1. Introduction
Colon cancer (CC) is the third-most prevalent type of cancer worldwide, with more than 1.1 million and 0.57 million new cases and cancer-related deaths, respectively, arising in 185 countries annually (1). Among the wide range of risk factors, the biological features of CC and tumour heterogeneity largely explain the different clinical outcomes. Advances in molecular pathology have increased the treatment options with anti-EGFR monoclonal antibodies for metastatic CC patients (stage IV) without RAS mutations (2). Nevertheless, the choice of treatment for the patients with loco-regional CC (stages II-III) is based on histopathological and clinical factors, considering chemotherapy in selected patients with bad prognostic factors (3). For this reason, there is an urgent need to refine these prognostic factors in order to help clinicians stratify stage II-III patients more effectively.
CC is categorised into four Consensus Molecular Subtypes (CMSs): CMS1 (MSI immune subtype), CMS2 (canonical subtype), CMS3 (metabolic subtype) and CMS4 (mesenchymal subtype) (4, 5). This classification is based on the differential expression, detected by microarrays, of crucial genes in cancer onset and progression. However, logistic and economic constraints render the use of DNA microarrays for routine classification unfeasible in most Pathology Departments. Nevertheless, an advance in this field was achieved using a new approach based on a surrogate immunohistochemistry (IHC) panel for application in routine clinical practice, using already available immunohistochemistry (IHC) assays (6). This panel comprises four immunohistochemical cases involved in crucial cell mechanisms: caudal-related homeobox 2 (CDX2), FERM domain-containing 6 (FRMD6), 5-hydroxytryptamine (Serotonin) receptor 2B, G protein-coupled (HTR2B) and zinc finger E-box binding homeobox 1 (ZEB1).
CDX2 is a homeobox transcription factor expressed in the nuclei of intestinal epithelial cells that plays an important role in the development and maintenance of the intestinal tract and is used as an IHC marker to distinguish between adenocarcinomas of colorectal origin and those arising in other organs (7). It inhibits Wnt signalling by inhibiting the activity of β-catenin/TCF-4 and consequently the epithelial–mesenchymal transition (EMT) (8); CDX2 hypermethylation is frequent in late stages of lung cancer (9) and plays an important role in the activation of lung cancer cell proliferation by suppressing Wnt signalling (10). This gene is also known to be hypermethylated in colorectal cancer (11), although few attempts have been made to determine its clinical value in colon cancer (11, 12).
The FRMD6 gene is also altered in cancer but its causes have not been thoroughly studied. FRMD6 is a protein that is crucial to the maintenance of the cell cytoarchitecture and that can bind to actin filaments and nectins, thereby regulating actomyosin contractility in the cytoskeleton and epithelial cell–cell junction complexes in order to maintain epithelial structure (13). FRMD6 has been identified as an upstream regulator of the Hippo signalling cascade regulating cell-contact inhibition, apoptosis, proliferation and tissue regeneration. Furthermore, Hippo pathway components are known to be deregulated in colon (14) and other (15) cancers.
HTR2B belongs to the G-protein coupled receptor 1 family. When HTR2B binds its ligand serotonin, it activates the G proteins GNAQ, GNA11 and GNA13, and participates in development, and cell proliferation and survival through the activation of a small number of signal-transduction pathways such as the phospholipase C, Janus kinase/signal transducer and activator of transcription (JAK/STAT) proteins, receptor tyrosine kinase (RTK)/phosphatidylinositol-4,5-bisphosphate-3-kinase (PI3K)/extracellular signal-regulated kinase (ERK)/mammalian target of rapamycin (mTOR) and RAF/mitogen-activated protein kinase kinase (MEK)/ERK pathways. The gene encoding the HTR2B receptor has been described as an oncogene in hepatocellular and prostate cancers and in uveal melanoma (16), and as a tumour suppressor gene in ovarian cancers.
ZEB1 belongs to the EMT-transcription factor (EMTIF) family, which is involved in promoting EMT in cancer, including CC (17); its expression is inhibited by miR200, which is activated by suppressor gene P53 (18); ZEB1 is known to be involved in regulating key factors in malignant cells at the invasive front of carcinomas, conferring cancer cells with a proinvasive and stem-like phenotype, as well as leading to a worse clinical prognosis in several human cancers (19).
Aberrant DNA methylation is the best-known epigenetic modification in human disease and is involved in regulating the expression of a great variety of genes that are critical in cancer (20). Despite the clinical utility of methylation in the early detection, prediction of prognosis and response to treatment in diverse cancers, as described for the response to temozolomide in glioma patients with MGMT hypermethylation (21), no aberrantly methylated genes with prognostic value have been exploited in clinical practice to treat CC patients (22). Therefore, and given the utility of CMS classification and the few studies of alterations in the subrogate genes, we analyse the presence of aberrant methylation in those genes and examine its clinical value in stage II-III CC patients, who are characterized by the lack of prognostic biomarkers useful in their clinical follow-up.
2. Materials and methods
2.1. Group of study
The group of patients studied consisted of 144 patients diagnosed with stage II (80 cases, 55.6%) and stage III (64 cases, 44.4%) CC between 2012 and 2013 in the Pathology Department of the Hospital Universitario de Navarra (Navarra Public Health System). All patients were operated upon and tumours were staged according to their size, lymph node involvement and distant metastasis, following the most recent recommendations (23). None of the patients had received radiation or chemotherapy before surgery. The study was approved by the Regional Clinical Research Ethics Committee (CEIC) (Pyto2017/51 Cod. MOL_CRC, 15 May 2018). The diagnosis of these tumours was confirmed following microscopic inspection by a certified pathologist with expertise and specialism in colon pathology (M.G.D.).
Tumours were classified by the subrogate IHC panel into CMS1 (MSI Immune), CMS2/CMS3 (Canonical/Metabolic) and CMS4 (Mesenchymal) in 18 (12.5%), 117 (81.3%) and 9 patients (6.3%), respectively, based on previously established criteria [
3].
Pathological and clinical characteristics are summarized in Table 1. Adjuvant chemotherapy was performed in 53 patients (37.9%), preferentially in stage III patients (81.1%) compared with stage II patients (18.9%), according to standard procedures. Follow-up included a physical and clinical examination every 4 months. During follow-up, 25 (17.4%) patients died of the disease and 26 (18.1%) died of other causes.
