Submitted:
30 June 2023
Posted:
06 July 2023
You are already at the latest version
Abstract

Keywords:
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Array-CGH Protocol
2.3. PFGE, Chromosome Blotting and Hybridization
2.4. Determination of Invertase Activity
2.5. Determination of the Activity of the AGT1 Permease
| Relevant Features or Sequence (5’ → 3’) | |
|---|---|
| Plasmids: | |
| pGRSd-AGT1 | ori ampr CEN6 URA3 PGPD-AGT1-TPGK [45] |
| pUG66 | ori ampr LoxP-BleR-LoxP [46] |
| Primers: 1 | |
| SUC100-F | GCGATAGACCTTTGGTCCAC |
| SUC1320-R | GGACCGTGGTAACTCTAAGG |
| V-SUC2F | GAAATTATCCGGGGGCGAAG |
| V2-SUC2F | GAGTTGTTGTCCTAGCGTAG |
| V2-SUC2R | TCCATTTCCCTCACTACTTC |
| V3-SUC2F | GCATCCACACGTCACAATCT |
| SUC100-R | GTGGACCAAAGGTCTATCGC |
| ssSUC2-F1 | ATGCTTTTGCAAGCTTTCCTTTTCCTTTTGGCTGGTTTTGCAGCCAAAATATCTGCATCAGCCAGCTGAAGCTTCGTACGC |
| 551SUC2-R1 | ATTCGTATTGGTAGCCTAAGAAACCTTCATTGGCAAATGCAGATTCTAGCTTCCAGGACTGCATAGGCCACTAGTGGATC |
| GPD-AGT1F | GCCATAGATTCTACTCGGTCTATCTATCATGTAACACTCCGTTGATGCGTACTAGAGAGTTTATCATTATCAATAC |
| AGT1-389R | GAAAAACTGGCAGGGCATAC |
| V-GPDF | CAACCATCAGTTCATAGGTC |
| AGT1-I505-R | ACGGGCCAGCACTATAGTCTTAGTTCTC |
| V-AGT1F1 | GAATTTTCGGTTGGTG |
2.6. Molecular Biology Techniques
2.7. Sucrose Batch Fermentations
3. Results
3.1. Microarray Karyotyping of Industrial Yeast Strains Used in Sugarcane-Based Fermentation Processes
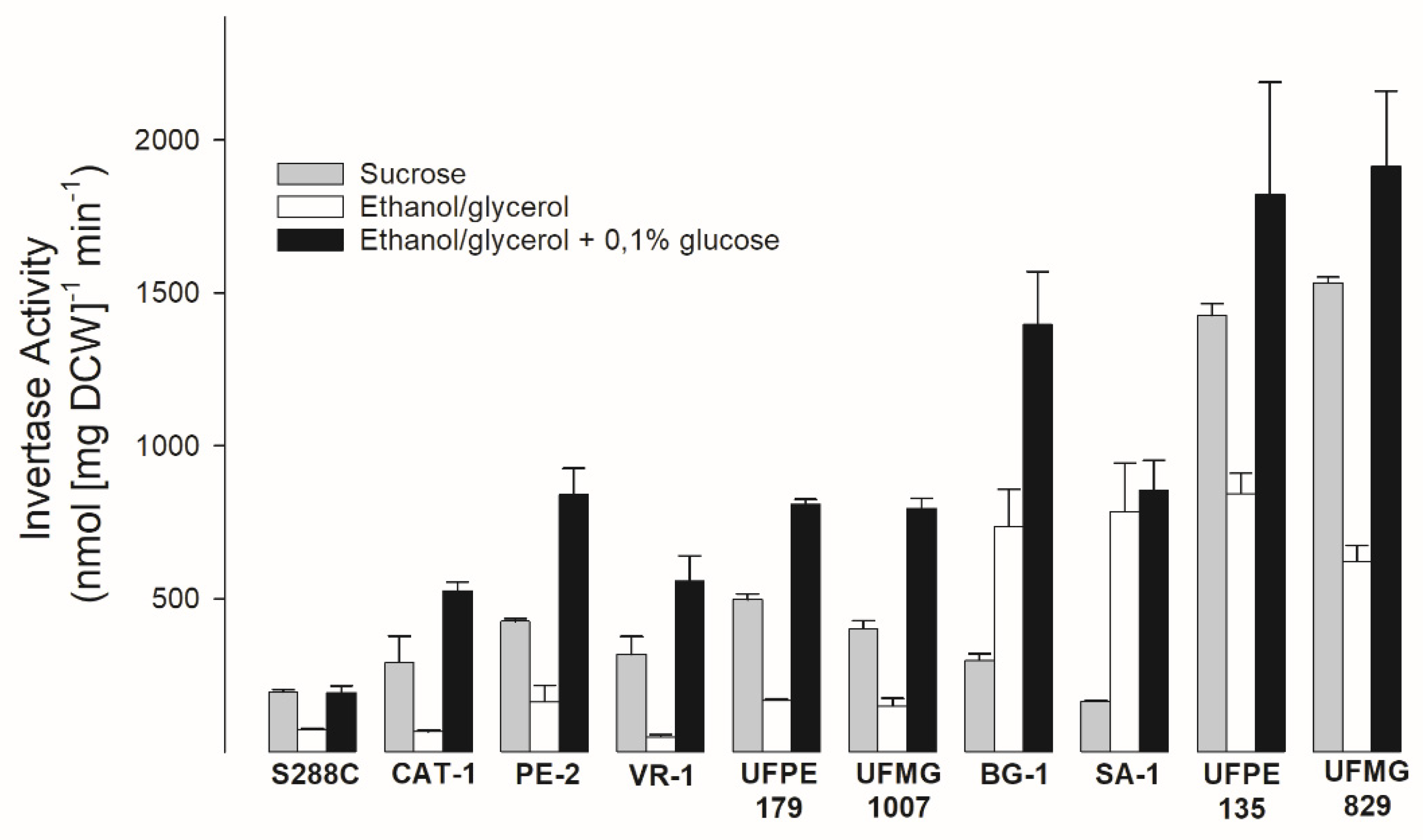
3.2. Invertase Activity of Industrial Yeast Strains Used in Sugarcane-Based Fermentation Processes
