1. Introduction
The presence of human-generated antibiotic resistances in surface waters is a fact of present day. Almost every resistance mechanism known from the clinical environment has been detected in rivers, lakes, oceans and even ground water [
1,
2,
3,
4].
Escherichia coli is one of the most common facultative pathogens in human medicine and is also a key factor in the study of water quality, especially in terms of fecal contamination. Studies have shown that a significant proportion of the
E. coli population found in surface water already exhibits at least one acquired resistance, and multi-resistant isolates are no longer a rarity [
3,
5,
6,
7,
8,
9,
10]. Antibiotic resistance in surface water does not appear to be just a short-term contamination from various sources. Populations in rivers and lakes are also subject to evolutionary selection. Therefore, it seems that certain strains, plasmids and resistance mechanisms are more dominant in waters than in humans or animals [
11,
12,
13]. However, knowledge about how this contributes to the problem of antimicrobial resistance is relatively limited.
The analysis of the
E. coli population bares some problems, especially in rivers. This is mainly due to the nature of the rivers, as it is difficult to distinguish between individuals that have only been in the river for a short time and those that have been there for a longer period, for example, as part of a biofilm. Biofilms are of high interest as a harborage for long-term bacterial colonizers. Unlike bacteria in open waters, bacteria in biofilms have more time to grow and have the opportunity to pass on genetic information [
14,
15]. The establishment and stabilization of antibiotic resistances in these biofilms is influenced by a number of factors that differ in their impact from those in open water [
16,
17]. Competition with other (environmental) bacteria and the burden of the additional genetic load of antibiotic resistance genes are factors that counteract the stability of the resistances. A higher concentration of toxic substances in the sediment and on other surface structures (e.g. stones) can have a positive effect on the selection of antimicrobial resistance [
18]. This does not have to happen directly due to the presence of antibiotics or their degradation products. Other substances can also contribute directly or indirectly to the stabilization of the antibiotic resistance mechanisms. In addition, the close contact of species in biofilms makes it easier to transfer genes, even to other strains or species that are much better adapted to life in the aquatic environment [
