Submitted:
26 June 2023
Posted:
26 June 2023
You are already at the latest version
Abstract
Keywords:
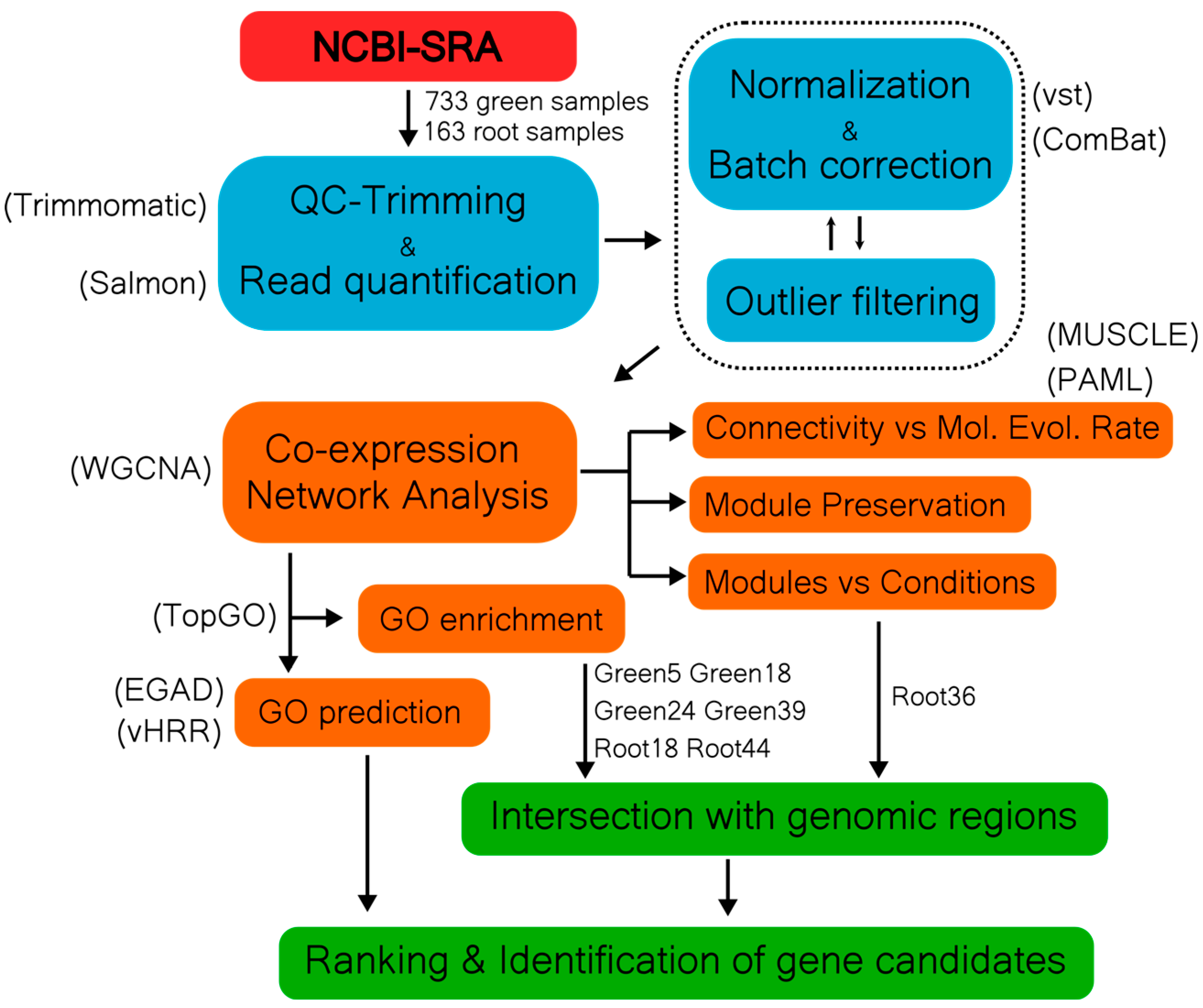
1. Introduction
2. Results
2.1. Construction of a Reference Weighted Gene Co-Expression Network from green healthy tissue
| Samples condition | Initially | Post filtering | |
|---|---|---|---|
| GreenGCN | Healthy | 733 | 686 |
| RootGCN | Healthy | 134 | 127 |
| V. dahliae infection | 14 | 14 | |
| R. irregulare infection | 6 | 6 | |
| O. cumana infection | 9 | 3 |
2.2. Construction of a weighted gene co-expression network from stressed and control root tissue
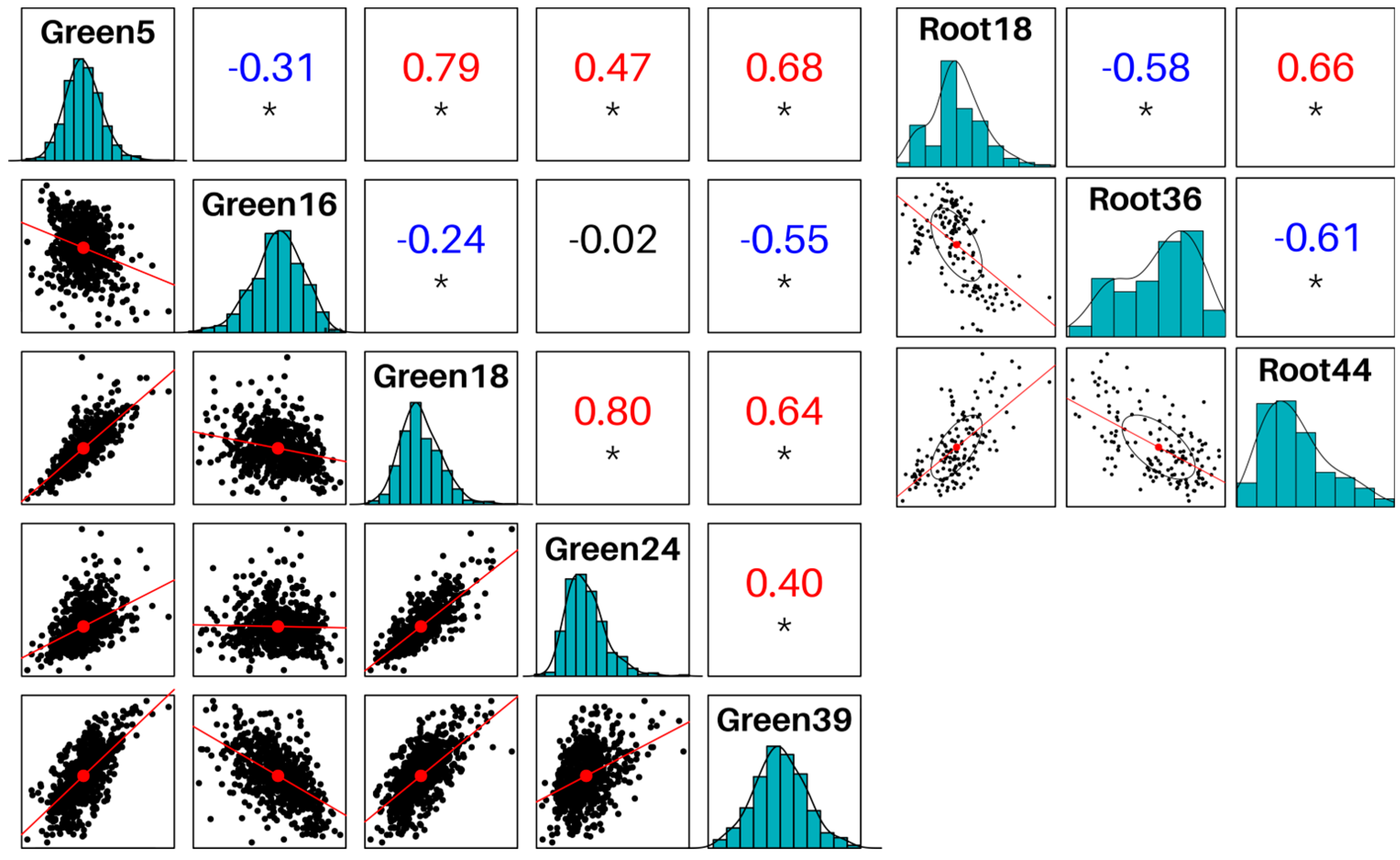
2.3. Identification of modules related to fungal resistance
| Sunflower gene | Closest homolog in A. thaliana | |||||
|---|---|---|---|---|---|---|
| Gene name | Module | IMK | Homolog | Identity % | Q-cov % | E-value |
| HanXRQChr10g0294651 | Green5 | 0.3 | AtWRKY70 * | 43.31 | 19 | 2E-23 |
| HanXRQChr16g0506841 | Green5 | 0.28 | AtWRKY6 | 54.02 | 81 | 8E-137 |
| HanXRQChr16g0508961 | Green5 | 0.26 | AtWRKY7 * | 49.72 | 95 | 1E-95 |
| HanXRQChr08g0211091 | Green5 | 0.21 | AtWRKY4 * | 53.71 | 97 | 2E-144 |
| HanXRQChr15g0480431 | Green5 / Root18 | 0.13 / 0.18 | AtWRKY28 * | 40.48 | 100 | 3E-61 |
| HanXRQChr11g0329641 | Green5 | 0.08 | AtWRKY41 * | 46.55 | 39 | 1E-29 |
| HanXRQChr10g0306731 | Green5 | 0.07 | AtWRKY1 * | 43.62 | 65 | 6E-68 |
