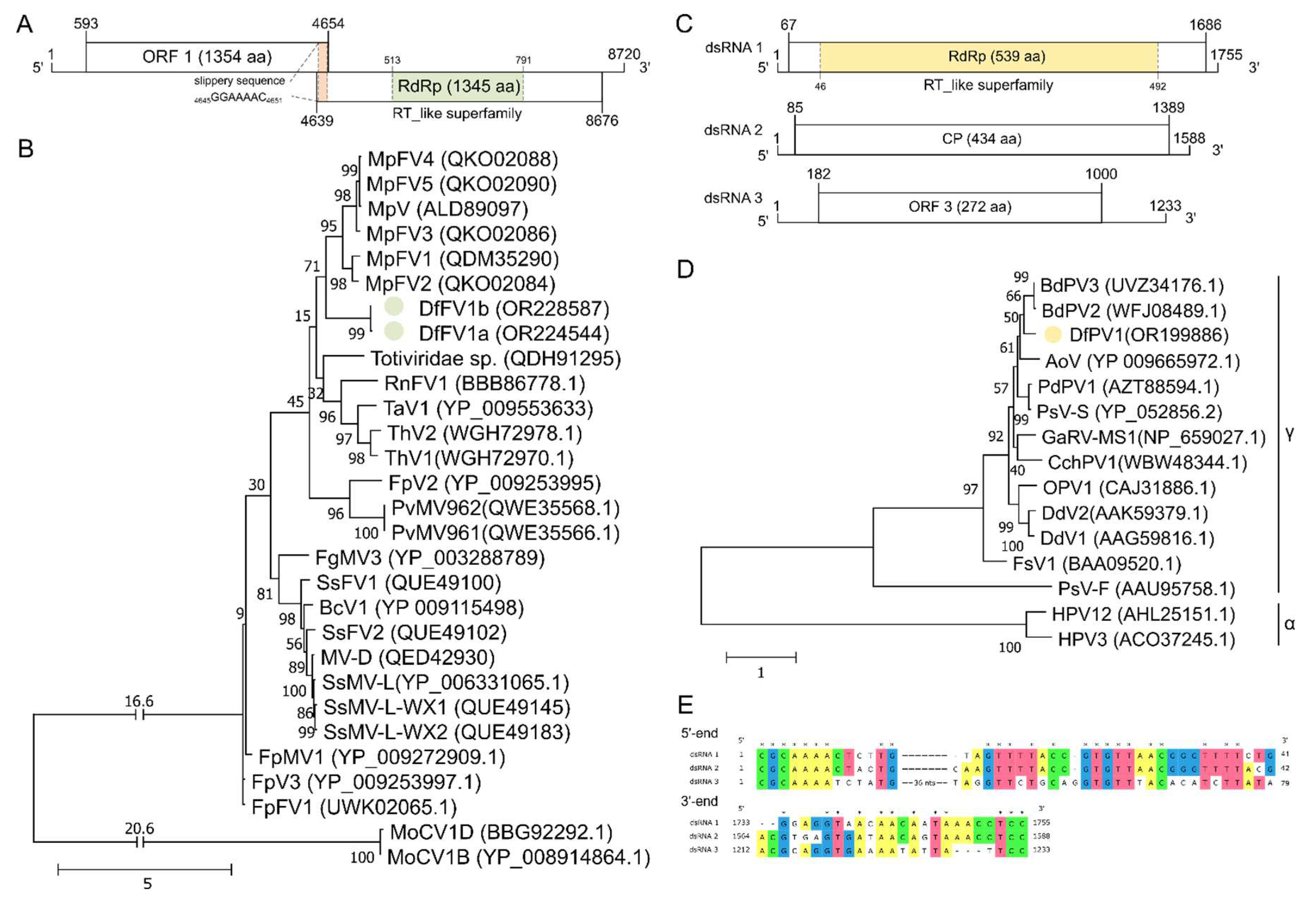
3.2. NW-FVA 1581 is Infected by a Novel Fusagravirus and NW-FVA 1706 Additionally by a Novel Partitivirus
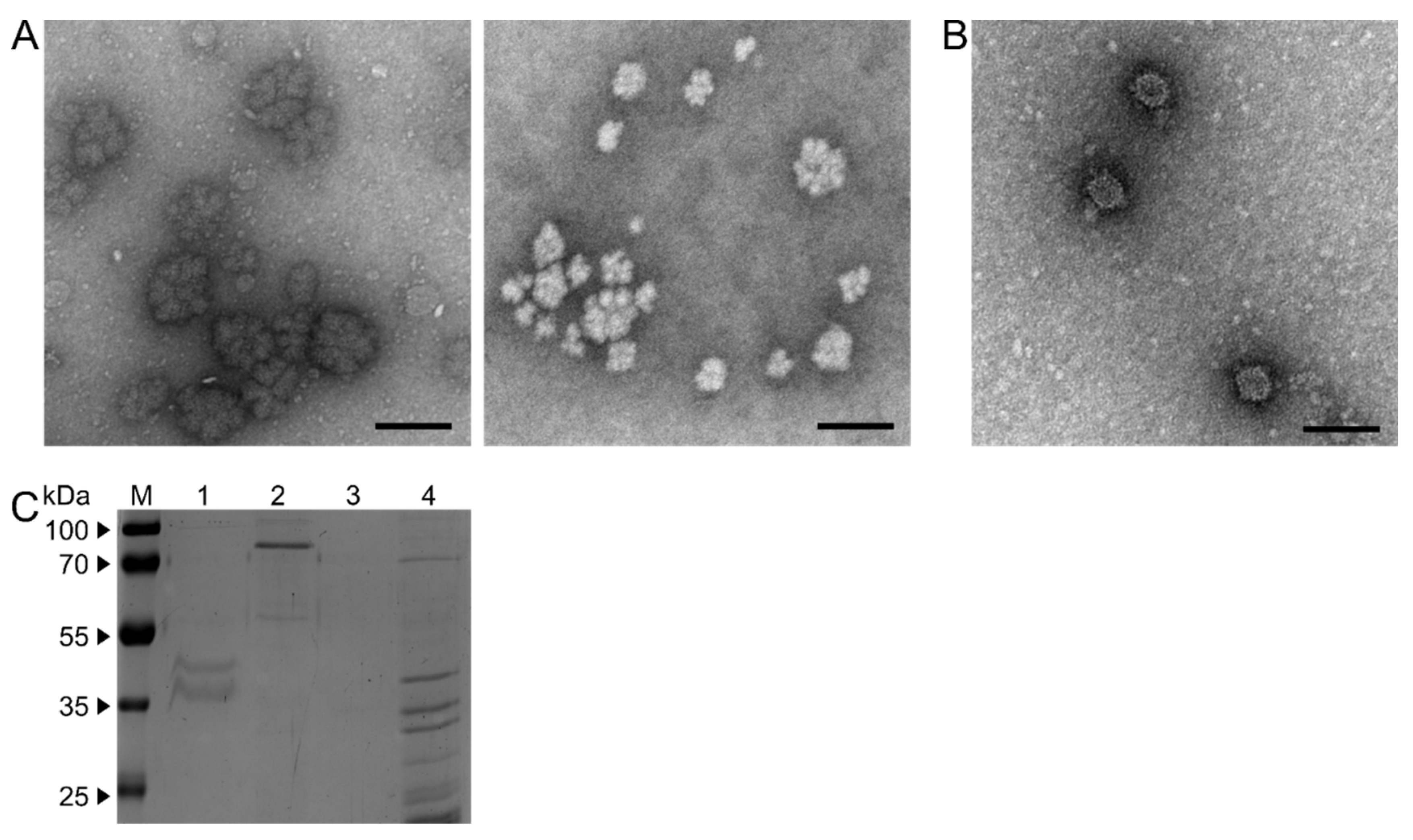
3.3. Single and Double Virus Infections Result in Different Ultra-Structures and dsRNA Patterns
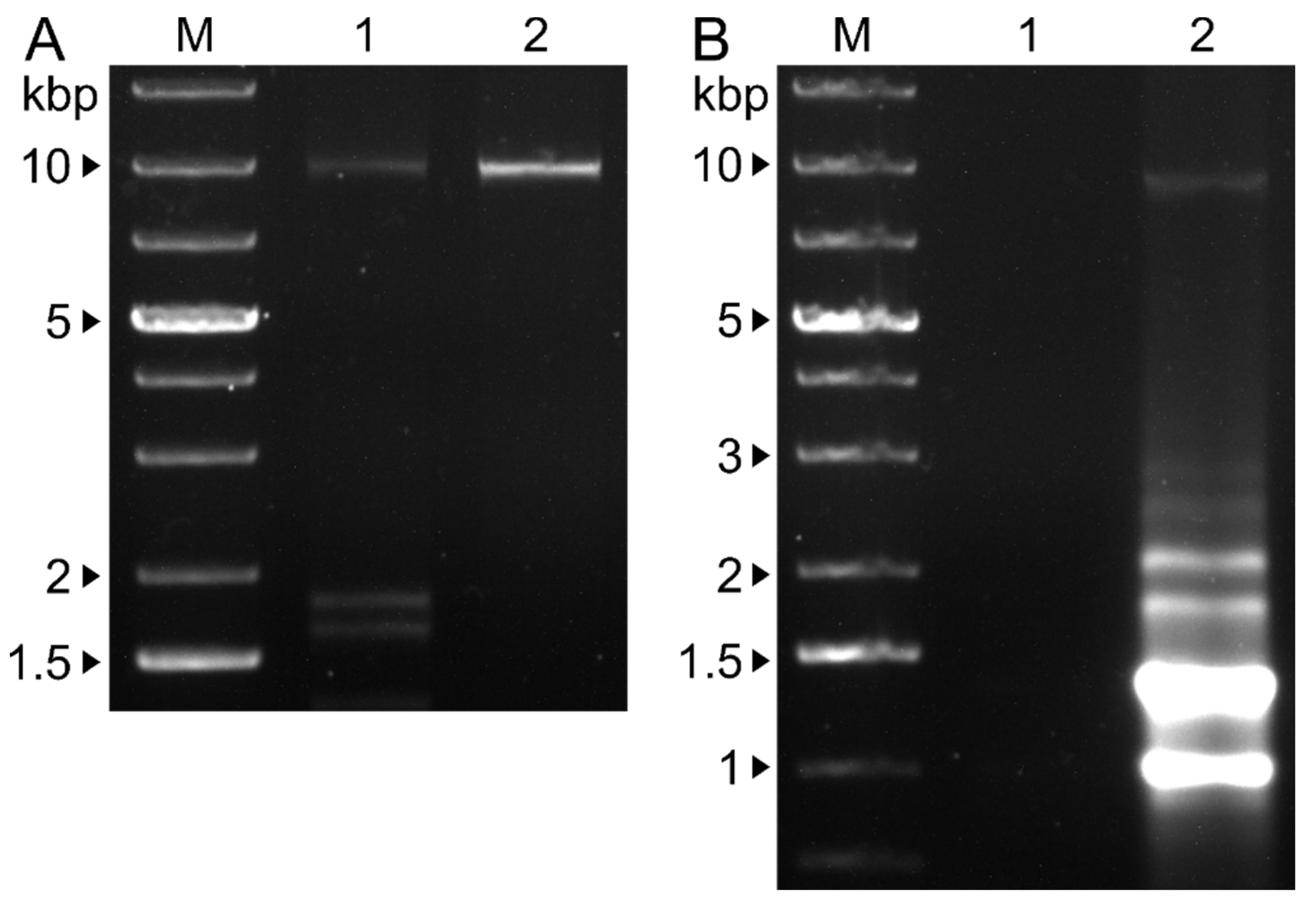
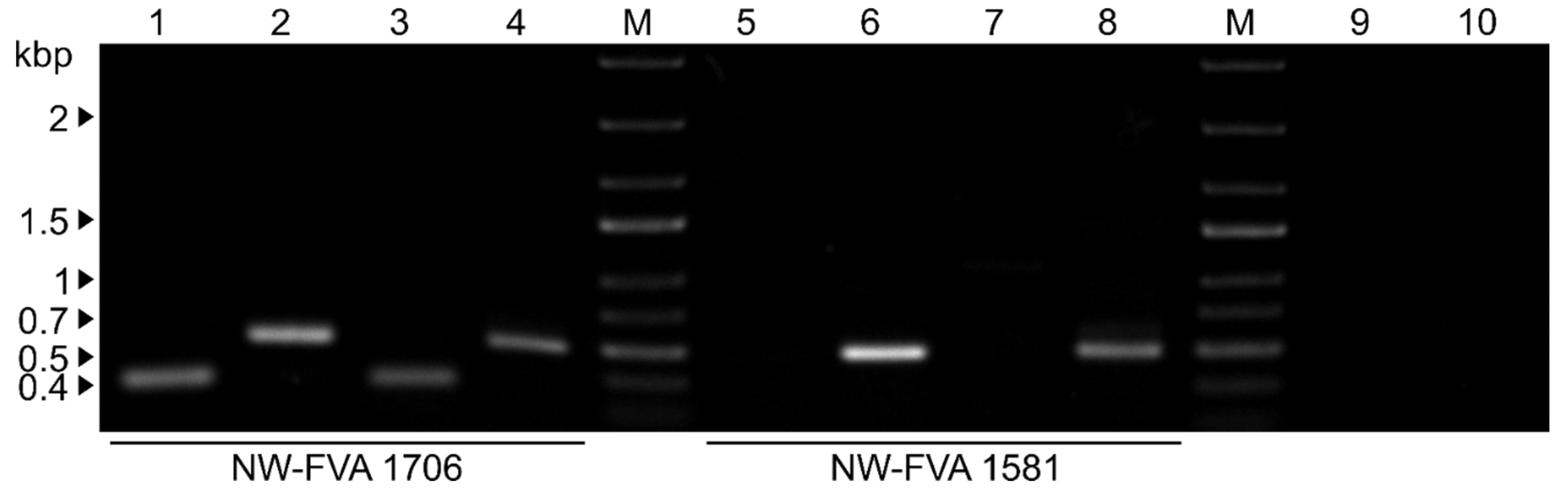
3.4. Single and Double Infections were Verified by RT-PCR from Mycelium and VLPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- George, J.-P.; Sanders, T.G.M.; Timmermann, V.; Potočić, N.; Lang, M. European-wide forest monitoring substantiate the neccessity for a joint conservation strategy to rescue European ash species (Fraxinus spp.). Sci. Rep. 2022, 12, 4764. [Google Scholar] [CrossRef] [PubMed]
- Linaldeddu, B.T.; Bottecchia, F.; Bregant, C.; Maddau, L.; Montecchio, L. Diplodia fraxini and Diplodia subglobosa: The Main Species Associated with Cankers and Dieback of Fraxinus excelsior in North-Eastern Italy. Forests 2020, 11, 883. [Google Scholar] [CrossRef]
- Przybyl, K. Fungi associated with necrotic apical parts of Fraxinus excelsior shoots. Forest Pathol 2002, 32, 387–394. [Google Scholar] [CrossRef]
- Bakys, R.; Vasaitis, R.; Barklund, P.; Thomsen, I.M.; Stenlid, J. Occurrence and pathogenicity of fungi in necrotic and non-symptomatic shoots of declining common ash (Fraxinus excelsior) in Sweden. Eur J Forest Res 2009, 128, 51–60. [Google Scholar] [CrossRef]
- Langer, G. Collar Rots in Forests of Northwest Germany Affected by Ash Dieback. Baltic Forestry 2017, 23, 4–19. [Google Scholar]