15].
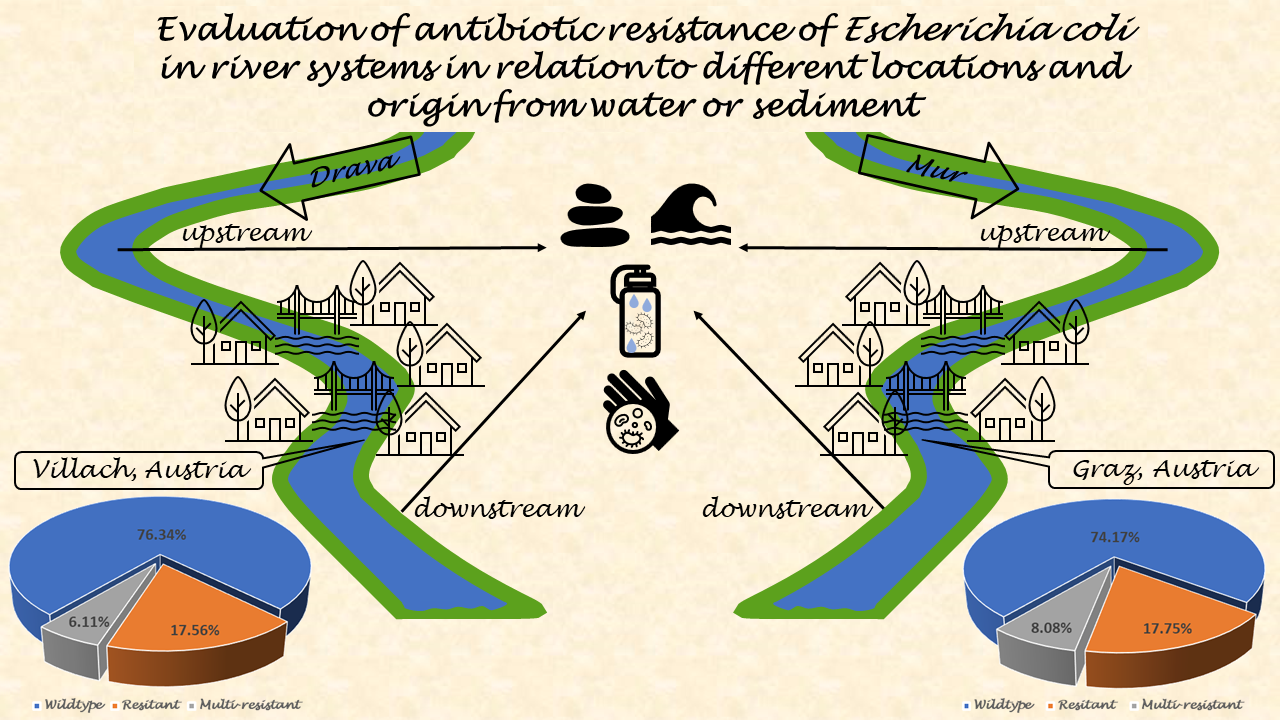
The aim of this study was to investigate the differences between antibiotic-resistant E. coli isolates from water and sediment. Two major rivers in southern Austria were chosen. Samples were taken from the river Mur, the main river in the state of Syria, where it flows through Graz. Samples from the Drava River, the main river in the state of Carinthia, were taken at its flow through Villach. E. coli were isolated from all samples, including resistant and multi-resistant isolates, and tested for their susceptibility to 21 antibiotics.
2. Results
In total, 831
E. coli were isolated from all samples of both rivers, 569 from Mur River and 262 from Drava River (
Table 1,
Supplementary Table S1).
E. coli isolates obtained from Mur water samples totaled 261. From Mur sediment samples, 308
E. coli isolates were collected. From the sampling point W0X (upstream of the WWTP) 144
E. coli were isolated from water and 113 from sediment samples. The obtained yield from the sampling point KD01 (downstream of the WWTP) was 117
E. coli from water and 195 from sediment samples (
Table 2).
E. coli isolates obtained from Drava water samples included 195
E. coli and 67
E. coli from sediment samples. From the sampling point DR01 (upstream of the WWTP), 90
E. coli were isolated from water and 33
E. coli from sediment samples. The achieved yield from the sampling point DK01 (downstream of the WWTP) was 105
E. coli from water and 34
E. coli from sediment samples (
Table 1).
Table 1.
Overview of the E. coli isolates isolated from all samples of the Mur and Drava rivers.
Table 1.
Overview of the E. coli isolates isolated from all samples of the Mur and Drava rivers.
| |
Mur water |
Mur sediment |
Sum of isolates Mur |
Drava water |
Drava sediment |
Sum of isolates Drava |
| upstream |
144 |
113 |
257 |
90 |
33 |
123 |
| downstream |
117 |
195 |
312 |
105 |
34 |
139 |
| Sum of isolates |
261 |
308 |
569 |
195 |
67 |
262 |
The
E. coli from Mur River water isolates showed a high proportion of wildtype
E. coli susceptible to all 21 tested antibiotics. Wildtype resistance was detected in 76.63% (200/261) of water isolates, while 16.48% (43/261) were resistant to one or two antibiotic classes. Only 6.90% (18/261) of the isolates were multi-resistant. (
Figure 1).