| HanXRQChr10g0281391 | Green5 | 0.05 | AtWRKY21 | 48.11 | 100 | 2E-106 |
| HanXRQChr03g0071411 | Green18 | 0.71 | AtWRKY51 * | 53.78 | 80 | 8E-39 |
| HanXRQChr14g0460611 | Green18 / Root44 | 0.41 / 0.78 | AtWRKY70 * | 42.94 | 54 | 2E-37 |
| HanXRQChr04g0113641 | Green18 | 0.09 | AtWRKY21 | 49.62 | 100 | 5E-113 |
| HanXRQChr16g0509771 | Green24 | 0.89 | AtWRKY33 * | 46.56 | 88 | 2E-117 |
| HanXRQChr09g0274431 | Green24 / Root18 | 0.69 / 0.97 | AtWRKY40 * | 53.17 | 99 | 8E-99 |
| HanXRQChr06g0166901 | Green24 | 0.67 | AtWRKY41 * | 37.25 | 100 | 3E-46 |
| HanXRQChr16g0499381 | Green24 / Root18 | 0.64 / 0.85 | AtWRKY40 * | 51.49 | 99 | 1E-93 |
| HanXRQChr03g0084521 | Green24 | 0.37 | AtWRKY11 * | 42.01 | 91 | 2E-46 |
| HanXRQChr03g0088861 | Green24 | 0.37 | AtWRKY6 | 47.67 | 93 | 7E-132 |
| HanXRQChr05g0142161 | Green24 | 0.28 | AtWRKY11 * | 37.70 | 95 | 2E-44 |
| HanXRQChr08g0216831 | Green24 / Root44 | 0.18 / 0,24 | AtWRKY70 * | 40.19 | 63 | 4E-37 |
| HanXRQChr08g0228641 | Green39 / Root18 | 0.79 / 0.38 | AtWRKY33 * | 46.65 | 91 | 3E-113 |
| HanXRQChr09g0264011 | Green39 / Root36 | 0.31 / 0.01 | AtWRKY4 * | 43.05 | 97 | 3E-107 |
| HanXRQChr16g0505941 | Green39 / Root18 | 0.29 / 0.05 | AtWRKY7 * | 47.15 | 94 | 1E-81 |
| HanXRQChr11g0348481 | Root18 | 0.17 | AtWRKY53 * | 40.96 | 98 | 1E-74 |
| HanXRQChr17g0544771 | Root36 | 0.71 | AtWRKY72 * | 38.29 | 81 | 6E-60 |