- Alves, A.; Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Phillips, A.J.L. The complex of Diplodia species associated with Fraxinus and some other woody hosts in Italy and Portugal. Fungal Diversity 2014, 67, 143–156. [Google Scholar] [CrossRef]
- Ayllón, M.A.; Vainio, E.J. Mycoviruses as a part of the global virome: Diversity, evolutionary links and lifestyle. Adv. Virus Res. 2023, 115, 1–86. [Google Scholar] [CrossRef]
- HOLLINGS, M. Viruses Associated with A Die-Back Disease of Cultivated Mushroom. Nature 1962, 196, 962–965. [Google Scholar] [CrossRef]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M.; Ictv, R.C. ICTV Virus Taxonomy Profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, J.; Yang, B.; Wang, Q.; Zhou, J.; Yu, W. Molecular Characterization of a Debilitation-Associated Partitivirus Infecting the Pathogenic Fungus Aspergillus flavus. Front. Microbiol. 2019, 10, 626. [Google Scholar] [CrossRef]
- Nerva, L.; Silvestri, A.; Ciuffo, M.; Palmano, S.; Varese, G.C.; Turina, M. Transmission of Penicillium aurantiogriseum partiti-like virus 1 to a new fungal host (Cryphonectria parasitica) confers higher resistance to salinity and reveals adaptive genomic changes. Environ. Microbiol. 2017, 19, 4480–4492. [Google Scholar] [CrossRef] [PubMed]
- Filippou, C.; Coutts, R.H.A.; Stevens, D.A.; Sabino, R.; Kotta-Loizou, I. Completion of the sequence of the Aspergillus fumigatus partitivirus 1 genome. Arch. Virol. 2020, 165, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wang, X.; Hua, H.; Cao, W.; Zhou, T.; Zhao, C.; Wu, X. Full genome sequence of a new three-segment gammapartitivirus from the phytopathogenic fungus Alternaria tenuissima on cotton in China. Arch. Virol. 