2.2. Immunohistochemical study
Three-μm sections of TMA blocks harbouring four tumour-carrying cores were placed on slides and then deparaffinized, hydrated and treated to block endogenous peroxidase activity using Vision Biosystems Bond-Max (Leica) and Bench-Mark XT Ventana (Roche) automatic immunostaining apparatus, as previously described (24). These slides were incubated with the appropriate primary antibodies against mismatch repair proteins-MMR (MLH1, MSH2, MSH6) and against proteins of the subrogate panel (CDX2, FRMD6, HTR2B and ZEB1) under the conditions summarised in Supplementary Table 1. A minimum of 500 tumour cells per tumour were counted by two independent pathologists with appropriate expertise. To evaluate the immunostaining pattern of the four proteins, we used the online test for CCR classification (
https://crcclassifier.shinyapps.io/appTesting/ ). Expression of nuclear CDX2 and cytoplasmic FRMD6 was evaluated by categorising counts of the number of positive tumour cells into three categories (null/low number of positive cells: no expression or expression in fewer than 25% of the cells; intermediate: expression in 26-55% of cells; high: expression in 56-100% of cells) and the intensity of the expression (low, intermediate and high). They were considered separate variables for the purpose of comparison. Diffuse CDX2 expression present in normal mucosa was used as an internal control and reference for the intensity of expression. Cytoplasmic HTR2B expression was evaluated in terms of its intensity, as in the case of CDX2/FRMD6. Nuclear ZEB1 was scored as its presence or absence. ZEB1 expression was also measured by IHC in complete sections of two groups of patients. The first group (16 cases) included eight completely unmethylated (0%) cases and eight highly methylated cases (>50% methylation). The second group consisted of undifferentiated high-grade tumours (tumours with <50% of glandular differentiation). PDL-1 and P53 proteins were also evaluated in TMAs, as previously described (25, 26).
2.3. DNA extraction from cell lines and tissue
DNA was extracted from 0.5 × 106 cells in the case of cell lines, while for tumoral/normal cases, it was obtained by QIAamp DNA FFPE Tissue kit (Qiagen, Hilden, Germany) from a representative area with >70% of tumoral cells in 5-μm-thick formalin-fixed, paraffin-embedded (FFPE) sections selected by the pathologist. DNA concentration was measured using an Invitrogen™ Qubit™ 3 Fluorometer (Thermo Scientific, USA).
2.4. Pyrosequencing of subrogate genes
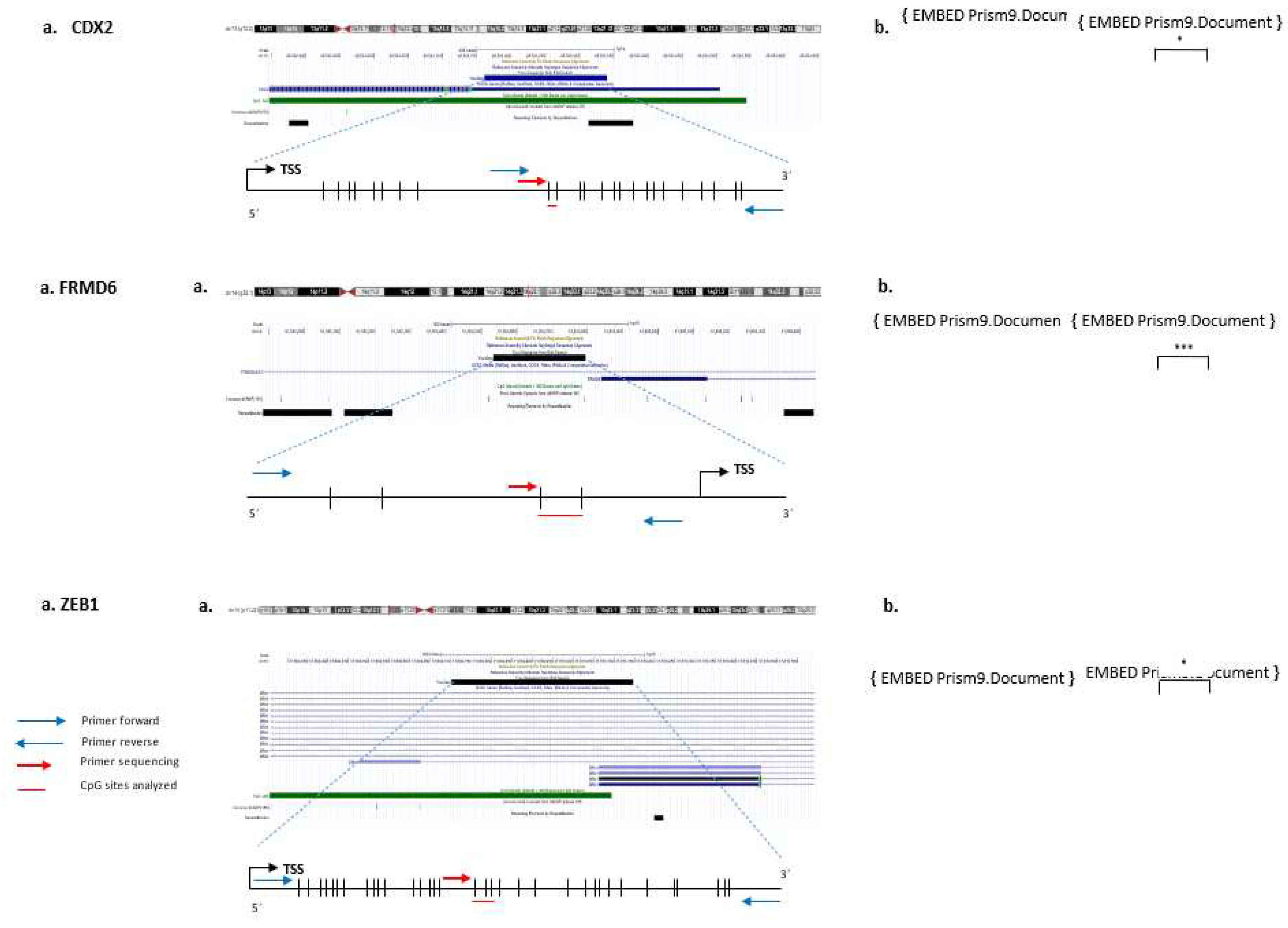
DNA methylation levels for CDX2, FRMD6 and ZEB1 genes were analysed in 144 cases and in 40 paired paraffin tumour-normal cases by bisulfite pyrosequencing. The sets of primers for PCR amplification of analysed CpGs and sequencing for each gene were designed using the specific software PyroMark assay design (version 2.0.01.15) (
Figure 1, Supplementary Table 2). It was not possible to design primers for HTR2B due to the high CpG density.
Bisulfite modification of DNA was performed with an EZ DNA methylation-gold kit (Zymo Research), following the manufacturer’s instructions. PCR amplification, pyrosequencing and methylation quantification were performed using PyroMark Q96 reagents in a PyroMark Q96 ID (Qiagen).
The survival of patients bearing these genes was analysed (see below) to test the clinical value of the aberrant gene methylation of subrogate genes.
2.5. In vitro studies
Additional in vitro and molecular studies were considered to test the biological value of ZEB1 hypermethylation as the most relevant finding in this report. A panel of seven cell lines derived from colon cancer (HCT116, HT29, LoVo, RKO, SW480, SW837 and T84) was used to study ZEB1 (kindly donated by Dr. Arozarena, Navarrabiomed, Spain). All these cell lines were grown in DMEM, supplemented with 10% foetal bovine serum and 1.0% penicillin/streptomycin (all from Life Technologies, Carlsbad, CA, USA) at 37°C in a humidified atmosphere with 5% CO2. The basal level of ZEB1 methylation was assessed in all cell lines.