| HanXRQChr11g0336511 | Root36 | 0.42 | AtWRKY6 | 36.49 | 94 | 1E-72 |
| HanXRQChr08g0209791 | Root44 | 0.32 | AtWRKY75 * | 92.13 | 35 | 2E-55 |
| Sunflower gene | Closest homolog in A. thaliana | |||||
|---|---|---|---|---|---|---|
| Name | Module | IMK | Homolog | Identity % | Q-cov % | E-value |
| HanXRQChr01g0014161 | Green5 | 1.0 | AT5G48380 | 60.54 | 90 | 0 |
| HanXRQChr04g0107431 | Green5/Root44 | 0.96 / 0.9 | AT1G34420 | 48.14 | 99 | 0 |
| HanXRQChr02g0045301 | Green5 | 0.82 | AT3G48090 | 38.99 | 98 | 1E-147 |
| HanXRQChr09g0248321 | Green24 / Root18 | 0.94 / 0.60 | AT5G12010 | 67.32 | 89 | 0 |
| HanXRQChr02g0040711 | Green24 | 0.89 | AT1G18740 | 60.21 | 98 | 5E-163 |
| HanXRQChr15g0496321 | Green24 | 0.89 | AT2G40140 | 51.39 | 94 | 1E-178 |
| HanXRQChr03g0086901 | Green24 | 0.88 | AT3G56880 | 34.54 | 99 | 2E-18 |
| HanXRQChr16g0504131 | Green24 | 0.86 | AT2G40140 | 51.25 | 97 | 0 |
| HanXRQChr13g0399921 | Green39 | 1 | AT3G09830 | 66.92 | 88 | 0 |
| HanXRQChr12g0366961 | Green39 | 0.81 | AT1G30755 | 49.69 | 100 | 0 |
| HanXRQChr10g0300021 | Root36 | 0.96 | AT5G01050 | 53.74 | 98 | 0 |
| HanXRQChr05g0161441 | Root36 | 0.94 | AT1G22400 | 52.50 | 98 | 0 |
| HanXRQChr07g0205991 | Root44 / Green18 | 1.0 / 0.39 | AT1G08450 | 70.12 | 95 | 0 |
| HanXRQChr17g0553831 | Root44 | 0.89 | AT5G42510 | 42.47 | 64 | 1E-38 |
| HanXRQChr01g0001251 | Root44 | 0.84 | AT3G54040 | 54.02 | 70 | 5E-58 |