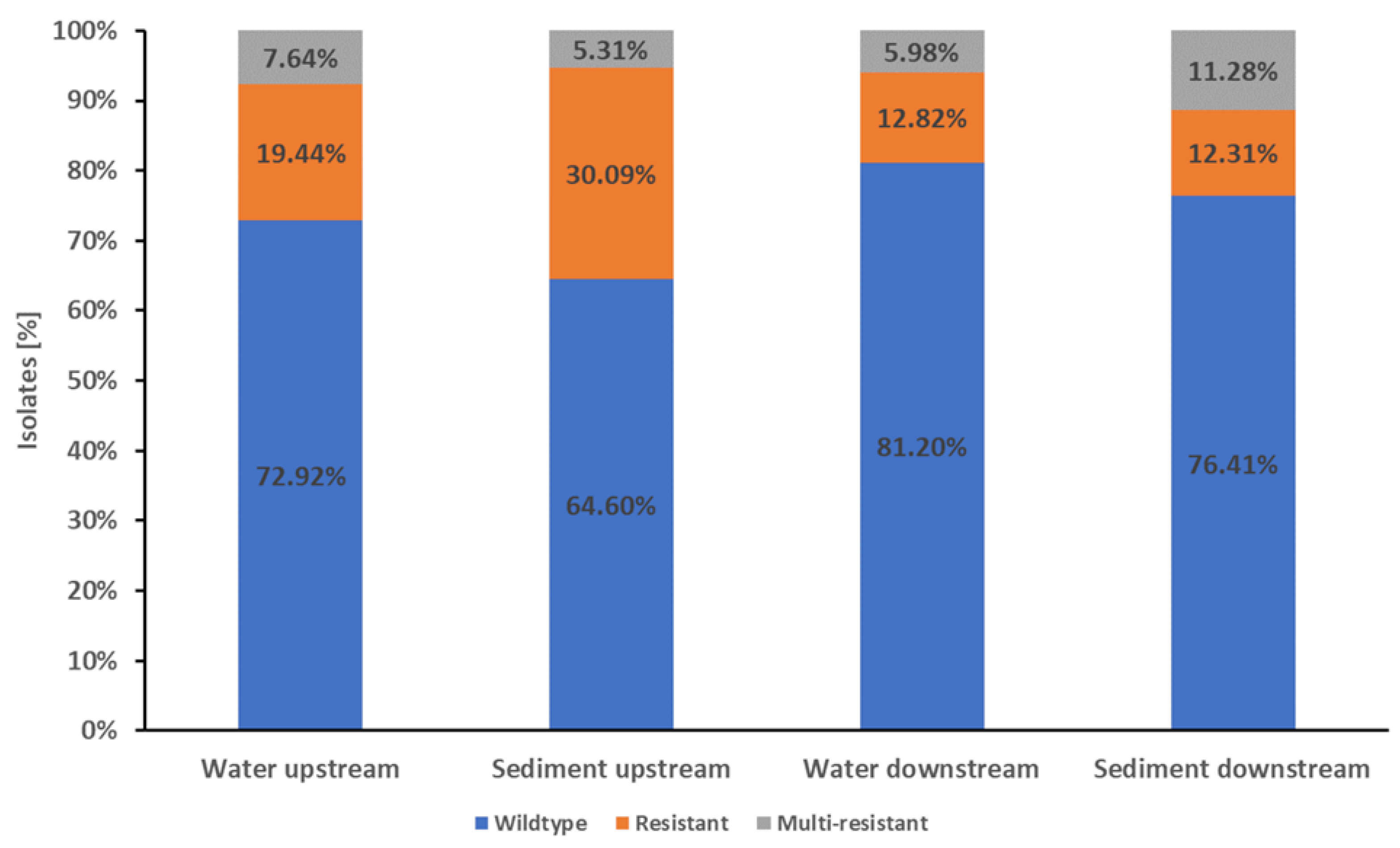
In the water samples from upstream the WWTP, 72.92% (105/144) of the isolates were wildtype, 19.44% (28/144) were resistant to one or two antibiotic classes, and 7.64% (11/144) were multi-resistant (
Figure 1). Of the isolates from downstream the WWTP, 81.20% (95/117) showed wildtype resistance, 12.82% (15/117) showed a resistance to one or two antibiotic classes, and 5.98% (7/117) were multi-resistant (
Figure 1).
The differences in resistance patterns of Mur isolates from up- and downstream were not significant (p-value ≥0.05).
In the Mur sediment samples, 72.1% (222/308) of the isolates were wildtype, 18.83% (58/308) were resistant and 11.07% (28/308) were multi-resistant (
Figure 1).