Two highly methylated cell lines (HCT116, HT29) and one demethylated (RKO) cell line were treated at low passage with the demethylating agent 5-aza-2′-deoxycytidine (AZA) and the histone deacetylase inhibitor trichostatin A (TSA) (both from Sigma-Aldrich, St Louis, MO, USA). Briefly, cells were seeded at a density of 1x105 cells/ml, allowed to attach overnight, and treated with 4 μM AZA for 72 h added freshly every 24 h, 300 nM TSA for 24 h, or the combination of the two drugs for the final 24 h, using PBS as a vehicle control.
2.6. RNA extraction and quantitative reverse-transcription PCR (qRT-PCR)
qRT-PCR was performed to check the restoration of ZEB1 expression in control and AZA+TSA-treated CC-derived cell lines. This analysis was also performed in 34 paired paraffin tumour-normal cases (17 methylated and 17 unmethylated cases) to check the differential expression of this marker in tissue.
To this end, total RNA was extracted and purified using the RecoverAll kit (ThermoFisher, Waltham, MA, USA) following the manufacturer’s instructions. 500 ng of total RNA were retrotranscribed using a PrimeScript™ RT Reagent Kit (TaKaRa, Otsu, Japan) at 37°C for 15 min and 85°C for 5 s. 1 μl of the resulting cDNA was placed in a 96-well plate with 0.5 μl TaqMan probes (ZEB1: Hs.PT.58.39178574; ZEB1-antisense: Hs.PT. Iñaki from IDT, Coralville, Iowa) and 19 μl of mix were included in the Premix ExTaq™ kit (TaKaRa, Otsu, Japan). PCR amplification was performed in triplicate using the Quant Studio 12K Flex (Life Technologies, Carlsbad, CA, USA) under thermal cycler conditions of 95°C for 30 s and 40 cycles at 95°C for 5 s and 60°C for 34 s. Cycle threshold (Ct) values were calculated using Quant Studio software (Life Technologies, Carlsbad, CA, USA), and the relative quantification (RQ) was calculated by the ΔCt method (RQ = 2−ΔCt). The reference housekeeping pseudogene-free ribosomal gene (18S rRNA: Hs.PT.39a.22214856.g, IDT), which shows little variation in basal expression in colon cancer (27, 28), was employed as a normalization standard for relative PCR quantification.
2.7. ZEB1 silencing in colon cancer cell lines
To check the expression of ZEB1 gene in CC, its expression was silenced in ZEB1-positive cell lines (RKO, SW620 and T84 cells). For shRNA construction, three sequences targeting ZEB1 (shZEB1_1, shZEB1_2, shZEB1_3), and one scramble sequence were used (Supplementary Table 3). After inserting shRNAs into the pHIV1-SIREN-PuroR plasmid (kindly provided by Dr. Escors, Navarrabiomed), BamHI and EcoRI restriction enzymes (Life Technologies, Carlsbad, CA, USA) and T4 DNA ligase enzyme (New England Biolabs, Ipswich, MA, USA), respectively, were used to digest and ligate the construction. XL1-Blue Competent cells were then transformed with these three shRNA constructions. Plasmids were purified using the Qiagen Plasmid Midi kit (Qiagen, Hilden, Germany) and sequenced to check the ligation. Since the plasmid contained the puromycin-resistance gene for mammalian cell selection, cell sensitivity to this antibiotic (ThermoFisher) was tested, and a concentration of 1 g/mL was chosen as optimal from a range of possibilities. 5x104 cells were seeded in six-well plates, allowed to attach overnight and then stably transfected with 1.2 g of the plasmid of interest and 1:3 (v/v) FuGene HD (Promega, Madison, WI, USA) containing scramble, shZEB1_1, shZEB1_2 and shZEB1_3 in 60 µl of DMEM (Lonza Biologics, Basel, Switzerland, Spain), as previously described (29).
2.8. Statistical analysis
Associations between gene hypermethylation and hypomethylation, expression, pathological and clinical variables of this retrospective study were assessed with the chi-square or Fisher’s exact test. Disease-free survival (DFS) and overall survival (OS) were analysed in all CC patients. Survival curves were calculated using the Kaplan–Meier method and compared by the log-rank test. Cox proportional hazard regression models were used for univariate analyses. The proportional hazard ratio and 95% confidence interval (95% CI) were calculated for each factor. Hazard risk was adjusted for tumour stage and patient age. Statistical significance was concluded for values of p<0.05 in all cases.
3. Results
3.1. Study of aberrant methylation in subrogate genes
CDX2, FRMD6 and ZEB1 methylation could not be analysed in 11.1%, 22.2% and 10.4% of the tumours, respectively, probably due to the effect of formalin fixation on the tissue (30). Median value was considered the cut-off for distinguishing statistically between the unmethylated and methylated
status of each of the CpG sites. In the evaluable cases, aberrant methylation was found for the three genes, with CDX2 and ZEB1 being hypermethylated in 32.8% and 32.6% of the cases, respectively, and FRMD6 being hypomethylated in 50.9% of the patients. Aberrant CDX2, FRMD6 and ZEB1 hypermethylation or hypomethylation was more frequent in tumoral than in normal tissue (p=0.04, p=0.0004, p=0.0024, respectively) (
Figure 1a, 1b, 1c).