| HanXRQChr04g0123531 | Root44 | 0.83 | AT3G60450 | 55.42 | 93 | 5E-93 |
2.4. Module preservation among GreenGCN and RootGCN
| Modules | Root18 | Root36 | Root44 |
|---|---|---|---|
| Green5 | 20* | 6 | 19* |
| Green16 | 1 | 0 | 0 |
| Green18 | 34* | 1 | 12* |
| Green24 | 24* | 0 | 1 |
| Green39 | 5 | 3 | 1 |
| Module | Number of shared genes | Zsummary | MedianRank |
|---|---|---|---|
| Green5 | 530 | 8.81 | 58 |
| Green16 | 186 | 3.12 | 61 |
| Green18 | 247 | 9.24 | 51 |
| Green24 | 181 | 12.67 | 39 |
| Green39 | 90 | 2.06 | 56 |
| Root18 | 209 | 8.58 | 61 |
| Root36 | 62 | 1.95 | 72 |
| Root44 | 42 | 12.65 | 10 |
2.5. Functional prediction of “unknown/uncharacterized” genes in defense modules
2.6. Functional prediction of candidate genes associated with resistance to V. dahliae and S. sclerotiorum
3. Discussion
4. Material and methods
4.1. Data acquisition
4.2. Quality Control and Mapping of Data
4.3. Co-expression network analysis
4.4. Guilt-by-Association network performance evaluation
4.5. Estimation of dN/dS ratios
4.6. GO enrichment analysis
4.7. Module-condition relationship
4.8. Module Preservation Analysis
4.9. Gene function prediction
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPPC Secretariat Scientific review of the impact of climate change on plant pests; FAO on behalf of the IPPC Secretariat, 2021; ISBN 978-92-5-134435-4.