3.3. Sucrose Batch Fermentation by the Industrial Yeast Strains Used in Sugarcane-Based Fermentation Processes
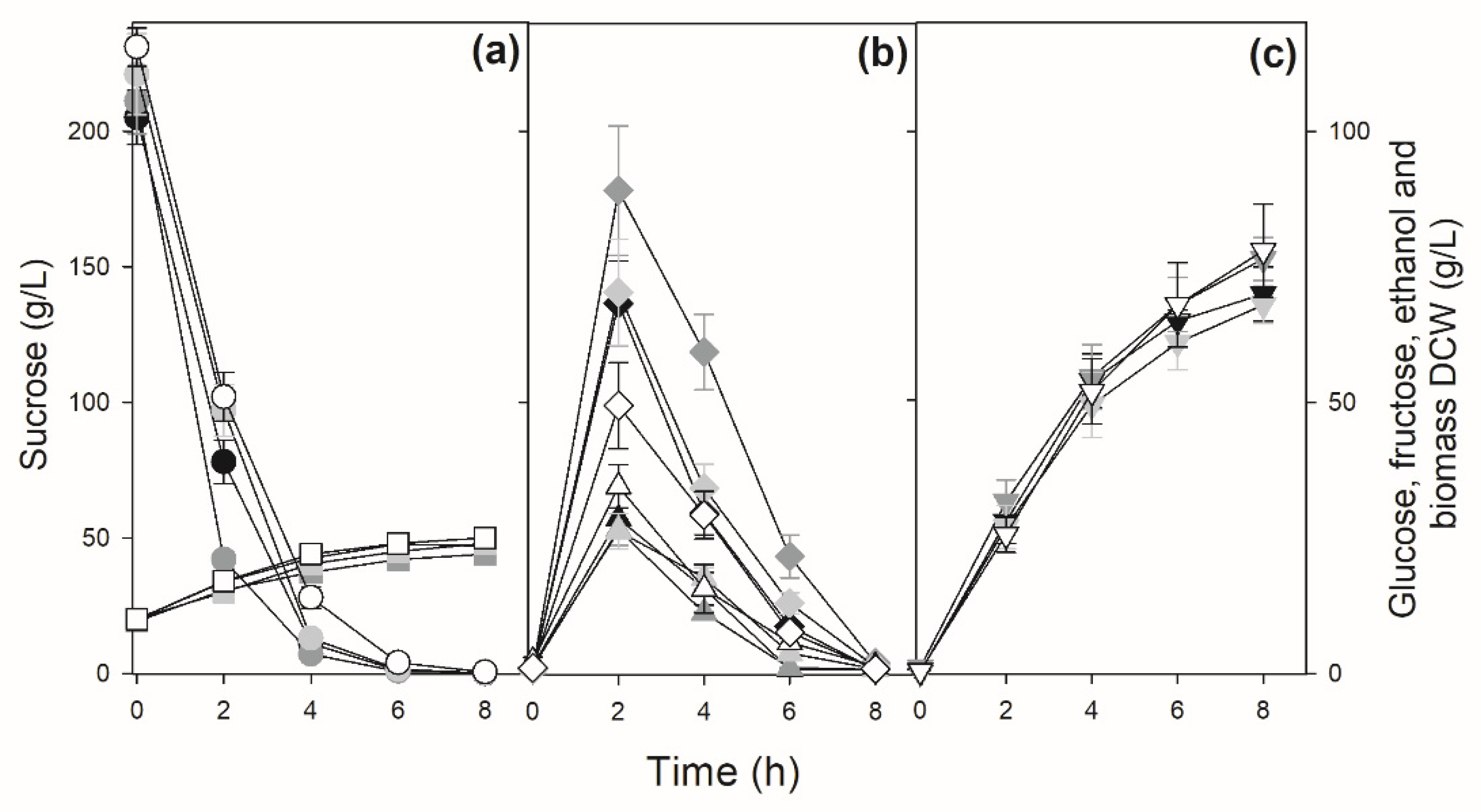
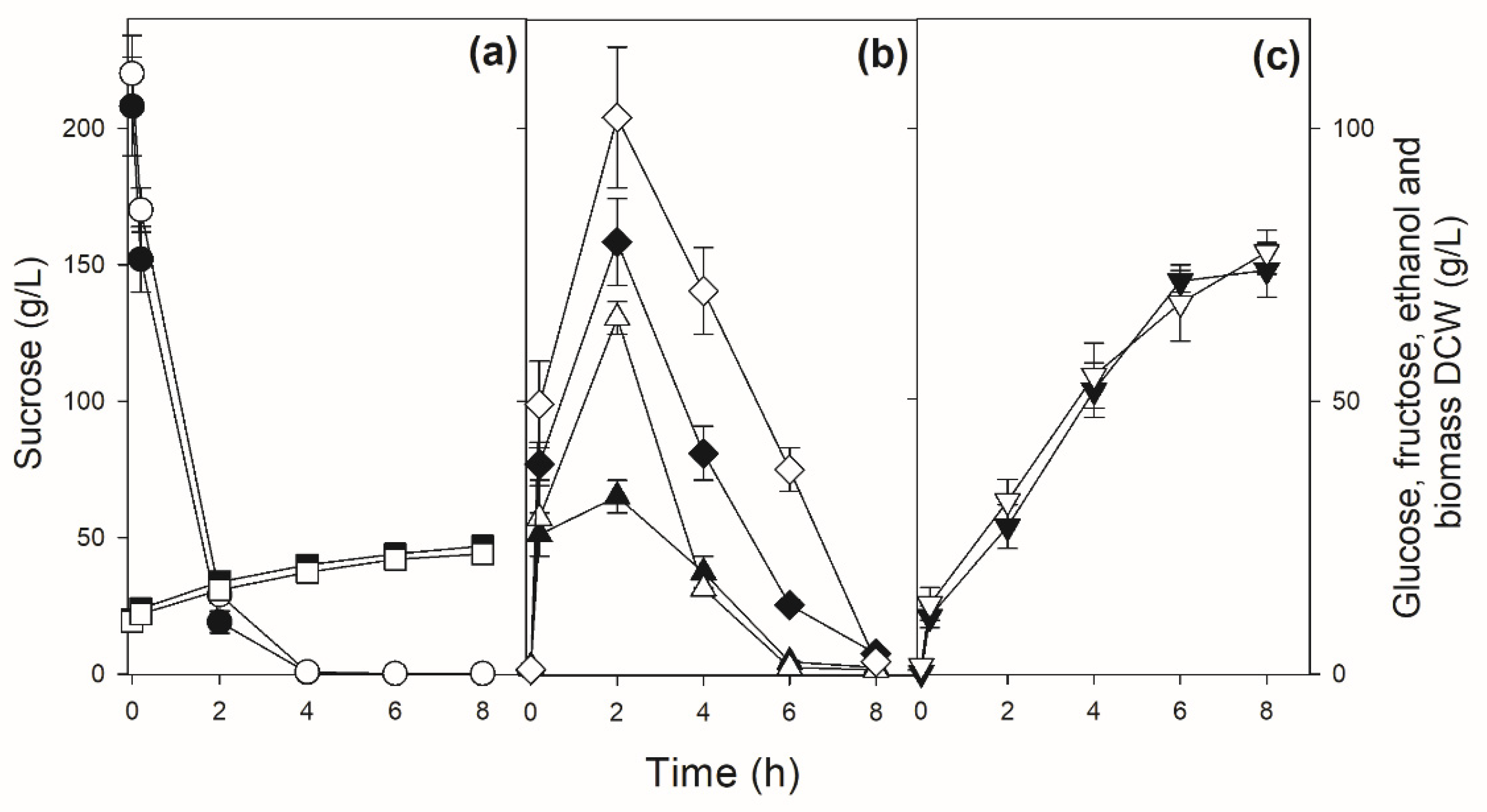
3.4. Modifying the Mode of Sucrose Fermentation by an Industrial Fuel-Ethanol Yeast Strain
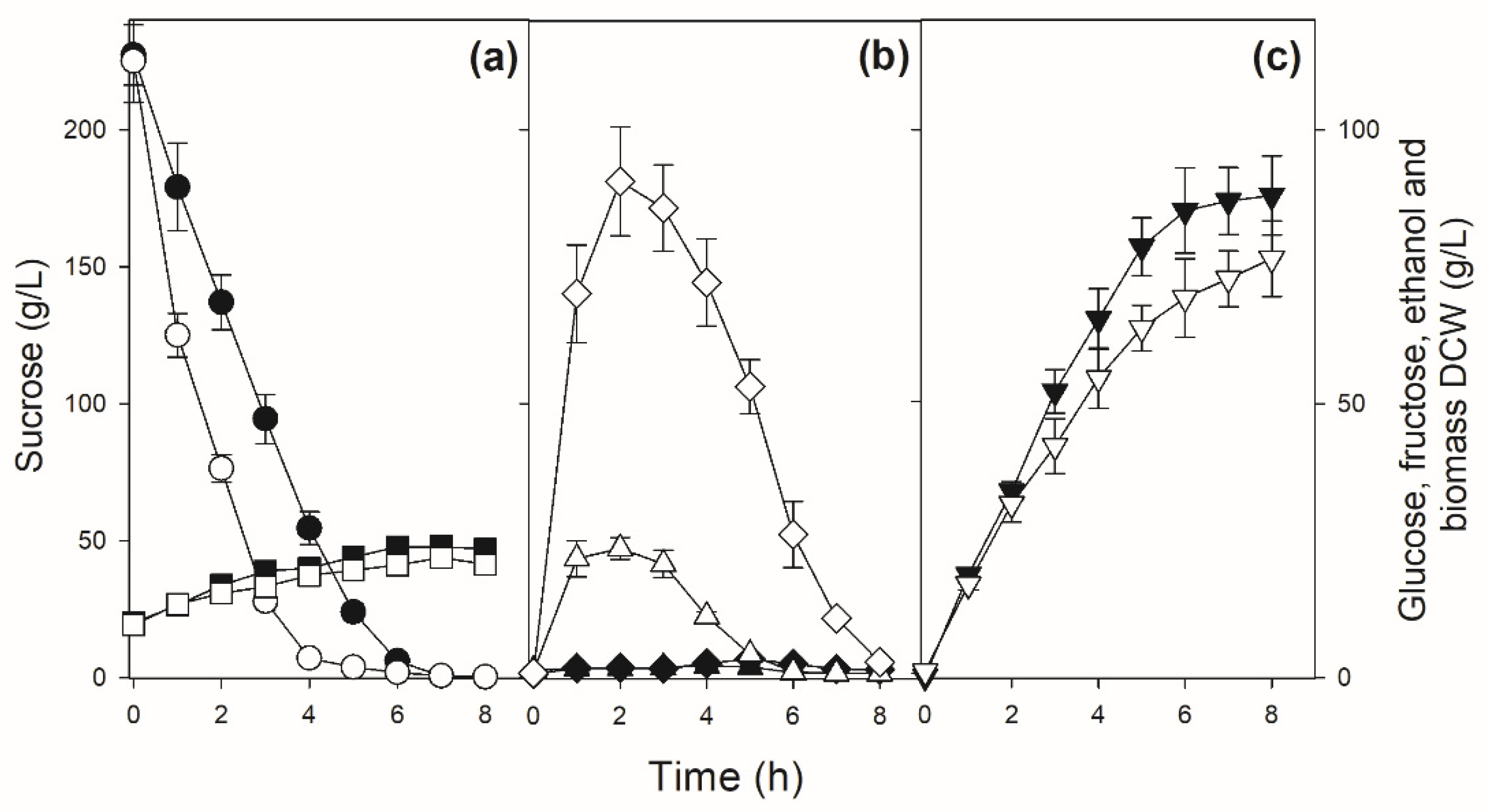
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldemberg J. Ethanol for a sustainable energy future. Science 2007, 315, 808-810. [CrossRef]
- CONAB – Companhia Nacional de Abastecimento - Acompanhamento da safra brasileira de cana-de-açúcar - Safra 2022/23. Available online: https://www.conab.gov.br/info-agro/safras/cana/boletim-da-safra-de-cana-de-acucar (accessed on 02 March 2023).