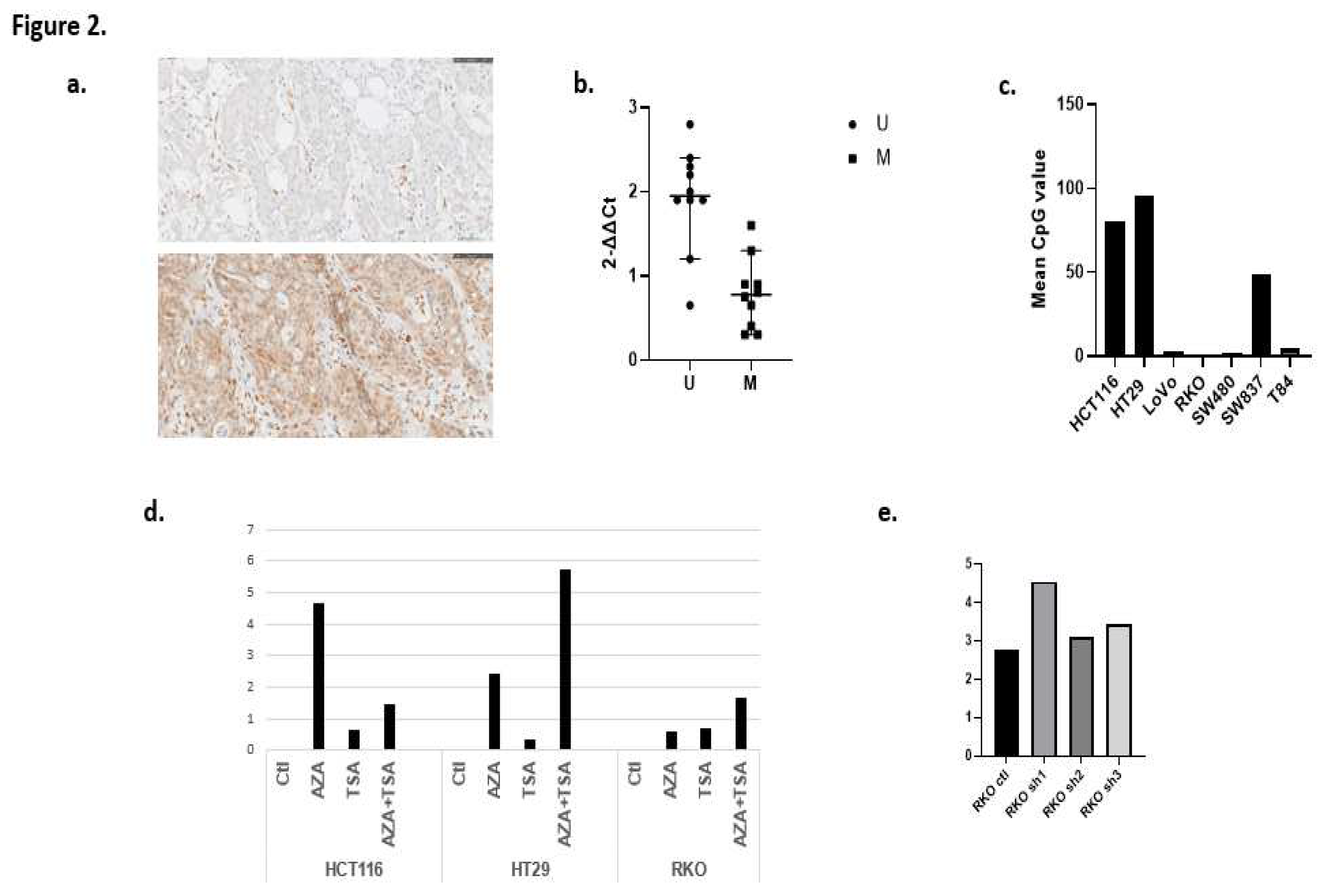
HCT116, HT29 and SW837 cells were clearly methylated for ZEB1 (80.5%, 96.0% and 49.0%, respectively). Conversely, ZEB1 was completely unmethylated in RKO cells (0.0%) and scarcely methylated in LoVo and SW480 cells (3.0% and 2.0%, respectively) (
Figure 2a).
3.2. Association between pathological and molecular parameters in CC
CC cases with absent/low CDX2 immune expression and a low percentage of positive cells (<25.0%) were associated with CDX2 hypermethylation (p=0.044 and p=0.048, respectively) and were preferentially of mesenchymal type and hMLH1/hPMS2 defective tumours (p<0.005). Absent/low expression is very frequent in stage III, less differentiated and right colon-sided CC tumours (p=0.024, p=0.006 and p=0.093, respectively).
FRMD6 hypomethylation was not associated with any of the variables included in the study, except for weaker PD-L1 expression (p=0.012). It is remarkable that four of the five cases (80%) of mesenchymal type were hypomethylated for this gene compared with 49.5% of the epithelial cases (p=0.182).
ZEB1 expression was well correlated with the findings obtained in TMAs, and produced no discordant results. Lymphocytes and mesenchymal cells were used as internal positive controls of expression (
Figure 2b). There were no cases with extensive positive ZEB1 expression, except isolated cell groups (<5-10% of the slide) in more differentiated tumoral areas in contrast to no/lower levels of expression in undifferentiated areas. One of three cases with a signet-ring phenotype characterized by its bad prognosis showed a low level of ZEB1 expression. ZEB1 hypermethylation was associated with focal ZEB1 expression (p=0.028). Additionally, ZEB1 expression detected by qPCR revealed that normalized ZEB1 expression of unmethylated cases was higher than in methylated cases (p=0.035) (
Figure 2c). Finally, ZEB1 hypermethylation was more frequent in the CMS1 subtype (p=0.072), with a clear association of this epigenetic alteration with the pathological (null) expression of hMLH1 and hPMS2 proteins (p=0.040 and p=0.022, respectively).
3.3. In vitro study
The highest levels of ZEB1 expression were detected mainly in AZA and AZA+TSA-treated HCT116 HT-29 cells, respectively (
Figure 2d).
The transfected RKO cells showed a significantly higher level of ZEB1 expression, mainly with shRNA_ZEB1 in comparison with shRNA_2 and shRNA_3, with the greatest difference compared with the control (
Figure 2e). SW620 and T84 cells did not re-express ZEB1, probably because ZEB1 expression is regulated by a different mechanism. It is notable that it was not possible to select transfected cells with these shRNAs because cells died as a consequence of ZEB1 silencing.
3.4. Survival analysis
The median follow-up for DFS and OS was 5.3 and 5.48 years, respectively. The univariate analyses confirmed that factors such as age, tumour size, stage, lymph node involvement, vessel invasion and perineural invasion were associated with worse prognosis as indicated by DFS (p<0.001, p<0.001, p=0.04, p=0.017, p=0.005 and p=0.034, respectively) and OS (p<0.001, p<0.001, p=0.042, p=0.03 and p=0.22, respectively). It is of particular note that CMS subtypes had differential prognoses (Supplementary Figure 1), that of CMS4 being the worst (p=0.014). CC cases with null/low levels of expression and a low percentage of positive cells were not of prognostic significance, as is the case with FRMD6 expression.