2021, 166, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Zhang, H.; Qiu, D.; Guo, L. Two Novel Relative Double-Stranded RNA Mycoviruses Infecting Fusarium poae Strain SX63. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- Das, S.; Hisano, S.; Eusebio-Cope, A.; Kondo, H.; Suzuki, N. A Transfectable Fusagravirus from a Japanese Strain of Cryphonectria carpinicola with Spherical Particles. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Cornejo, C.; Hisano, S.; Bragança, H.; Suzuki, N.; Rigling, D. A New Double-Stranded RNA Mycovirus in Cryphonectria naterciae Is Able to Cross the Species Barrier and Is Deleterious to a New Host. J. Fungi (Basel) 2021, 7. [Google Scholar] [CrossRef]
- Sá Antunes, T.F.; Amaral, R.J.V.; Ventura, J.A.; Godinho, M.T.; Amaral, J.G.; Souza, F.O.; Zerbini, P.A.; Zerbini, F.M.; Fernandes, P.M.B. The dsRNA Virus Papaya Meleira Virus and an ssRNA Virus Are Associated with Papaya Sticky Disease. PLoS One 2016, 11, e0155240. [Google Scholar] [CrossRef]
- Spear, A.; Sisterson, M.S.; Yokomi, R.; Stenger, D.C. Plant-feeding insects harbor double-stranded RNA viruses encoding a novel proline-alanine rich protein and a polymerase distantly related to that of fungal viruses. Virology 2010, 404, 304–311. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, Y.; Zhao, H.; Ni, Y.; Liu, X.; Zhao, X.; Wang, G.; Xiao, X.; Liu, H. A novel double-stranded RNA mycovirus that infects Macrophomina phaseolina. Arch. Virol. 2019, 164, 2411–2416. [Google Scholar] [CrossRef]
- Brierley, I.; Pennell, S.; Gilbert, R.J.C. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007, 5, 598–610. [Google Scholar] [CrossRef]