The isolates from the Mur sediment samples from upstream the WWTP showed 64.60% (73/113) wildtype characteristics, with 30.09% (34/113) of the isolates resistant to one or two antibiotic classes and 5.31% (6/113) being multi-resistant. In the sediment samples from downstream the WWTP, 76.41% (149/195) of the isolates were wildtype, 12.31% (24/195) were resistant to one or two antibiotic classes and 11.28% (22/195) were multi-resistant (
Figure 1).
The proportion of wildtype isolates was significant higher (p-value <0.05) in the downstream samples compared the upstream samples. Conversely, the proportion of isolates resistant to one or two antibiotic classes was significant higher (p-value <0.05) in the upstream samples. The sediment samples from downstream had a higher proportion of multi-resistant isolates than the upstream samples. However, this difference in multi-resistance was not significant (p-value ≥0.05).
There were no significant differences in resistance patterns of E. coli between combined water and combined sediment samples from the Mur River (p-value ≥0.05).
Highlighting the most important species
E. coli and
K. pneumoniae in comparison (
Figure 1) resistance patterns show a high similarity with the exception of the ß-Lactamase inhibitors and combinations (AMC, TZP) and GM. Generally,
E. coli showed a lower resistant proportion in tested isolates.
Figure 1.
Proportions of wildtype, resistance and multi-resistance in E. coli Mur River isolates. The stapled columns from left to right represent the proportions of isolates showing the respective phenotype in the water and sediment samples from upstream and downstream of Graz and its WWTP. Isolates were classified as wildtype (indicated as blue part of columns) when showing no resistance to the tested antibiotics. Isolates with resistance to one or two classes of the tested antibiotics were classified as resistant (indicated as orange part of columns). Resistance to three or more classes of the tested antibiotics was classified as multi-resistant (indicated as grey part of columns).
Figure 1.
Proportions of wildtype, resistance and multi-resistance in E. coli Mur River isolates. The stapled columns from left to right represent the proportions of isolates showing the respective phenotype in the water and sediment samples from upstream and downstream of Graz and its WWTP. Isolates were classified as wildtype (indicated as blue part of columns) when showing no resistance to the tested antibiotics. Isolates with resistance to one or two classes of the tested antibiotics were classified as resistant (indicated as orange part of columns). Resistance to three or more classes of the tested antibiotics was classified as multi-resistant (indicated as grey part of columns).