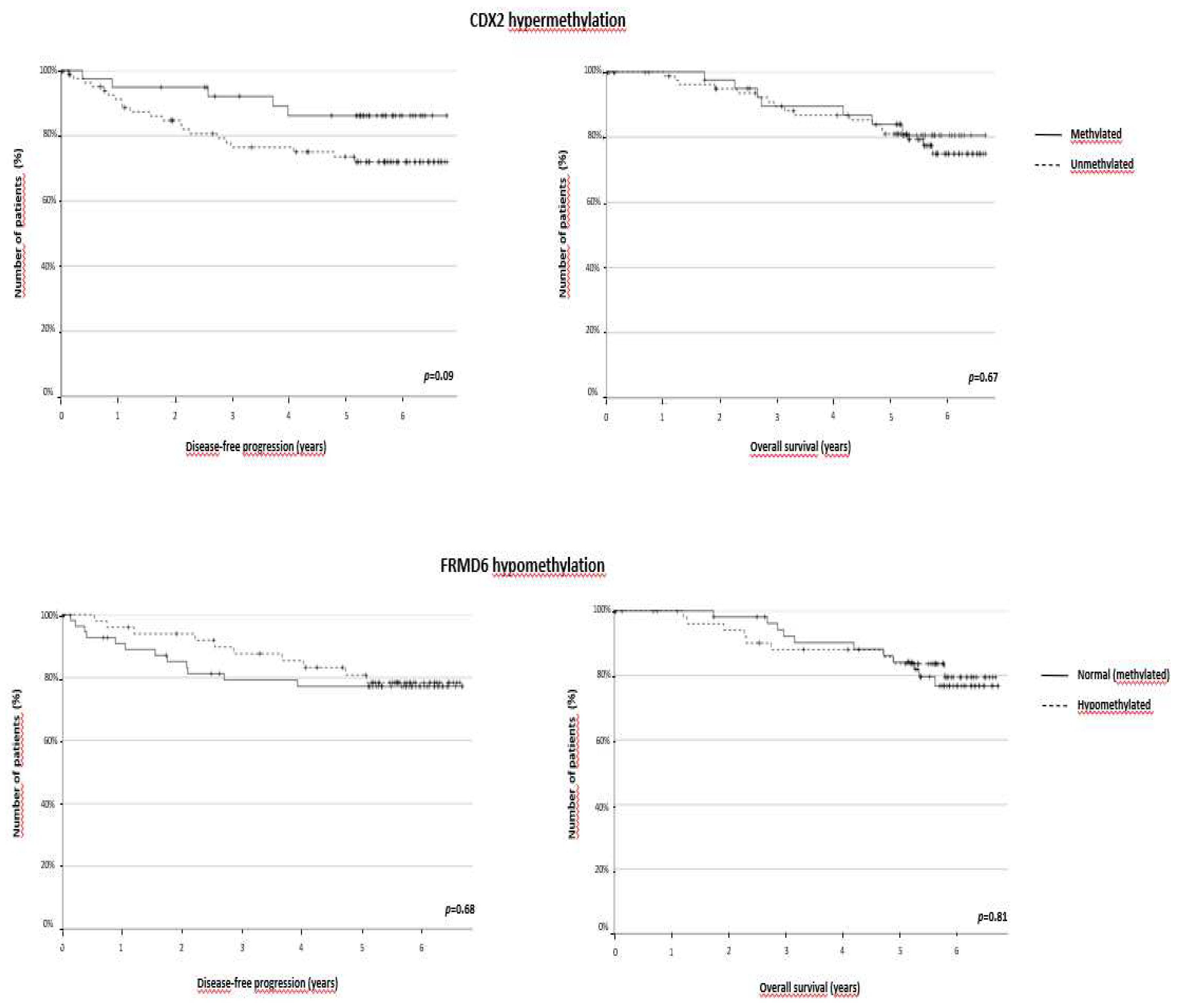
It is very striking that ZEB1 hypermethylation was clearly associated with longer DFS and OS (p=0.017 and p=0.007, respectively), whereas the other genes were not of prognostic significance (
Figure 3). This prognostic role was maintained in the case of OS (p=0.030) considering the median instead of the mean estimates, although DFS was also longer for patients with methylated ZEB1. Therefore, the independent impact of ZEB1 hypermethylation on DFS and OS, regardless of significant clinicopathological variables, was tested in a Cox regression model. ZEB1 hypermethylation was still significantly associated with longer DFS (p=0.015) and OS (p=0.006), irrespective of age, tumour size, stage, and blood vessel and perineural invasion (Table 2). The prognostic role of this alteration was maintained in the CM2/3 subtypes (DFS: p=0.023; OS: p=0.035) (Supplementary Figure 2).
4. Discussion
CC heterogeneity highlights the importance of undertaking studies to find new molecular markers. MMR proteins currently help distinguish between MSI tumours and MSS, with better and prognosis and response to treatment for the first group (31). The incorporation of the subrogate IHC panel to classify CC into CMS to detect tumours of mesenchymal type characterized by their bad prognosis is an easy task (32). Nevertheless, the subrogate panel is clearly incapable of distinguishing CMS2 from CMS3 cases, which is a problem, given that CMS2/CMS3 is the most numerous group (81.0% of our series). In this context, the discovery of key molecular alterations would allow new, clinically useful biomarkers to be proposed for the management of CC patients.
The detection of epigenetic alterations such as hypermethylation and hypomethylation of regulatory regions could explain the patterns of expression in tumours and may be clinically significant, as we described previously in brain tumours, and breast and cervical cancer (33-35). It is worth noting that there have been no studies of epigenetic alterations of the FRMD6 and ZEB1 genes, which encode the proteins included in the panel. Even less is known about the clinical role of these alterations in CC.
In the group of patients studied here, loss or absence of CDX2 expression was much more frequent in the CMS1 subtype, although it was not of prognostic significance (36), consistent with the findings of Baba et al. in sporadic CC (7). The association between CDX2 hypermethylation and lower levels of CDX2 expression is consistent with the first description of CDX2 hypermethylation in CC, which was detected by the less-informative methylation-specific PCR assay (37).
The clinical importance of the lack of CDX2 expression, measured as the level of mRNA or protein, has already been described in two reports (32, 38). Less information is available about the clinical role of CDX2 hypermethylation (12, 39). In the group studied here, CDX2 expression was not correlated with clinicopathological variables except for a non-significant tendency for patients with methylated tumours (mainly in CMS2/3 patients) to display longer disease-free survival. This contrasts with the study by Jiang et al., which reported that CDX2 hypermethylation was a bad prognostic factor (39). It is worth noting that all the stages were included in the previous report, whereas our present study examined only stage II/III tumours.
Very little is known about the FRMD6 gene, expect that it is crucial to the Hippo pathway and therefore also to the EMT pathway. There is no agreement about what suppressor or oncogenic role FRMD6 alteration might play in cancer. In keeping with its suppressor role, FRMD6 mutations dysregulate the Hippo pathway by translocating the YAP/TAZ complex into the nucleus and thus activating the expression of genes affecting key EMT genes (ZEB1, Snail/Slug, Twist) (40). In line with these findings, low levels of FRMD6 expression are associated with worse prognosis in prostate cancer (41), and inhibition of the gene is directly related to progression of hepatocellular carcinoma (42). Nevertheless, FRMD6 expression also contributes to cancer progression by activating the mTOR signalling pathway, similar to what occurs in lung cancer (43).
In the case of CC, FRMD6 is more frequently expressed in colorectal cancer subtype 3 (CCS3) equivalent to CMS4. This expression is associated with worse prognosis (44). FRMD6 was also reported to be upregulated in the poor survival CRC group by unknown causes (45) and is also one of the panel of five key biomarkers of poor prognosis expressed in gastric cancer (46). The mechanism underlying FRMD6 upregulation has not yet been determined. To our knowledge, our study is the first to report that FRMD6 is highly methylated in normal colon tissue and hypomethylated in tumours. Overexpression could be mediated, at least in part, by DNA hypomethylation; in our study group, the lack of association between FRMD6 expression and hypomethylation could be related to components of post-transcriptional regulation of FRMD6 expression, such as phosphorylation events, or other epigenetic modifications (e.g., DNA methylation, histone acetylation, miRNA expression) (47).
In our study, neither FRMD6 hypomethylation nor IHC expression was associated with any clinicopathological variable, except for a clear association with a lower level of PD-L1 expression, which has not been reported elsewhere. Few studies have addressed the involvement of FMRD6 protein in the immune response; it is thought to be a neoantigen directly associated with the expression of HLA A, and B and T cell activation characteristic of immune activated basal-like breast cancers with favourable prognosis (48).