- Jaiswal, D.; de Souza, A.P.; Larsen, S.; LeBauer, D.S.; Miguez, F.E.; Sparovek, G.; Bollero, G.; Buckeridge, M.S.; Long, S.P. Brazilian sugarcane ethanol as an expandable green alternative to crude oil use. Nat. Clim. Change 2017, 7, 788-792. [CrossRef]
- Jacobus, A.P.; Gross, J.; Evans, J.H.; Ceccato-Antonini, S.R.; Gombert, A.K. Saccharomyces cerevisiae strains used industrially for bioethanol production. Essays Biochem. 2021, 65, 147-161. [CrossRef]
- do Nascimento e Silva, J.H.; Verruma-Bernardi, M.R.; de Oliveira, A.L Cachaça production in Brazil and its main contaminant (ethyl carbamate). Sci. Agric. 2018, 77, e20180135. [CrossRef]
- Amorim, H.V.; Lopes, M.L.; Oliveira, J.V.C.; Buckeridge, M.S.; Goldman, G.H. Scientific challenges of bioethanol production in Brazil. Appl. Microbiol. Biotechnol. 2011, 91, 1267-1275. [CrossRef]
- Lopes, M.L.; Paulillo, S.C.; Godoy, A.; Cherubin, R.A., Lorenzi, M.S., Giometti, F.H., Bernardino, C.D., Amorim Neto, H.B., Amorim, H.V. Ethanol production in Brazil: a bridge between science and industry. Braz. J. Microbiol. 2016, 47(Suppl 1), 64-76. [CrossRef]
- Badotti, F.; Gomes, F.C.O.; Rosa, C.A. Brazilian cachaça: fermentation and production. In: Handbook of Plant-Based Fermented Food and Beverage Technology, Second ed.; Hui, Y.H.; Evranuz, E.Ö. Eds; CRC Press, Florida, USA; 2012; pp 639–648.
- Guerra, J.B.; Araújo, R.A.; Pataro, C.; Franco, G.R.; Moreira, E.S.; Mendonça-Hagler, L.C.; Rosa, C.A. Genetic diversity of Saccharomyces cerevisiae strains during the 24 h fermentative cycle for the production of the artisanal Brazilian cachaça. Lett. Appl. Microbiol. 2001, 33, 106-111. [CrossRef]
- da Silva-Filho, E.; Santos, S.K.; Resende, A.M.; Morais, J.O.; de Morais-Jr, M.A.; Simões, D.A. Yeast population dynamics of industrial fuel-ethanol fermentation process assessed by PCR-fingerprinting. Antonie van Leeuwenhoeck 2005, 88, 13-23. [CrossRef]
- Badotti, F.; Rosa, C.A.; Stambuk, B.U. Biochemical and molecular characterization of Saccharomyces cerevisiae strains isolated from artisan-type fermentations of sugarcane and molasses in the production of sugarcane brandy in Florianópolis-SC, Brazil. Braz. J. Food Technol. 2010, 13, 205-213. [CrossRef]
- Basso, L.C.; de Amorim, H.V.; de Oliveira, A.J.; Lopes, M.L. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 2008, 8, 1155-1163. [CrossRef]
- Gomes, F.C.; Silva, C.L.; Marini, M.M.; Oliveira, E.S.; Rosa, C.A. Use of selected indigenous Saccharomyces cerevisiae strains for the production of the traditional cachaça in Brazil. J. Appl. Microbiol. 2007, 103, 2438-2447. [CrossRef]
- Campos, C.R.; Silva, C.F.; Dias, D.R.; Basso, L.C.; Amorim, H.V.; Schwan, R.F. Features of Saccharomyces cerevisiae as a culture starter for the production of the distilled sugar cane beverage, cachaça in Brazil. J. Appl. Microbiol. 2010, 108, 1871-1879. [CrossRef]
- Marques, W.L.; Raghavendran, V.; Stambuk, B.U.; Gombert, A.K. Sucrose and Saccharomyces cerevisiae: A relationship most sweet. FEMS Yeast Res. 2016, 16, fov107. [CrossRef]
- Korshunova, I.V.; Naumova, E.S.; Naumov, G.I. Comparative molecular genetic analysis of β-fructosidases of yeasts Saccharomyces. Mol. Biol. 2005, 39, 413-419. [CrossRef]
- Naumova, E.S.; Sadykova, A.Z.; Martynenko, N.N.; Naumov, G.I. Molecular polymorphism of β-fructosidase SUC genes in the Saccharomyces yeasts. Mol. Biol. 2014, 48, 573–582. [CrossRef]
- Codón, A.C.; Benítez, T.; Korhola, M. Chromosomal polymorphism and adaptation to specific industrial environments of Saccharomyces strains. Appl. Microbiol. Biotechnol. 1998, 49, 154-163. [CrossRef]
- Naumova, E.S.; Sadykova, A.Z.; Martynenko, N.N.; Naumov, G.I. Molecular genetic characteristics of Saccharomyces cerevisiae distillers’ yeasts. Microbiology 2013, 82, 175–185. [CrossRef]
- Carlson, M.; Botstein, D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 1982, 28, 145-154. [CrossRef]
- Herwing, C.; Doerries, C.; Marison, I.; von Stockar, U. Quantitative analysis of the regulation scheme of invertase expression in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001, 76, 247-258. [CrossRef]
- Belinchón, M.M.; Gancedo J.M. Different signaling pathways mediate glucose induction of SUC2, HXT1 and pyruvate decarboxylase in yeast. FEMS Yeast Res. 2007, 7, 40-47. [CrossRef]
- Takeshige, K.; Ouchi, K. Effects of yeast invertase on ethanol production in molasses. J. Ferment. Bioeng. 1995, 79, 513-515. [CrossRef]
- Myers, D.K.; Lawlor, D.T.M.; Attfield, P.V. Influence of invertase activity and glycerol synthesis and retention on fermentation of media with a high sugar concentration by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997, 63, 145-150. [CrossRef]
- Greig, D.; Travisano, M. The Prisoner’s Dilemma and polymorphism in yeast SUC genes. Proc. Biol. Sci. 2004, 271(Suppl 3), S25-6. [CrossRef]