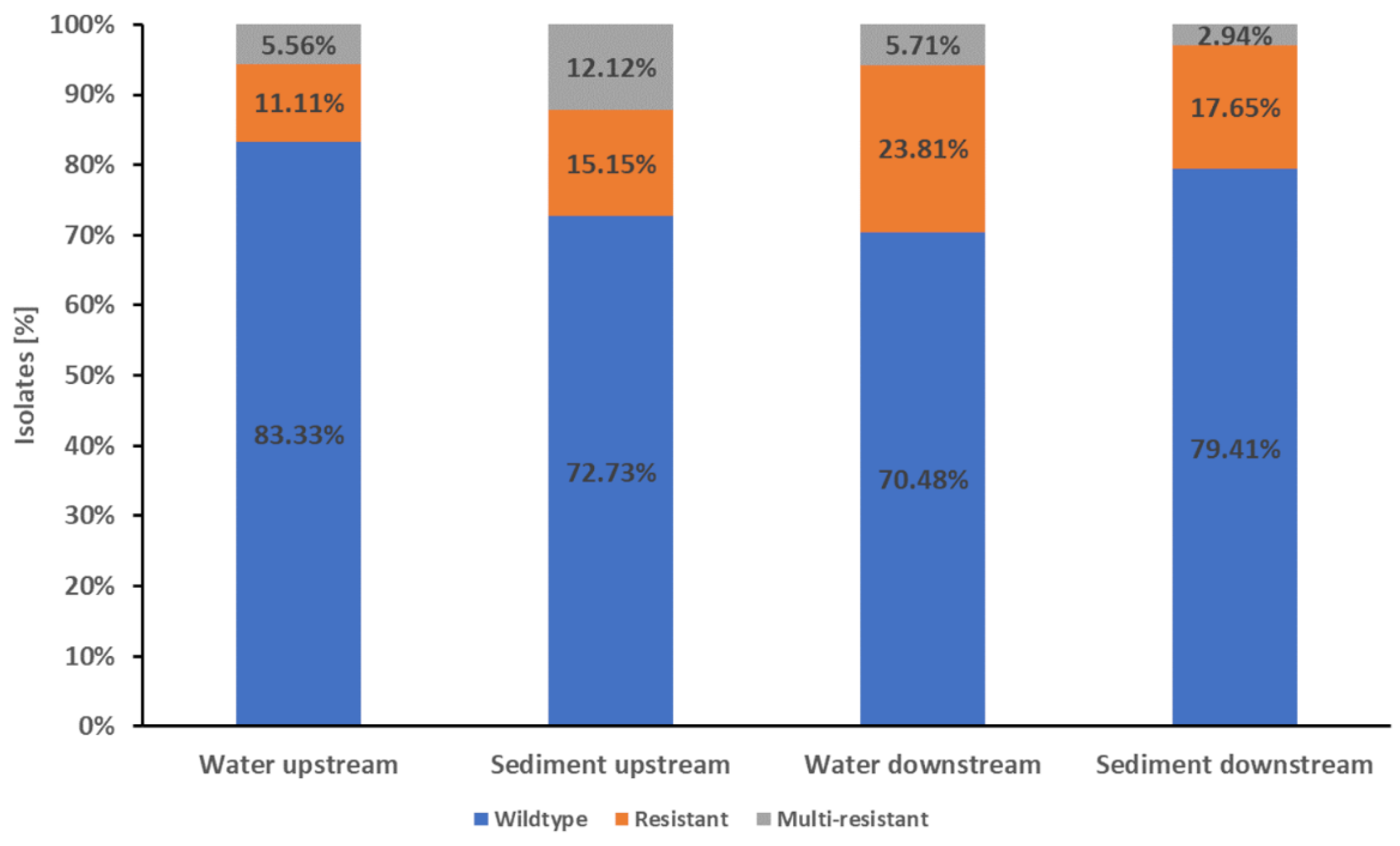
The water isolates from the Drava Rivers showed that 76.41% (149/195)had a wildtype resistance pattern, while 17.95% (35/195) were resistant to one or two antibiotic classes and 5.64% (11/195) were multi-resistant (
Figure 2).
The water samples from upstream the WWTP showed 83.33% (75/90) of wildtype
E. coli, while 11.11% (10/90) of the isolates were resistant to one or two antibiotic classes. The multi-resistant isolates reached a proportion of 5.56% (5/90). The water isolates from downstream the WWTP showed a wildtype resistance pattern of 70.48% (74/105), while 23.81% (25/105) of the isolates were resistant to one or two antibiotic classes and 5.71% (6/105) were multi-resistant (
Figure 2).
Thus, the water isolates from upstream showed a significant higher proportion of isolates with wildtype characteristics compared to downstream (83.33% vs. 70.48%, p-value <0.05). However, the downstream samples showed a significant higher proportion of resistant isolates than the upstream samples (23.81%vs. 11.11%, p-value <0.05).
The proportion of multi-resistant isolates was similar in the up- and downstream samples. In total, about three quarters (76.12%, 51/67) of the isolates from Drava sediment samples showed wildtype characteristics while 16.42% (11/67) were resistant. The proportion of multi-resistant isolates was 7.46% (5/67) (
Figure 2).
Figure 2.
Proportions of wildtype, resistance and multi-resistance in E. coli Drava River isolates. The stapled columns from left to right represent the proportions of isolates showing the respective phenotype in the water and sediment samples from upstream downstream of Villach and its WWTP. Isolates were classified as wildtype (indicated as blue part of columns) when showing no resistance to the tested antibiotics. Isolates with resistance to one or two classes of the tested antibiotics were classified as resistant (indicated as orange part of columns). Resistance to three or more classes of the tested antibiotics was classified as multi-resistant (indicated as grey part of columns).