HTR2B was recently described as being a suppressor gene whose mutations are related to prognosis of squamous lung cancer (49) and metastasis in uveal melanoma (50). Conversely, it has been described as an oncogene in CC whose aberrant activation promotes the TGFbeta pathway and metastasis (51). There is no information about the presence of epigenetic alterations in this gene; it was not possible to study this here because the targeted CpG-rich region is very dense and the design of primers without CpGs in their sequence cannot be implemented. This is a frequent drawback in the analysis of FFPE samples, in which the starting material of study is so highly fragmented that the optimal amplicon length is restricted, thereby further limiting the options for primer placement (52).
ZEB1 is a crucial transcriptional repressor of the transformation from epithelial phenotype to mesenchymal phenotype that promotes invasion, intravasation and dissemination to distant sites (53). ZEB1 is upregulated in colorectal cancer, alongside other types of cancer such as those of the bladder, breast, stomach, pancreas and prostrate, and endometrial adenocarcinoma, oesophageal squamous cell carcinoma, head and neck squamous cell carcinoma, hepatocarcinoma, leiomyosarcoma and lung carcinoma (54-56). Consistent with the protective role of this alteration, it was also identified as one of seven candidates associated with better preoperative chemoradiation therapy responses in rectal cancer (57).
In the group studied here, ZEB1 hypermethylation, which was associated with a lower level of ZEB1 expression, was clearly associated with better prognosis, as indicated by DFS and OS, independently of other significant variables. This important finding also pertains to the CM2/3 subgroup, which could enable clinicians to stratify this heterogeneous group of patients into groups with different degrees of risk of relapse or of death. Additionally, this epigenetic alteration is associated with the CMS1 subtype, characterized by its high degree of immune infiltration and better prognosis confirmed in our sample. It is notable that the only patient with the CMS1 subtype who died displayed an unmethylated promoter. The influence of ZEB1 expression in immune infiltration has been well studied, with reports on the inhibition of immune response exerted by this protein in melanoma, triple-negative breast cancer and lung cancer (58-60). The role of ZEB1 hypermethylation in this context should therefore be investigated further.
In summary, to our knowledge this is the first report of aberrant methylation of the subrogate genes CDX2, FRMD6 and ZEB1 being used for CMS classification. The role of ZEB1 hypermethylation is crucial for the better prognosis of colon cancer patients, as represented by disease-free survival and overall survival.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary Figure 1. Kaplan–Meier plots stratified by CMS subtype. Curves for disease-free survival (left) and overall survival (right). Supplementary Figure 2. Kaplan–Meier plots stratified by ZEB1 promoter hypermethylation presence or absence in CMS2/3 patients. Curves for disease-free survival (left) and overall survival (right).
Author Contributions
Conceptualization, I.F.DL.R., D.G.S. and M.G.D.; methodology, I.F.DL.R., I.M.S., A.F.F., M.F., L.A., B.F.M., P.A.S., J.S., D.G.S. and M.G.D.; validation, I.F.DL.R.,D.G.S., M.G.D. and A.C.; formal analysis, D.G.S.; investigation, I.F.DL.R., D.G.S. and M.G.D.; resources, D.G.S. and M.G.D.; data curation, I.F.DL.R., D.G.S., M.G.D. and J.S.; writing—original draft preparation, D.G.S. and I.F.DL.R.; supervision, D.G.S.; project administration, D.G.S.; funding acquisition, D.G.S. and M.G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by European Union Regional Development Fund (17-20, RefBioII, Trans-Pyrenean cooperation network program 2016–2018, INTERREG-POCTEFA) https://ec.europa.eu/regional_policy/en/funding/erdf/.
Institutional Review Board Statement
The study was approved by the Regional Clinical Research Ethics Committee (CEIC) Pyto2017/51 Cod. MOL_CRC, 15/05/2018.
Informed Consent Statement
Patient consent was waived due to usage of stored tumor samples for research purposes in compliance with the current Spanish and European Union legislation (resolution 1387/2017 (08/11) and resolution 193/2018 (06/03) of the Navarra Health Service—Osasunbidea).
Acknowledgments
We are grateful to all the patients and the clinicians who collaborated in the project.
Disclosure of Potential Conflicts of Interest
The authors have no potential conflicts of interest to disclose.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, E.; Ciardiello, D.; Martini, G.; Troiani, T.; Cardone, C.; Vitiello, P.; Normanno, N.; Rachiglio, A.; Maiello, E.; Latiano, T.; et al. Implementing anti-epidermal growth factor receptor (EGFR) therapy in metastatic colorectal cancer: challenges and future perspectives. Ann. Oncol. 2020, 31, 30–40. [Google Scholar] [CrossRef]
- Center NCC. Colon Cancer ( Version 3.2022) 2022.
- Available from: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf.
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350-6.
- Trinh A, Trumpi K, De Sousa EMF, Wang X, de Jong JH, Fessler E, et al. Practical and Robust Identification of Molecular Subtypes in Colorectal Cancer by Immunohistochemistry. Clin Cancer Res. 2017;23(2):387-98.
- Baba, Y.; Nosho, K.; Shima, K.; Freed, E.; Irahara, N.; Philips, J.; Meyerhardt, J.A.; Hornick, J.L.; Shivdasani, R.A.; Fuchs, C.S.; et al. Relationship of CDX2 Loss with Molecular Features and Prognosis in Colorectal Cancer. Clin. Cancer Res. 2009, 15, 4665–4673. [Google Scholar] [CrossRef]
- Kim JH, Kang GH. Molecular and prognostic heterogeneity of microsatellite-unstable colorectal cancer. World J Gastroenterol. 2014;20(15):4230-43.
- Selamat, S.A.; Galler, J.S.; Joshi, A.D.; Fyfe, M.N.; Campan, M.; Siegmund, K.D.; Kerr, K.M.; Laird-Offringa, I.A. DNA Methylation Changes in Atypical Adenomatous Hyperplasia, Adenocarcinoma In Situ, and Lung Adenocarcinoma. PLOS ONE 2011, 6, e21443. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, X.; Zhan, Q.; Brock, M.V.; Herman, J.G.; Guo, M. CDX2 serves as a Wnt signaling inhibitor and is frequently methylated in lung cancer. Cancer Biol. Ther. 2012, 13, 1152–1157. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Li, W.; Liu, S.; Han, B. Methylation of promoter region of CDX2 gene in colorectal cancer. Oncol. Lett. 2016, 12, 3229–3233. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Li, W.; Liu, S.; Han, B. Methylation of CDX2 gene promoter in the prediction of treatment efficacy in colorectal cancer. Oncol. Lett. 2018, 16, 195–198. [Google Scholar] [CrossRef]
- Moleirinho, S.; Tilston-Lunel, A.; Angus, L.; Gunn-Moore, F.; Reynolds, P.A. The expanding family of FERM proteins. Biochem. J. 2013, 452, 183–193. [Google Scholar] [CrossRef]
- Yin, F.; Dong, J.; Kang, L.-I.; Liu, X. Hippo-YAP signaling in digestive system tumors. 2021, 11, 2495–2507.