- Gore, J.; Youk, H.; Oudenaarden, A. Snowdrift game dynamics and facultative cheating in yeast. Nature 2009, 459, 253-256. [CrossRef]
- Stambuk, B.U.; Batista, A.S.; de Araujo, P.S. Kinetics of active sucrose transport in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2000, 89, 212-214. [CrossRef]
- Batista, A.S.; Miletti, L.C.; Stambuk, B.U. Sucrose fermentation by Saccharomyces cerevisiae lacking hexose transport. J. Mol. Microbiol. Biotechnol. 2004, 8, 26-33. [CrossRef]
- Badotti, F.; Dário, M.G.; Alves-Jr, S.L.; Cordioli, M.L.; Miletti, L.C.; de Araujo, P.S.; Stambuk, B.U. Switching the mode of sucrose utilization by Saccharomyces cerevisiae. Microb. Cell. Fact. 2008, 7, 4. [CrossRef]
- Brown, C.A.; Murray, A.W.; Verstrepen, K.J. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 2010, 20, 895-903. [CrossRef]
- Marques, W.L.; Mans, R.; Marella, E.R.; Cordeiro, R.L.; van den Broek, M.; Daran, J.G.; Pronk, J.T.; Gombert, A.K.; van Maris, A.J. Elimination of sucrose transport and hydrolysis in Saccharomyces cerevisiae: a platform strain for engineering sucrose metabolism. FEMS Yeast Res. 2017, 17, fox006. [CrossRef]
- Basso, T.O.; de Kok, S.; Dario, M.; do Espirito-Santo, J.C.; Müller, G.; Schlölg, P.S.; Silva, C.P.; Tonso, A.; Daran, J.M.; Gombert, A.K.; van Maris, A.J.; Pronk, J.T.; Stambuk, B.U. Engineering topology and kinetics of sucrose metabolism in Saccharomyces cerevisiae for improved ethanol yield. Metab. Eng. 2011, 13, 694-703. [CrossRef]
- Gombert, A.K.; van Maris, A.J. Improving conversion yield of fermentable sugars into fuel ethanol in 1st generation yeast-based production processes. Curr. Opin. Biotechnol. 2015, 33, 81-86. [CrossRef]
- van Aalst, A.C.A.; de Valk, S.C.; van Gulik, W.M.; Jansen, M.L.A.; Pronk, J.T.; Mans, R. Pathway engineering strategies for improved product yield in yeast-based industrial ethanol production. Synth. Syst. Biotechnol. 2022, 7, 554-566. [CrossRef]
- de Figueiredo, C.M.; Hock, D.H.; Trichez, D.; Magalhães, M.d.L.B.; Lopes, M.L.; de Amorim, H.V.; Stambuk, B.U. High foam phenotypic diversity and variability in flocculant gene observed for various yeast cell surfaces present as industrial contaminants. Fermentation 2021, 7, 127. [CrossRef]
- Stambuk, B.U.; Dunn, B.; Alves-Jr, S.L.; Duval, E.H.; Sherlock, G. Industrial fuel ethanol yeasts contain adaptive copy number changes in genes involved in vitamin B1 and B6 biosynthesis. Genome Res. 2009, 19, 2271-2278. [CrossRef]
- Oliveira, E.S.; Rosa, C.A.; Morgano, M.A.; Fermentation characteristics as criteria for selection of cachaça yeast. World J. Microbiol. Biotechnol, 2004, 20, 19–24. [CrossRef]
- Marini, M.M.; Gomes, F.C.O.; Silva, C.L.C.; Cadete, R.M.; Badotti, F.; Oliveira, E.S.; Cardoso, C.R.; Rosa, C.A. The use of selected starter Saccharomyces cerevisiae strains to produce traditional and industrial cachaça: a comparative study. World J. Microbiol. Biotechnol. 2009, 25, 235–242. [CrossRef]
- Duval, E.H.; Alves-Jr, S.L.; Dunn, B.; Sherlock, G.; Stambuk, B.U. Microarray karyotyping of maltose-fermenting Saccharomyces yeasts with differing maltotriose utilization profiles reveals copy number variation in genes involved in maltose and maltotriose utilization. J. Appl. Microbiol. 2010, 109, 248-259. [CrossRef]
- DeRisi, J.L.; Iyer, V.R.; Brown, P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 1997, 278, 680-686. [CrossRef]
- Wang, P.; Kim, Y.; Pollack, J.; Narasimhan, B.; Tibshirani, R. A method for calling gains and losses in array CGH data. Biostatistics 2005, 6, 45–58. [CrossRef]
- Silveira, M.C.; Carvajal, E.; Bom, E.P. Assay for in vivo yeast invertase activity using NaF. Anal. Biochem. 1996, 238, 26-28. [CrossRef]
- Stambuk, B.U. A simple experiment illustrating metabolic regulation: induction versus repression of yeast α-glucosidase, Biochem. Educ. 1999, 27, 177-180. [CrossRef]
- Hollatz, C.; Stambuk, B.U. Colorimetric determination of active α-glucoside transport in Saccharomyces cerevisiae. J. Microbiol. Methods. 2001, 46, 253-259. [CrossRef]
- Jules, M.; Guillou, V.; François, J.; Parrou, J.L. Two distinct pathways for trehalose assimilation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2004, 70, 2771-2778. [CrossRef]
- Petracek, M.E.; Longtine, M.S. PCR-based engineering of yeast genome. Methods Enzymol. 2002, 350, 445-469. [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Short Protocols in Molecular Biology, 3rd ed.; John Wiley & Sons: New York, USA, 1995.
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [CrossRef]
- Badotti, F.; Batista, A.S.; Stambuk, B.U. Sucrose active transport and fermentation by Saccharomyces cerevisiae. Braz. Arch. Biol. Techn. 2006, 49(Supl 1), 115–123.