- Dreher, T.W.; Miller, W.A. Translational control in positive strand RNA plant viruses. Virology 2006, 344, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.L.; Atkins, J.F.; Gesteland, R.F. Programmed ribosomal frameshifting: much ado about knotting! Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 14177–14179. [Google Scholar] [CrossRef] [PubMed]
- Kozlakidis, Z.; Hacker, C.V.; Bradley, D.; Jamal, A.; Phoon, X.; Webber, J.; Brasier, C.M.; Buck, K.W.; Coutts, R.H.A. Molecular characterisation of two novel double-stranded RNA elements from Phlebiopsis gigantea. Virus Genes 2009, 39, 132–136. [Google Scholar] [CrossRef]
- Liu, H.; Fu, Y.; Xie, J.; Cheng, J.; Ghabrial, S.A.; Li, G.; Peng, Y.; Yi, X.; Jiang, D. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol. Biol. 2012, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Xiang, J.; Zhang, M.; Fu, M.; Yang, Z.; Hong, N.; Wang, G. Characterization of a novel double-stranded RNA mycovirus conferring hypovirulence from the phytopathogenic fungus Botryosphaeria dothidea. Virology 2016, 493, 75–85. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, H.; Cao, J.; Yin, X.; Guo, Y.; Guo, L.; Wu, H.; Zhang, M. Characterization of a Novel Mycovirus from the Phytopathogenic Fungus Botryosphaeria dothidea. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Ding, Z.; Zhou, T.; Guo, L.-Y. Characterization of a novel strain of Botryosphaeria dothidea chrysovirus 1 from the apple white rot pathogen Botryosphaeria dothidea. Arch. Virol. 2017, 162, 2097–2102. [Google Scholar] [CrossRef]