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Benhassine, M.; Guérin, S.L. Transcription of the Human 5-Hydroxytryptamine Receptor 2B (HTR2B) Gene Is under the Regulatory Influence of the Transcription Factors NFI and RUNX1 in Human Uveal Melanoma. Int. J. Mol. Sci. 2018, 19, 3272. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Semenov, O.; Daks, A.; Fedorova, O.; Shuvalov, O.; Barlev, N.A. Opposing Roles of Wild-type and Mutant p53 in the Process of Epithelial to Mesenchymal Transition. Front. Mol. Biosci. 2022, 9, 928399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, L.; Li, A.; Han, X. The roles of ZEB1 in tumorigenic progression and epigenetic modifications. BioMedicine 2019, 110, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Martisova, A.; Holcakova, J.; Izadi, N.; Sebuyoya, R.; Hrstka, R.; Bartosik, M. DNA Methylation in Solid Tumors: Functions and Methods of Detection. Int. J. Mol. Sci. 2021, 22, 4247. [Google Scholar] [CrossRef]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997-1003.
- Essa, H.Y.S.; Kusaf, G.; Yuruker, O.; Kalkan, R. Epigenetic Alteration in Colorectal Cancer: A Biomarker for Diagnostic and Therapeutic Application. Glob. Med Genet. 2022, 09, 258–262. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. International Agency for Research on Cancer WHO classification of tumours of the digestive system, 5th edn. Lyon: International Agency for Research on Cancer; 2019.
- Azcue, P.; Setas, D.G.; Encío, I.; Ibáñez-Beroiz, B.; Mercado, M.; Vera, R.; Gómez-Dorronsoro, M.L. A Novel Prognostic Biomarker Panel for Early-Stage Colon Carcinoma. Cancers 2021, 13, 5909. [Google Scholar] [CrossRef]
- Azcue, P.; Encío, I.; Setas, D.G.; Alecha, J.S.; Galbete, A.; Mercado, M.; Vera, R.; Gomez-Dorronsoro, M.L. PD-L1 as a Prognostic Factor in Early-Stage Colon Carcinoma within the Immunohistochemical Molecular Subtype Classification. Cancers 2021, 13, 1943. [Google Scholar] [CrossRef]
- Kim, K.M.; Ahn, A.-R.; Park, H.S.; Jang, K.Y.; Moon, W.S.; Kang, M.J.; Ha, G.W.; Lee, M.R.; Chung, M.J. Clinical significance of p53 protein expression and TP53 variation status in colorectal cancer. BMC Cancer 2022, 22, 1–17. [Google Scholar] [CrossRef]
- E Ahmed, F.; Ahmed, N.C.; Vos, P.W.; Bonnerup, C.; Atkins, J.N.; Casey, M.; Nuovo, G.J.; Naziri, W.; E Wiley, J.; Mota, H.; et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genom. - Proteom. 2013, 10. [Google Scholar]
- Korenkova, V.; Slyskova, J.; Novosadova, V.; Pizzamiglio, S.; Langerova, L.; Bjorkman, J.; Vycital, O.; Liska, V.; Levy, M.; Veskrna, K.; et al. The focus on sample quality: Influence of colon tissue collection on reliability of qPCR data. Sci. Rep. 2016, 6, 29023. [Google Scholar] [CrossRef]
- Mendaza, S.; Ulazia-Garmendia, A.; Monreal-Santesteban, I.; Córdoba, A.; de Azúa, Y.R.; Aguiar, B.; Beloqui, R.; Armendáriz, P.; Arriola, M.; Martín-Sánchez, E.; et al. ADAM12 is A Potential Therapeutic Target Regulated by Hypomethylation in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2020, 21, 903. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Ye, W.; Zhou, L.; Collins, L.B.; Chen, X.; Gold, A.; Ball, L.M.; Swenberg, J.A. Structural Characterization of Formaldehyde-Induced Cross-Links Between Amino Acids and Deoxynucleosides and Their Oligomers. J. Am. Chem. Soc. 2010, 132, 3388–3399. [Google Scholar] [CrossRef] [PubMed]
- Taieb, J.; Svrcek, M.; Cohen, R.; Basile, D.; Tougeron, D.; Phelip, J.-M. Deficient mismatch repair/microsatellite unstable colorectal cancer: Diagnosis, prognosis and treatment. Eur. J. Cancer 2022, 175, 136–157. [Google Scholar] [CrossRef] [PubMed]
- Pilati, C.; Taieb, J.; Balogoun, R.; Marisa, L.; de Reyniès, A.; Laurent-Puig, P. CDX2 prognostic value in stage II/III resected colon cancer is related to CMS classification. Ann. Oncol. 2017, 28, 1032–1035. [Google Scholar] [CrossRef]
- Guerrero-Setas, D.; Pérez-Janices, N.; Blanco-Fernandez, L.; Ojer, A.; Cambra, K.; Berdasco, M.; Esteller, M.; Maria-Ruiz, S.; Torrea, N.; Guarch, R. RASSF2 hypermethylation is present and related to shorter survival in squamous cervical cancer. Mod. Pathol. 2013, 26, 1111–1122. [Google Scholar] [CrossRef]
- Perez-Janices, N.; Blanco-Luquin, I.; Torrea, N.; Liechtenstein, T.; Escors, D.; Cordoba, A.; Vicente-Garcia, F.; Jauregui, I.; De La Cruz, S.; Illarramendi, J.J.; et al. Differential involvement of RASSF2 hypermethylation in breast cancer subtypes and their prognosis. Oncotarget 2015, 6, 23944–23958. [Google Scholar] [CrossRef]
- Perez-Janices, N.; Blanco-Luquin, I.; Tuñón, M.T.; Barba-Ramos, E.; Ibáñez, B.; Zazpe-Cenoz, I.; Martinez-Aguillo, M.T.; Hernandez, B.; Martínez-Lopez, E.; Fernández, A.F.; et al. EPB41L3, TSP-1 and RASSF2 as new clinically relevant prognostic biomarkers in diffuse gliomas. Oncotarget 2015, 6, 368–380. [Google Scholar] [CrossRef]
- Konukiewitz, B.; Schmitt, M.; Silva, M.; Pohl, J.; Lang, C.; Steiger, K.; Halfter, K.; Engel, J.; Schlitter, A.M.; Boxberg, M.; et al. Loss of CDX2 in colorectal cancer is associated with histopathologic subtypes and microsatellite instability but is prognostically inferior to hematoxylin–eosin-based morphologic parameters from the WHO classification. Br. J. Cancer 2021, 125, 1632–1646. [Google Scholar] [CrossRef]
- Kawai, H.; Tomii, K.; Toyooka, S.; Yano, M.; Murakami, M.; Tsukuda, K.; Shimizu, N. Promoter methylation downregulates CDX2 expression in colorectal carcinomas. Oncol. Rep. 2005, 13, 547–551. [Google Scholar] [CrossRef]
- den Uil SH, de Wit M, Slebos RJC, Delis-van Diemen PM, Sanders J, Piersma SR, et al. Quantitative analysis of CDX2 protein expression improves its clinical utility as a prognostic biomarker in stage II and III colon cancer. Eur J Cancer. 2021;144:91-100.