- Maron, L.G.; Piñeros, M.A.; Kochian, L.V.; McCouch, S.R. Redefining “stress resistance genes”, and why it matters. J. Exp. Bot. 2016, 67, 5588–5591. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijevic, A.; Horn, R. Sunflower hybrid breeding: from markers to genomic selection. Front. Plant Sci. 2017, 8, 2238. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shi, H.; Yu, H.; Ma, Y.; Hu, H.; Han, Z.; Zhang, Y.; Zhen, Z.; Yi, L.; Hou, J. Combined GWAS and transcriptome analyses provide new insights into the response mechanisms of sunflower against drought stress. Front. Plant Sci. 2022, 13, 847435. [Google Scholar] [CrossRef]
- Guo, S.; Zuo, Y.; Zhang, Y.; Wu, C.; Su, W.; Jin, W.; Yu, H.; An, Y.; Li, Q. Large-scale transcriptome comparison of sunflower genes responsive to Verticillium dahliae. BMC Genomics 2017, 18, 42. [Google Scholar] [CrossRef]
- Ramu, V.S.; Paramanantham, A.; Ramegowda, V.; Mohan-Raju, B.; Udayakumar, M.; Senthil-Kumar, M. Transcriptome Analysis of Sunflower Genotypes with Contrasting Oxidative Stress Tolerance Reveals Individual- and Combined- Biotic and Abiotic Stress Tolerance Mechanisms. PLoS ONE 2016, 11, e0157522. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; Legrand, L.; Gill, N.; Kane, N.C.; Bowers, J.E.; Hubner, S.; Bellec, A.; Bérard, A.; Bergès, H.; Blanchet, N.; Boniface, M.-C.; Langlade, N.B. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef]
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef]
- Mason, C.M.; Bowsher, A.W.; Crowell, B.L.; Celoy, R.M.; Tsai, C.-J.; Donovan, L.A. Macroevolution of leaf defenses and secondary metabolites across the genus Helianthus. New Phytol. 2016, 209, 1720–1733. [Google Scholar] [CrossRef]
- Cowger, C.; Brown, J.K.M. Durability of quantitative resistance in crops: greater than we know? Annu. Rev. Phytopathol. 2019, 57, 253–277. [Google Scholar] [CrossRef]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. ; AmiGO Hub; Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Mundodi, S.; Reiser, L.; Huala, E.; Garcia-Hernandez, M.; Zhang, P.; Mueller, L.A.; Yoon, J.; Doyle, A.; Lander, G.; Moseyko, N.; Yoo, D.; Xu, I.; Zoeckler, B.; Montoya, M.; Miller, N.; Weems, D.; Rhee, S.Y. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004, 135, 745–755. [Google Scholar] [CrossRef]
- Depuydt, T.; Vandepoele, K. Multi-omics network-based functional annotation of unknown Arabidopsis genes. Plant J. 2021, 108, 1193–1212. [Google Scholar] [CrossRef]
- Di Persia, L.; Lopez, T.; Arce, A.; Milone, D.H.; Stegmayer, G. exp2GO: improving prediction of functions in the Gene Ontology with expression data. IEEE/ACM Trans Comput Biol Bioinform. [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Mahood, E.H.; Kruse, L.H.; Moghe, G.D. Machine learning: A powerful tool for gene function prediction in plants. Appl. Plant Sci. 2020, 8, e11376. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Jiang, Y.; Bergquist, T.R.; Lee, A.J.; Kacsoh, B.Z.; Crocker, A.W.; Lewis, K.A.; Georghiou, G.; Nguyen, H.N.; Hamid, M.N.; Davis, L.; Dogan, T.; Atalay, V.; Rifaioglu, A.S.; Dalkıran, A.; Cetin Atalay, R.; Zhang, C.; Hurto, R.L.; Freddolino, P.L.; Zhang, Y.; Friedberg, I. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol. 2019, 20, 244. [Google Scholar] [CrossRef]
- Mutwil, M.; Usadel, B.; Schütte, M.; Loraine, A.; Ebenhöh, O.; Persson, S. Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 2010, 152, 29–43. [Google Scholar] [CrossRef]
- Rao, X.; Dixon, R.A. Co-expression networks for plant biology: why and how. Acta Biochim Biophys Sin (Shanghai) 2019, 51, 981–988. [Google Scholar] [CrossRef]
- Montecchia, J.F.; Fass, M.I.; Cerrudo, I.; Quiroz, F.J.; Nicosia, S.; Maringolo, C.A.; Di Rienzo, J.; Troglia, C.; Hopp, H.E.; Escande, A.; González, J.; Álvarez, D.; Heinz, R.A.; Lia, V.V.; Paniego, N.B. On-field phenotypic evaluation of sunflower populations for broad-spectrum resistance to Verticillium leaf mottle and wilt. Sci. Rep. 2021, 11, 11644. [Google Scholar] [CrossRef]
- Mathew, F.; Harveson, R.; Block, C.; Gulya, T.; Ryley, M.; Thompson, S.; Markell, S. Sclerotinia diseases of sunflower. PHI 2020. [CrossRef]
- Gulya, T.; Rashid, K.Y.; Marisevic, S.M. ; Gulya K Y Rashid, S N Masirevic, T.J. Sunflower diseases. In Sunflower Technology and Production; (ed.), I. A. A. S., Ed.; ASA, CSSA, and SSSA, Madison, WI., 1997; Vol. 35, pp. 263–379, ISBN 9780891182276.