- Khan, H.A.; Telengech, P.; Kondo, H.; Bhatti, M.F.; Suzuki, N. Mycovirus Hunting Revealed the Presence of Diverse Viruses in a Single Isolate of the Phytopathogenic Fungus Diplodia seriata From Pakistan. Front. Cell. Infect. Microbiol. 2022, 12, 913619. [Google Scholar] [CrossRef]
- Leach, J.; Lang, B.R.; Yoder, O.C. Methods for Selection of Mutants and In Vitro Culture of Cochliobolus heterostrophus. Microbiology 1982, 128, 1719–1729. [Google Scholar] [CrossRef]
- Damm, U.; Mostert, L.; Crous, P.W.; Fourie, P.H. Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia 2008, 20, 87–102. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal and RNA Genes for Phylogenetics. In PCR protocols: A guide to methods and applications, [Nachdr.]; Innis, M.A., Ed.; Acad. Press: San Diego, Calif., 1994; ISBN 9780123721808. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Lutz, T.; Petersen, J.M.; Yanık, C.; de Oliveira, C.; Heinze, C. Processing of the capsid proteins of the Betachrysovirus Fusarium graminearum virus-China 9 (FgV-ch9). Virology 2021, 563, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Pang, X.D.; Zhu, H.J.; Da Gao, B.; Huang, W.K.; Zhou, Q. Molecular Characterization of a Trisegmented Mycovirus from the Plant Pathogenic Fungus Colletotrichum gloeosporioides. Viruses 2016, 8. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004, 5, 113. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Higashiura, T.; Katoh, Y.; Urayama, S.-I.; Hayashi, O.; Aihara, M.; Fukuhara, T.; Fuji, S.-I.; Kobayashi, T.; Hase, S.; Arie, T.; et al. Magnaporthe oryzae chrysovirus 1 strain D confers growth inhibition to the host fungus and exhibits multiform viral structural proteins. Virology 2019, 535, 241–254. [Google Scholar] [CrossRef]