Figure 2.
Proportions of wildtype, resistance and multi-resistance in E. coli Drava River isolates. The stapled columns from left to right represent the proportions of isolates showing the respective phenotype in the water and sediment samples from upstream downstream of Villach and its WWTP. Isolates were classified as wildtype (indicated as blue part of columns) when showing no resistance to the tested antibiotics. Isolates with resistance to one or two classes of the tested antibiotics were classified as resistant (indicated as orange part of columns). Resistance to three or more classes of the tested antibiotics was classified as multi-resistant (indicated as grey part of columns).
The isolates of sediment samples from upstream the WWTP showed 72.73% (24/33) with wildtype characteristics, while 15.15% (5/33) were resistant to one or two antibiotic classes and 12.12% (4/33) were multi-resistant. The sediment samples from downstream the WWTP showed 79.41% (27/34) wildtype isolates, while 17.65% (6/37) were resistant and 2.94% (1/37) were multi-resistant (
Figure 2). None of the differences in resistance patterns were significant (p-value ≥0.05).
There were no significant differences between the combined water and combined sediment samples from the Drava River (p-value ≥0.05).
The most common resistance in
E. coli water and sediment isolates in both rivers was the resistance to the ß-lactam antibiotic ampicillin. The highest number of ampicillin resistant isolates was discovered in the Mur sediment isolates with a proportion of 15.26% (47/308) (Table 3,
Supplementary Tables S2–S5).
The greatest differences in the proportion of resistance were found between the Mur water and sediment isolates with regard to resistance to nalidixic acid. Of the Mur sediment isolates, 15.91% (49/308) were resistant to nalidixic acid, compared to 10.34% (27/261) of the water isolates. In the Drava, a high proportion of isolates were also resistant to nalidixic acid: 9.23% (18/195) of the water isolates were resistant to the antibiotic, in contrast to only 5.97% (4/67) of the sediment isolate samples (
Table 2,
Supplementary Tables S2–S5).
When the resistances to nalidixic acid in the E. coli isolates from the two different rivers are compared, the proportion distribution in the water and sediment isolates is exactly the opposite. The Drava water isolates showed a higher proportion of resistance to nalidixic acid than the Drava sediment isolates. Conversely, the results from the Mur showed a higher proportion of isolates resistant to nalidixic acid in sediment than in water samples.
A high proportion of isolates resistant to tetracycline was detected in the Mur: 10.34% (27/261) of the water isolates and 10.39% (32/308) of the sediment isolates (
Table 2).
In addition, a resistance to ciprofloxacin was strongly detected in Drava sediment isolates. In the Dur samples, 8.96% (6/67) of the isolates were resistant to the fluoroquinolone, while the Mur sediment sample’s isolates reached only 5.84% (18/308) (
Table 2).
Resistance to aminoglycosides (amikacin and gentamycin) was rare in both rivers, but one isolate from the Drava showed a resistance to the last line antibiotic amikacin. All isolates were susceptible to colistin, tigecycline and carbapenems, with one exception: one isolate from the Mur sediment showed resistance to meropenem and imipenem (
Table 2).
Table 2.
Antibiotic resistances of E. coli isolates in comparison between water and sediment samples from the rivers Mur and Drava: The proportions and, in parentheses, the number of isolates resistant to each antibiotic. All classes of antimicrobial resistance are given with the corresponding p-values of the statistical tests. Isolates were classified as wildtype when showing no resistance to the tested antibiotics. Isolates with resistance to one or two classes of the tested antibiotics were classified as resistant. Resistance to three or more classes of the tested antibiotics was classified as multi-resistant. P-values <0.05 were considered statistically significant. P-values with more than four decimal places containing a value of nine were rounded to one.
Table 2.