- Bastos, R.W.; Pedroso, S.H.; Vieira, A.T.; Moreira, L.M.; França, C.S.; Cartelle, C.T.; Arantes, R.M.; Generoso, S.V.; Cardoso, V.N.; Neves, M.J.; Nicoli, J.R.; Martins, F.S. Saccharomyces cerevisiae UFMG A-905 treatment reduces intestinal damage in a murine model of irinotecan-induced mucositis. Benef. Microbes. 2016, 7, 549-557. [CrossRef]
- Miranda, V.C.; Santos, S.S.; Assis, H.C.; Faria, A.M.C.; Quintanilha, M.F.; Morão, R.P.; Nicoli, J.R.; Cara, D.C.; Martins, F.S. Effect of Saccharomyces cerevisiae UFMG A-905 in a murine model of food allergy. Benef. Microbes. 2020, 11, 255-268. [CrossRef]
- Saldanha, A.J. Java Treeview - extensible visualization of microarray data. Bioinformatics 2004, 20, 3246-3248. [CrossRef]
- Reifenberger, E.; Freidel, K.; Ciriacy, M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol. 1995, 16, 157-167. [CrossRef]
- Diderich, J.A.; Schepper, M.; van Hoek, P.; Luttik, M.A.; van Dijken, J.P.; Pronk, J.T.; Klaassen, P.; Boelens, H.F.; de Mattos, M.J.; van Dam, K.; Kruckeberg, A.L. Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 15350-15359. [CrossRef]
- Ozcan, S.; Johnston, M. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 1999, 63, 554-569. [CrossRef]
- Jordan, P.; Choe, J.Y.; Boles, E.; Oreb, M. Hxt13, Hxt15, Hxt16 and Hxt17 from Saccharomyces cerevisiae represent a novel type of polyol transporters. Sci. Rep. 2016, 6, 23502. [CrossRef]
- Donnini, C.; Lodi, T.; Ferrero, I.; Algeri, A.; Puglisi, P.P. Allelism of IMP1 and GAL2 genes of Saccharomyces cerevisiae. J. Bacteriol. 1992, 174, 3411-3415. [CrossRef]
- Paxhia, M.D.; Downs, D.M. SNZ3 encodes a PLP synthase involved in thiamine synthesis in Saccharomyces cerevisiae. G3: Gen. Genom. Genet. 2019, 9, 335-344. [CrossRef]
- Jacobus, A.P.; Stephens, T.G.; Youssef, P.; González-Pech, R.; Ciccotosto-Camp, M.M.; Dougan, K.E.; Chen, Y.; Basso, L.C.; Frazzon, J.; Chan, C.X.; Gross, J. Comparative genomics supports that Brazilian bioethanol Saccharomyces cerevisiae comprise a unified group of domesticated strains related to cachaça spirit yeasts. Front. Microbiol. 2021, 12, 644089. [CrossRef]
- McIlwain, S.J.; Peris, D.; Sardi, M.; Moskvin, O.V.; Zhan, F.; Myers, K.S.; Riley, N.M.; Buzzell, A.; Parreiras, L.S.; Ong, I.M.; Landick, R.; Coon, J.J.; Gasch, A.P.; Sato, T.K.; Hittinger, C.T. Genome sequence and analysis of a stress-tolerant, wild-derived strain of Saccharomyces cerevisiae used in biofuels research. G3: Gen. Genom. Genet. 2016, 6, 1757-1766. [CrossRef]
- Nosaka, K. Recent progress in understanding thiamin biosynthesis and its genetic regulation in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2006, 72, 30-40. [CrossRef]
- Mojzita, D.; Hohmann, S. Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics. 2006, 276, 147-161. [CrossRef]
- Labuschagne, P.; Divol, B. Thiamine: a key nutrient for yeasts during wine alcoholic fermentation. Appl. Microbiol. Biotechnol. 2021, 105, 953-973. [CrossRef]