- Urayama, S.-I.; Sakoda, H.; Takai, R.; Katoh, Y.; Minh Le, T.; Fukuhara, T.; Arie, T.; Teraoka, T.; Moriyama, H. A dsRNA mycovirus, Magnaporthe oryzae chrysovirus 1-B, suppresses vegetative growth and development of the rice blast fungus. Virology 2014, 448, 265–273. [Google Scholar] [CrossRef]
- Kashif, M.; Hyder, R.; de Vega Perez, D.; Hantula, J.; Vainio, E.J. Heterobasidion wood decay fungi host diverse and globally distributed viruses related to Helicobasidium mompa partitivirus V70. Virus Res. 2015, 195, 119–123. [Google Scholar] [CrossRef]
- Vainio, E.J.; Korhonen, K.; Tuomivirta, T.T.; Hantula, J. A novel putative partitivirus of the saprotrophic fungus Heterobasidion ecrustosum infects pathogenic species of the Heterobasidion annosum complex. Fungal Biol. 2010, 114, 955–965. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Forgia, M.; Ciuffo, M.; Chitarra, W.; Chiapello, M.; Vallino, M.; Varese, G.C.; Turina, M. The mycovirome of a fungal collection from the sea cucumber Holothuria polii. Virus Res. 2019, 273, 197737. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.; Correia, A.; Phillips, A. Multi-gene genealogies and morphological data support Diplodia cupressi sp. nov., previously recognized as D. pinea f. sp. cupressi, as a distinct species. Fungal Diversity 2006, 23, 1–15. [Google Scholar]
- The Subviral Agents. Virus Taxonomy; Elsevier, 2012; pp. 1211–1219. [Google Scholar]
- Lutz, T.; Japić, E.; Bien, S.; Langer, G.J.; Heinze, C. Characterization of a novel alternavirus infecting the fungal pathogen Fusarium solani. Virus Res. 2022, 317, 198817. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Sun, X.; Cheng, J.; Fu, Y.; Liu, H.; Jiang, D.; Ghabrial, S.A.; Xie, J. Characterization of a Novel Megabirnavirus from Sclerotinia sclerotiorum Reveals Horizontal Gene Transfer from Single-Stranded RNA Virus to Double-Stranded RNA Virus. J. Virol. 2015, 89, 8567–8579. [Google Scholar] [CrossRef]