- Zubryzcki, J.; Fusari, C.; Maringolo, C.; Dirienzo, J.; Cervigni, G.; Nishinakamasu, V.; Filippi, C.; Troglia, C.; Quiroz, F.; Alvarez, D.; Escande, A.; Hopp, H.E.; Heinz, R.; Lia, V.; Paniego, N. Biparental QTL and Association Mapping for Sclerotinia head rot resistance in cultivated sunflower. In Asagir Mar Del Plata {&} Balcarce 2012; Argentina; Mar del Plata, Argentina, 2012; Vol. II.
- Filippi, C.V.; Zubrzycki, J.E.; Di Rienzo, J.A.; Quiroz, F.J.; Puebla, A.F.; Alvarez, D.; Maringolo, C.A.; Escande, A.R.; Hopp, H.E.; Heinz, R.A.; Paniego, N.B.; Lia, V.V. Unveiling the genetic basis of Sclerotinia head rot resistance in sunflower. BMC Plant Biol. 2020, 20, 322. [Google Scholar] [CrossRef]
- Talukder, Z.I.; Hulke, B.S.; Qi, L.; Scheffler, B.E.; Pegadaraju, V.; McPhee, K.; Gulya, T.J. Candidate gene association mapping of Sclerotinia stalk rot resistance in sunflower (Helianthus annuus L.) uncovers the importance of COI1 homologs. Theoretical and Applied Genetics 2014. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008, 9, 559. [Google Scholar] [CrossRef]
- Somssich, M.; Khan, G.A.; Persson, S. Cell wall heterogeneity in root development of arabidopsis. Front. Plant Sci. 2016, 7, 1242. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Filippi, C.V.; Corro Molas, A.; Dominguez, M.; Colombo, D.; Heinz, N.; Troglia, C.; Maringolo, C.; Quiroz, F.; Alvarez, D.; Lia, V.; Paniego, N. Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding. Genes (Basel) 2022, 13. [Google Scholar] [CrossRef]
- Zubrzycki, J.E.; Maringolo, C.A.; Filippi, C.V.; Quiróz, F.J.; Nishinakamasu, V.; Puebla, A.F.; Di Rienzo, J.A.; Escande, A.; Lia, V.V.; Heinz, R.A.; Hopp, H.E.; Cervigni, G.D.L.; Paniego, N.B. Main and epistatic QTL analyses for Sclerotinia Head Rot resistance in sunflower. PLoS ONE 2017, 12, e0189859. [Google Scholar] [CrossRef]
- Talukder, Z.I.; Underwood, W.; Misar, C.G.; Seiler, G.J.; Cai, X.; Li, X.; Qi, L. Genomic Insights Into Sclerotinia Basal Stalk Rot Resistance Introgressed From Wild Helianthus praecox Into Cultivated Sunflower (Helianthus annuus L.). Front. Plant Sci. 2022, 13, 840954. [Google Scholar] [CrossRef] [PubMed]
- Liesecke, F.; De Craene, J.-O.; Besseau, S.; Courdavault, V.; Clastre, M.; Vergès, V.; Papon, N.; Giglioli-Guivarc’h, N.; Glévarec, G.; Pichon, O.; Dugé de Bernonville, T. Improved gene co-expression network quality through expression dataset down-sampling and network aggregation. Sci. Rep. 2019, 9, 14431. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Zhang, L.; Yi, X.; Zhang, K.; Yao, D.; Zhang, X.; Wang, Q.; Zhao, X.; Ling, Y.; Xu, W.; Li, F.; Su, Z. Co-expression network analyses identify functional modules associated with development and stress response in Gossypium arboreum. Sci. Rep. 2016, 6, 38436. [Google Scholar] [CrossRef]
- Gupta, C.; Pereira, A. Recent advances in gene function prediction using context-specific coexpression networks in plants. [version 1; peer review: 2 approved]. F1000Res. 2019, 8. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Liu, X.; Zhou, J.; Deng, H.; Zhang, G.; Xiao, Y.; Tang, W. WGCNA Analysis Identifies the Hub Genes Related to Heat Stress in Seedling of Rice (Oryza sativa L.). Genes (Basel) 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.; Shi, H.; Hu, H.; Yi, L.; Hou, J. Time-course transcriptome and WGCNA analysis revealed the drought response mechanism of two sunflower inbred lines. PLoS ONE 2022, 17, e0265447. [Google Scholar] [CrossRef]
- Du, J.; Wang, S.; He, C.; Zhou, B.; Ruan, Y.-L.; Shou, H. Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 2017, 68, 1955–1972. [Google Scholar] [CrossRef]