- Ozcan, S.; Vallier, L.G.; Flick, J.S.; Carlson, M.; Johnston, M. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 1997, 13, 127-137. [CrossRef]
- Babrzadeh, F.; Jalili, R.; Wang, C.; Shokralla, S.; Pierce, S.; Robinson-Mosher, A.; Nyren, P.; Shafer, R.W.; Basso, L.C.; de Amorim, H.V.; de Oliveira, A.J.; Davis, R.W.; Ronaghi, M.; Gharizadeh, B.; Stambuk, B.U. Whole-genome sequencing of the efficient industrial fuel-ethanol fermentative Saccharomyces cerevisiae strain CAT-1. Mol. Genet. Genomics 2012, 287, 485-494. [CrossRef]
- Vidgren, V.; Kankainen, M.; Londesborough. J.; Ruohonen, L. Identification of regulatory elements in the AGT1 promoter of ale and lager strains of brewer’s yeast. Yeast 2011, 28, 579-594. [CrossRef]
- Alves-Jr, S.L.; Thevelein, J.M.; Stambuk, B.U. Extracellular maltotriose hydrolysis by Saccharomyces cerevisiae cells lacking the AGT1 permease. Lett. Appl. Microbiol. 2018, 67, 377-383. [CrossRef]
- Vidgren, V.; Huuskonen, A.; Virtanen, H.; Ruohonen, L.; Londesborough, J. Improved fermentation performance of a lager yeast after repair of its AGT1 maltose and maltotriose transporter genes. Appl. Environ. Microbiol. 2009, 75, 2333-2345. [CrossRef]
- Legras, J.L.; Merdinoglu, D.; Cornuet, J.M.; Karst, F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 2007, 16, 2091–2102. [CrossRef]
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; Tsai, I.J.; Bergman, C.M.; Bensasson, D.; O’Kelly, M.J.; van Oudenaarden, A.; Barton, D.B.; Bailes, E.; Nguyen, A.N.; Jones, M.; Quail, M.A.; Goodhead, I.; Sims, S.; Smith, F.; Blomberg, A.; Durbin, R.; Louis, E.J. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341. [CrossRef]
- Peter, J.; de Chiara, M.; Friedrich, A.; Yue, J.X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; Cruaud, C.; Labadie, K.; Aury, J.M.; Istace, B.; Lebrigand, K.; Barbry, P.; Engelen, S.; Lemainque, A.; Wincker, P.; Liti, G.; Schacherer, J. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339-344. [CrossRef]
- Liu, Z.L.; Huang, X. Copy number variants impact phenotype-genotype relationships for adaptation of industrial yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2022, 106, 6611-6623. [CrossRef]
- Barbosa, R.; Pontes, A.; Santos, R.O.; Montandon, G.G.; de Ponzzes-Gomes, C.M.; Morais, P.B.; Gonçalves, P.; Rosa, C.A.; Sampaio, J.P. Multiple rounds of artificial selection promote microbe secondary domestication-The case of cachaça yeasts. Genome Biol. Evol. 2018, 10, 1939-1955. [CrossRef]
- Rego-Costa, A.; Huang, I.T.; Desai, M.M.; Gombert, A.K. Yeast population dynamics in Brazilian bioethanol production. G3: Gen. Genom. Genet. 2023, 13, kad104. [CrossRef]
- Alves-Jr, S.L.; Herberts, R.A.; Hollatz, C.; Miletti, L.C.; Stambuk, B.U. Maltose and maltotriose active transport and fermentation by Saccharomyces cerevisiae. J. Am. Soc. Brew. Chem. 2007, 65, 99-104. [CrossRef]
- Pereira, L.F.; Lucatti, E.; Basso, L.C.; de Morais-Jr, M.A. The fermentation of sugarcane molasses by Dekkera bruxellensis and the mobilization of reserve carbohydrates. Antonie Van Leeuwenhoek 2014, 105, 481-489. [CrossRef]