- Jiang, G.; Luo, C.; Sun, M.; Zhao, Z.; Li, W.; Chen, K.; Fan, T.; Kondkar, A.A.; Mousa, A.; Azad, T.A.; et al. Methylation of CDX2 as a Predictor in Poor Clinical Outcome of Patients with Colorectal Cancer. Genet. Test. Mol. Biomarkers 2016, 20, 710–714. [Google Scholar] [CrossRef]
- Akrida, I.; Bravou, V.; Papadaki, H. The deadly cross-talk between Hippo pathway and epithelial–mesenchymal transition (EMT) in cancer. Mol. Biol. Rep. 2022, 49, 10065–10076. [Google Scholar] [CrossRef] [PubMed]
- Haldrup, J.; Strand, S.H.; Cieza-Borrella, C.; Jakobsson, M.E.; Riedel, M.; Norgaard, M.; Hedensted, S.; Dagnaes-Hansen, F.; Ulhoi, B.P.; Eeles, R.; et al. FRMD6 has tumor suppressor functions in prostate cancer. Oncogene 2020, 40, 763–776. [Google Scholar] [CrossRef]
- Guan, C.; Chang, Z.; Gu, X.; Liu, R. MTA2 promotes HCC progression through repressing FRMD6, a key upstream component of hippo signaling pathway. Biochem. Biophys. Res. Commun. 2019, 515, 112–118. [Google Scholar] [CrossRef]
- Wang, T.; Guo, H.; Zhang, L.; Yu, M.; Li, Q.; Zhang, J.; Tang, Y.; Zhang, H.; Zhan, J. FERM domain-containing protein FRMD6 activates the mTOR signaling pathway and promotes lung cancer progression. Front. Med. 2023, 1–15. [Google Scholar] [CrossRef]
- Melo, F.D.S.E.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; De Rooij, L.P.M.H.; De Jong, J.H.; de Boer, O.J.; Van Leersum, R.; Bijlsma, M.F.; et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Abdul Aziz NA, Mokhtar NM, Harun R, Mollah MM, Mohamed Rose I, Sagap I, et al. A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genomics. 2016;9(1):58.
- Liu, D.; Zhou, B.; Liu, R. A transcriptional co-expression network-based approach to identify prognostic biomarkers in gastric carcinoma. PeerJ 2020, 8, e8504. [Google Scholar] [CrossRef]
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22-36.
- Noblejas-López, M.d.M.; Nieto-Jiménez, C.; García, S.M.; Pérez-Peña, J.; Nuncia-Cantarero, M.; Andrés-Pretel, F.; Galán-Moya, E.M.; Amir, E.; Pandiella, A.; Győrffy, B.; et al. Expression of MHC class I, HLA-A and HLA-B identifies immune-activated breast tumors with favorable outcome. OncoImmunology 2019, 8, e1629780. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Zhang, Y.; Wang, J.; Wang, H.; Dong, N.; Su, X.; Xu, M.; Wang, X. Variations of chromosome 2 gene expressions among patients with lung cancer or non-cancer. Cell Biol. Toxicol. 2016, 32, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Chen, L.; Zhang, J. Expression analysis of genes and pathways associated with liver metastases of the uveal melanoma. BMC Med Genet. 2014, 15, 29–29. [Google Scholar] [CrossRef]
- Mao, L.; Xin, F.; Ren, J.; Xu, S.; Huang, H.; Zha, X.; Wen, X.; Gu, G.; Yang, G.; Cheng, Y.; et al. 5-HT2B-mediated serotonin activation in enterocytes suppresses colitis-associated cancer initiation and promotes cancer progression. Theranostics 2022, 12, 3928–3945. [Google Scholar] [CrossRef] [PubMed]
- Candiloro, I.L.M.; Mikeska, T.; Dobrovic, A. Assessing alternative base substitutions at primer CpG sites to optimise unbiased PCR amplification of methylated sequences. Clin. Epigenetics 2017, 9, 1–9. [Google Scholar] [CrossRef]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97-110.
- Chai, H.; Sun, C.; Liu, J.; Sheng, H.; Zhao, R.; Feng, Z. The Relationship Between ZEB1-AS1 Expression and the Prognosis of Patients With Advanced Gastric Cancer Receiving Chemotherapy. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef]
- Ruan, L.; Chen, W.; Zhao, X.; Fang, N.; Li, T. Predictive Potentials of ZEB1-AS1 in Colorectal Cancer Prognosis and Their Correlation with Immunotherapy. J. Oncol. 2022, 2022, 1–13. [Google Scholar] [CrossRef]
- Sánchez-Tilló, E.; Liu, Y.; de Barrios, O.; Siles, L.; Fanlo, L.; Cuatrecasas, M.; Darling, D.S.; Dean, D.C.; Castells, A.; Postigo, A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell. Mol. Life Sci. 2012, 69, 3429–3456. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.J.; Kim, C.W.; Roh, S.A.; Cho, D.H.; Park, J.L.; Kim, S.Y.; Kim, J.H.; Choi, E.K.; Kim, Y.S.; Kim, J.C. Epigenetic Regulation of KLHL34 Predictive of Pathologic Response to Preoperative Chemoradiation Therapy in Rectal Cancer Patients. Int. J. Radiat. Oncol. 2015, 91, 650–658. [Google Scholar] [CrossRef]
- Plaschka, M.; Benboubker, V.; Grimont, M.; Berthet, J.; Tonon, L.; Lopez, J.; Le-Bouar, M.; Balme, B.; Tondeur, G.; de la Fouchardière, A.; et al. ZEB1 transcription factor promotes immune escape in melanoma. J. Immunother. Cancer 2022, 10, e003484. [Google Scholar] [CrossRef] [PubMed]
- Dhaou, M.O.; Kossai, M.; Morel, A.-P.; Devouassoux-Shisheboran, M.; Puisieux, A.; Penault-Llorca, F.; Radosevic-Robin, N. Zeb1 expression by tumor or stromal cells is associated with spatial distribution patterns of CD8+ tumor-infiltrating lymphocytes: a hypothesis-generating study on 113 triple negative breast cancers. Am. J. Cancer Res. 2020, 10, 3370–3381. [Google Scholar]
- Wang, Z.; Zhang, L.; Xu, W.; Li, J.; Liu, Y.; Zeng, X.; Zhong, M.; Zhu, Y. The Multi-Omics Analysis of Key Genes Regulating EGFR-TKI Resistance, Immune Infiltration, SCLC Transformation in EGFR-Mutant NSCLC. J. Inflamm. Res. 2022, ume 15, 649–667. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).