Antibiotic resistances of E. coli isolates in comparison between water and sediment samples from the rivers Mur and Drava: The proportions and, in parentheses, the number of isolates resistant to each antibiotic. All classes of antimicrobial resistance are given with the corresponding p-values of the statistical tests. Isolates were classified as wildtype when showing no resistance to the tested antibiotics. Isolates with resistance to one or two classes of the tested antibiotics were classified as resistant. Resistance to three or more classes of the tested antibiotics was classified as multi-resistant. P-values <0.05 were considered statistically significant. P-values with more than four decimal places containing a value of nine were rounded to one.
| |
Mur water |
Mur sediment |
p-value |
Drava water |
Drava sediment |
p-value |
| |
(261 isolates) |
(308 isolates) |
|
(195 isolates) |
(67 isolates) |
|
| β-Lactams |
|
|
|
|
|
|
| Ampicillin |
10.73% (28) |
15.26% (47) |
0.14 |
14.87% (29) |
14.93% (10) |
1 |
| Amoxicillin/clavulanic acid |
3.83% (10) |
5.19% (16) |
0.55 |
11.28% (22) |
13.43% (9) |
0.66 |
| Cefalexin |
3.45% (9) |
3.25% (10) |
1 |
2.56% (5) |
0% (0) |
0.33 |
| Cefuroxime |
3.07% (8) |
2.92% (9) |
1 |
2.05% (4) |
0% (0) |
0.58 |
| Cefoxitin |
1.15% (3) |
0.97% (3) |
1 |
1.03% (2) |
0% (0) |
1 |
| Cefotaxime |
2.68% (7) |
2.92% (9) |
1 |
1.54% (3) |
0% (0) |
0.57 |
| Piperacillin/tazobactam |
1.53% (4) |
1.62% (5) |
1 |
0% (0) |
2.99% (2) |
0.06 |
| Ceftazidime |
2.3% (6) |
2.6% (8) |
1 |
1.03% (2) |
0% (0) |
1 |
| Cefepime |
2.68% (7) |
3.25% (10) |
0.81 |
1.54% (3) |
0% (0) |
0.57 |
| Imipenem |
0% (0) |
0.32% (1) |
1 |
0% (0) |
0% (0) |
1 |
| Meropenem |
0% (0) |
0.32% (1) |
1 |
0% (0) |
0% (0) |
1 |
| Quinolones |
| Moxifloxacin |
5.75% (15) |
4.22% (13) |
0.44 |
5.64% (11) |
2.99% (2) |
0.53 |
| Ciprofloxacin |
6.13% (16) |
5.84% (18) |
1 |
6.15% (12) |
8.96% (6) |
0.41 |
| Nalidixic acid |
10.34% (27) |
15.91% (49) |
0.06 |
9.23% (18) |
5.97% (4) |
0.61 |
| Tetracyclines |
|
|
|
|
|
|
| Tetracycline |
10.34% (27) |
10.39% (32) |
1 |
6.67% (13) |
1.49% (1) |
0.13 |
| Tigecycline |
0% (0) |
0% (0) |
1 |
0% (0) |
0% (0) |
1 |
| Aminoglycosides |
|
|
|
|
|
|
| Gentamicin |
0.38% (1) |
2.6% (8) |
0.04 |
0.51% (1) |
2.99% (2) |
0.16 |
| Amikacin |
0% (0) |
0% (0) |
1 |
0.51% (1) |
0% (0) |
1 |
| Antifolate |
|
|
|
|
|
|
| Trimethoprim/sulfamethoxazole |
8.43% (22) |
6.82% (21) |
0.53 |
9.74% (19) |
5.97% (4) |
0.46 |
| Polymyxins |
|
|
|
|
|
|
| Colistin |
0% (0) |
0% (0) |
1 |
0% (0) |
0% (0) |
1 |
| Chloramphenicols |
|
|
|
|
|
|
| Chloramphenicol |
2.3% (6) |
3.25% (10) |
0.61 |
2.05% (4) |
2.99% (2) |
0.65 |
Phenotypic differentiation of all isolates by evaluation of metabolic reactions was performed using the PhenePlate (PhP) system.
For the Mur River, differentiation resulted in 87 PhP clusters consisting of 75.92% (432/569) of all isolates, and 24.08% (137/569) singletons (
Supplementary Table S1).