- Erasmus, D.J.; van der Merwe, G.K.; van Vuuren, H.J. Genome-wide expression analyses: Metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res. 2003, 3, 375-399. [CrossRef]
- Parmar, J.H.; Bhartiya, S.; Venkatesh, K.V. Characterization of the adaptive response and growth upon hyperosmotic shock in Saccharomyces cerevisiae. Mol. Biosyst. 2011, 7, 1138-1148. [CrossRef]
- Petelenz-Kurdziel, E.; Kuehn, C.; Nordlander, B.; Klein, D.; Hong, K.K.; Jacobson, T.; Dahl, P.; Schaber, J.; Nielsen, J.; Hohmann, S.; Klipp, E. Quantitative analysis of glycerol accumulation, glycolysis and growth under hyper osmotic stress. PLoS Comput. Biol. 2013, 9, e1003084. [CrossRef]
- Blomberg, A. Yeast osmoregulation - glycerol still in pole position. FEMS Yeast Res. 2022, 22, foac035. [CrossRef]
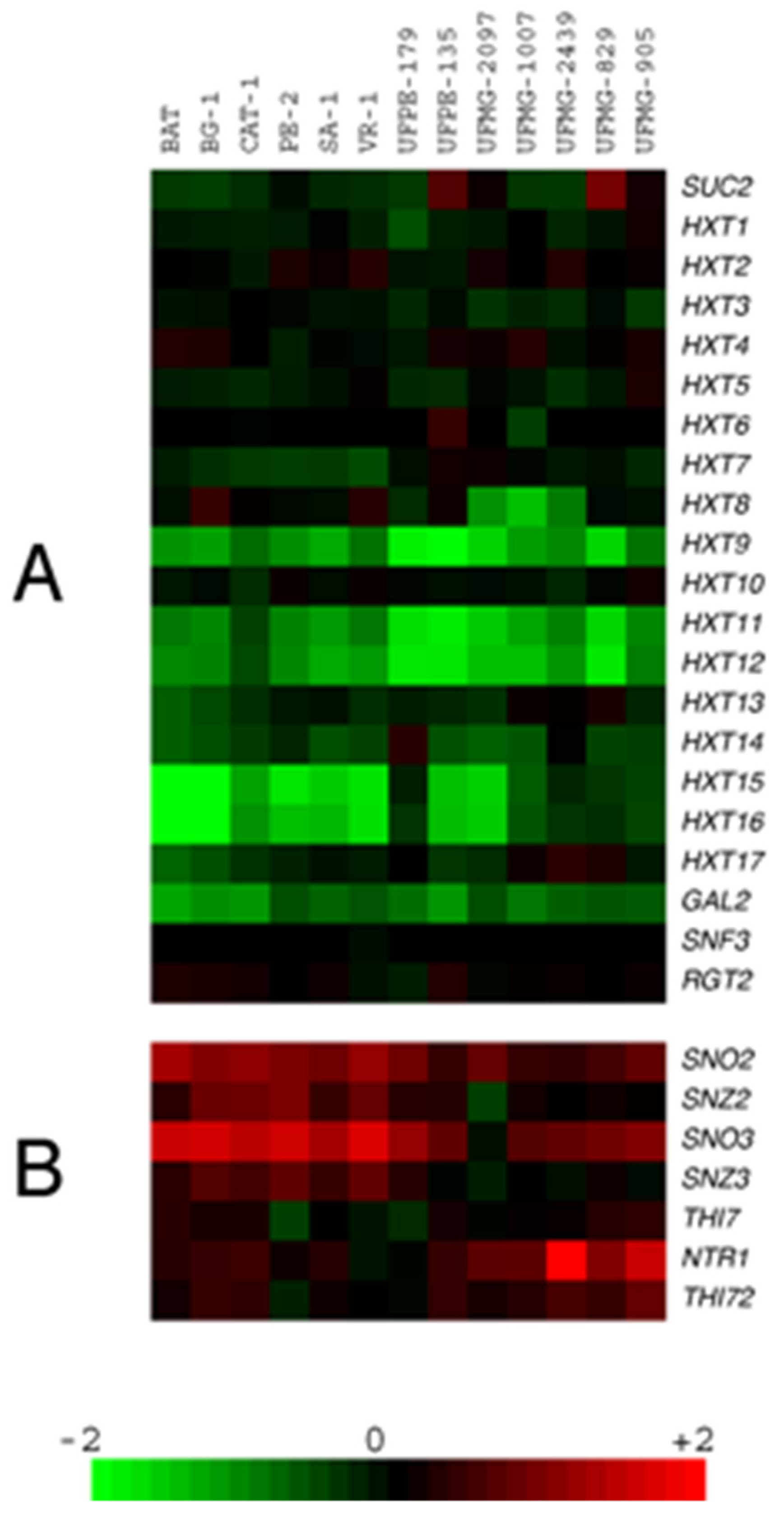
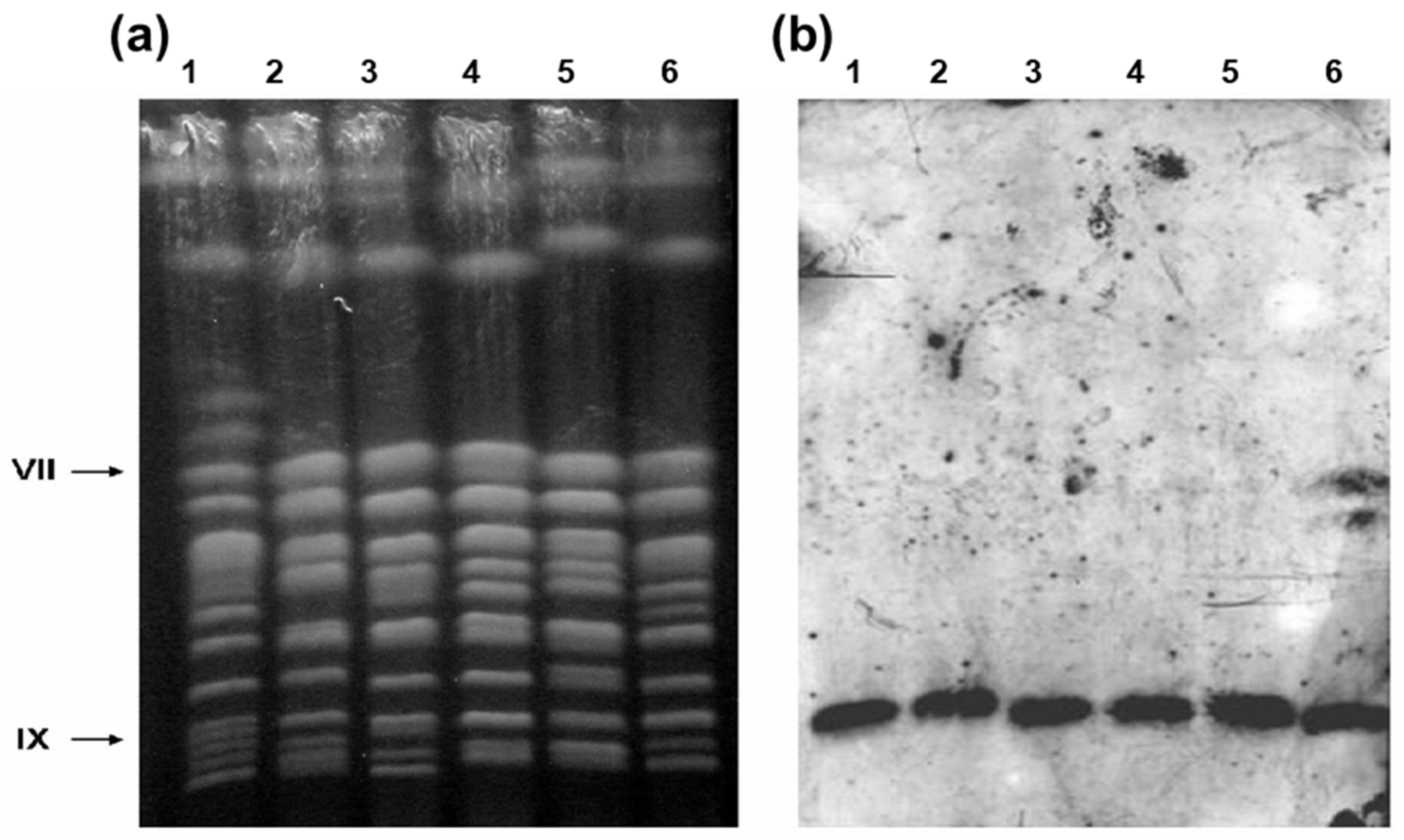
| Yeast strains | Relevant features | Source |
|---|---|---|
| BAT | Industrial fuel ethanol strain isolated in 2011 from Usina Batatais, São Paulo, Brazil. | [35] |
| BG-1 | Industrial fuel ethanol strain isolated in 1989/1990 from Usina Barra Grande, Sao Paulo, Brazil. | [36] |
| CAT-1 | Industrial fuel ethanol strain isolated in 1998/1999 from Usina VO Catanduva, São Paulo, Brazil. | [12,36] |
| PE-2 | Industrial fuel ethanol strain isolated in 1993/1994 from Usina da Pedra, Sao Paulo, Brazil. | [12,36] |
| SA-1 | Industrial fuel ethanol strain isolated in 1989/1990 from Usina Santa Adelia, Sao Paulo, Brazil. | [36] |
| S288C | MATα mal gal2 mel flo1 flo8-1 hap1 SUC2 | [35] |
| BSY21-34B3 | MATa ura3-52 trp-289 kanMX-PADH1::iSUC2 | [32] |
| UFMG-829 | Cachaça strain isolated in 1996 from a distillery in Porto Firme, Minas Gerais, Brazil. | [13] |
| UFMG-905 | Cachaça strain isolated in 1996 from a distillery in Nova União, Minas Gerais, Brazil. | [13,37] |
| UFMG-1007 | Cachaça strain isolated in 1996 from a distillery in Salinas, Minas Gerais, Brazil. | [13] |
| UFMG-2097 | Cachaça strain isolated in 1999 from a distillery in Salinas, Minas Gerais, Brazil. | [38] |
| UFMG-2439 | Cachaça strain isolated in 1999 from a distillery in Salinas, Minas Gerais, Brazil. | [38] |
| UFPE-135 | Industrial fuel ethanol strain isolated in 1998/1999 from Japungu distillery, Paraíba, Brazil | [10] |
| UFPE-179 | Industrial fuel ethanol strain isolated in 1998/1999 from Miriri distillery, Paraíba, Brazil | [10] |
| VR-1 | Industrial fuel ethanol strain isolated in 1993/1994 from Usina Vale do Rosario, Sao Paulo, Brazil | [12,36] |
| GMY08 | Isogenic to CAT-1, but kanMX-PADH1::iSUC2 / suc2Δ::BleR PTDH3::AGT1 / AGT1 | This work |
| Strain | Activity1 (nmol of product [mg DCW]-1 min-1) | Glycerol produced at the end of fermentation2 (g/L) | ||
|---|---|---|---|---|
| Extracellular invertase | Intracellular invertase | pNPαG transport | ||
| CAT-1 | 295 ± 70 | 74 ± 6 | 0.1 ± 0.1 | 10.5 ± 1.2 |
| GMY08 | 56 ± 2 | 2,223 ± 45 | 4.5 ± 0.3 | 5.4 ± 0.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