- Zainal-Abidin, R.-A.; Harun, S.; Vengatharajuloo, V.; Tamizi, A.-A.; Samsulrizal, N.H. Gene Co-Expression Network Tools and Databases for Crop Improvement. Plants 2022, 11. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, Article17. [Google Scholar] [CrossRef]
- Giacomelli, J.I.; Ribichich, K.F.; Dezar, C.A.; Chan, R.L. Expression analyses indicate the involvement of sunflower WRKY transcription factors in stress responses, and phylogenetic reconstructions reveal the existence of a novel clade in the Asteraceae. Plant Sci. 2010, 178, 398–410. [Google Scholar] [CrossRef]
- Peluffo, L. Caracterización de los mecanismos de defensa a Sclerotinia sclerotiorum, agente causal de la Podredúmbre Húmeda de Girasol, a través del estudio de perfiles metabólicos y transcripcionales. Doctoral dissertation, Instituto de Biotecnología, CICVyA, INTA-Castelar, 2010.
- Filippi, C.V. Diversidad genómica y mapeo por asociación para la resistencia a la podredumbre húmeda del capítulo causada por Sclerotinia sclerotiorum en girasol. Doctoral dissertation, Instituto Nacional de Tecnología Agropecuaria (INTA) Castelar. Instituto de Biotecnología. Centro de Investigación en Ciencias Veterinarias y Agronómicas (CICVyA), 2015.
- Liu, A.; Liu, C.; Lei, H.; Wang, Z.; Zhang, M.; Yan, X.; Yang, G.; Ren, J. Phylogenetic analysis and transcriptional profiling of WRKY genes in sunflower (Helianthus annuus L.): Genetic diversity and their responses to different biotic and abiotic stresses. Industrial Crops and Products 2020, 148, 112268. [Google Scholar] [CrossRef]
- Jones, A.M.E.; Thomas, V.; Bennett, M.H.; Mansfield, J.; Grant, M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006, 142, 1603–1620. [Google Scholar] [CrossRef]
- Fass, M.I.; Rivarola, M.; Ehrenbolger, G.F.; Maringolo, C.A.; Montecchia, J.F.; Quiroz, F.; García-García, F.; Blázquez, J.D.; Hopp, H.E.; Heinz, R.A.; Paniego, N.B.; Lia, V.V. Exploring sunflower responses to Sclerotinia head rot at early stages of infection using RNA-seq analysis. Sci. Rep. 2020, 10, 13347. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Jiang, Y.; Ritchie, E.S.; Macho, A.P.; Yu, F.; Wu, D. Manipulation of plant metabolism by pathogen effectors: more than just food. FEMS Microbiol. Rev. 2023, 47. [Google Scholar] [CrossRef] [PubMed]
- Morkunas, I.; Ratajczak, L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant. 2014, 36, 1607–1619. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data.
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. BioRxiv 2014. [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Ballouz, S.; Weber, M.; Pavlidis, P.; Gillis, J. EGAD: ultra-fast functional analysis of gene networks. Bioinformatics 2017, 33, 612–614. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–12. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. 2019.
- Alexa, A.; Rahnenführer, J. Gene set enrichment analysis with topGO. 2023.
- Langfelder, P.; Horvath, S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 2007, 1, 54. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiarizadeh, M.R.; Hosseinpour, B.; Shahhoseini, M.; Korte, A.; Gifani, P. Weighted Gene Co-expression Network Analysis of Endometriosis and Identification of Functional Modules Associated With Its Main Hallmarks. Front. Genet. 2018, 9, 453. [Google Scholar] [CrossRef]
- Langfelder, P.; Luo, R.; Oldham, M.C.; Horvath, S. Is my network module preserved and reproducible? PLoS Comput. Biol. 2011, 7, e1001057. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