- Hillman, B.I.; Cai, G. The family narnaviridae: simplest of RNA viruses. Adv. Virus Res. 2013, 86, 149–176. [Google Scholar] [CrossRef]
- Wilkinson, M.; Yllanes, D.; Huber, G. Polysomally protected viruses. Phys. Biol. 2021, 18. [Google Scholar] [CrossRef]
- Matsuo, Y.; Ikeuchi, K.; Saeki, Y.; Iwasaki, S.; Schmidt, C.; Udagawa, T.; Sato, F.; Tsuchiya, H.; Becker, T.; Tanaka, K.; et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017, 8, 159. [Google Scholar] [CrossRef]
- Dinan, A.M.; Lukhovitskaya, N.I.; Olendraite, I.; Firth, A.E. A case for a negative-strand coding sequence in a group of positive-sense RNA viruses. Virus Evol. 2020, 6, veaa007. [Google Scholar] [CrossRef]
- Sato, Y.; Das, S.; Velasco, L.; Turina, M.; Osaki, H.; Kotta-Loizou, I.; Coutts, R.H.A.; Kondo, H.; Sabanadzovic, S.; Suzuki, N.; et al. ICTV Virus Taxonomy Profile: Yadokariviridae 2023. J. Gen. Virol. 2023, 104. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Kim, D.-H. Co-infection of a novel fusagravirus and a partitivirus in a Korean isolate of Rosellinia necatrix KACC40168. Virus Genes 2021, 57, 121–126. [Google Scholar] [CrossRef] [PubMed]
| Taxon | Isolate | Acc. ID ITS | Acc. ID TEF1-α | Acc. ID TUB |
|---|---|---|---|---|
| D. fraxini | NW-FVA 1581 | OR050980 | OR079892 | OR079888 |
| D. fraxini | NW-FVA 1706 | OR050981 | OR079893 | OR079889 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