Over half the clusters, 55 of the 87, occurred in more than one sample. Fifteen of these clusters had only members from upstream water and sediment, and 23 clusters consisted exclusively of downstream isolates. Three clusters were water isolates only (up and downstream), and one consisted of sediment isolates only. There were 11 clusters with members from three samples, and two clusters (M-14 and M-18) had members from all four Mur samples (
Supplementary Table S1).
Cluster M-18 was also the largest Mur cluster with 27 isolates. In total, nine clusters consisted of ten or more isolates, and all of them had members of at least two different samples (always with a water and a sediment sample) (
Supplementary Table S1).
For the Drava River differentiation resulted in 33 clusters, including 75.19% (197/262) of all isolates, and 65 (24.8%) singletons.
29 of the 33 clusters occurred in more than one sample. Five of these clusters had only members from upstream water and sediment, and another five clusters only members from downstream the WWTP. Seven clusters consisted of water isolates only (up and downstream), 11 clusters had members of three samples, and one cluster (D-01) had members from all Drava samples (
Supplementary Table S1).
With a total of seven isolates, cluster D-01 was the smallest by far. Six clusters consisted of at least 10 isolates. Only one of these large clusters, D-27, consisted exclusively of isolates from the downstream sediment, while the other five clusters included isolates from more than one sample. The largest cluster was D-30 with 45 total isolates, all from upstream. Therefore, the influence of clusters D-27 and D-30 must be given special consideration in the further analyses of diversity (
Supplementary Table S1).
The overall diversity of the Mur River isolates was 2.33 isolates per cluster, and 2.7 isolates per cluster for the Drava River.
The diversity between the water samples was lower (i.e., more isolates per cluster) at2.05 (Mur) and 2.50 (Drava) than in the sediment samples, which were 1.75 (Mur) and 1.67 (Drava). Also, in both rivers the upstream population had a lower diversity than the downstream one. In this comparison, the highest difference was detected between upstream Mur isolates with 2.82 and downstream isolates with only 1.75. The isolates from Drava River revealed 2.51 isolates per cluster upstream and 2.09 downstream (Table 3 and
Table 4).
Perhaps due to the relatively small number of Drava sediment isolates, including a large cluster of 12 isolates (D-27), this sample set does not follow the general trend and these values should therefore be taken with caution (
Table 4).
Table 4.
Average number of isolates per cluster (including singletons) for the sample set from the Mur River.
Table 4.
Average number of isolates per cluster (including singletons) for the sample set from the Mur River.
| Mur River |
Water isolates |
Sediment isolates |
Total isolates |
| Upstream |
2.25 |
2.22 |
2.82 |
| Downstream |
1.60 |
1.37 |
1.75 |
| Total course |
2.06 |
1.75 |
2.33 |
Table 5.
Average number of isolates per cluster (including singletons) for the sample set from the Drava River.
Table 5.
Average number of isolates per cluster (including singletons) for the sample set from the Drava River.
| Drava River |
Water isolates |
Sediment isolates |
Total isolates |
| Upstream |
2.50 |
1.27 |
2.51 |
| Downstream |
1.81 |
1.95 |
2.09 |
| Total coures |
2.50 |
1.67 |
2.70 |
In total, 13 E. coli isolates with a presumptive ESBL phenotype according to susceptibility testing were confirmed via CLSI-test as ESBL. These ESBL-isolates were present in all four sampling locations in the water samples, but could only be isolated from sediment samples from the Mur River. In addition, one of these E. coli isolates (KD01EC110) revealed resistance to imipenem and meropenem. After a Rosco test, it was suspected of being a KPC-producer (Table 5).
The genes of these phenotypes were then analyzed by sequencing. Detected ESBL genes were eight blaCTX-M-15, three blaCTX-M-1, one blaCTX-M-14 and one blaSHV-12. Genes for the non ESBL-ß-lactamase TEM-1 were detected in six E. coli isolates.
The carbapenemase
blaKPC-2 was genetically confirmed in the isolate KD01EC110 in combination with
blaCTX-M-1 and
blaTEM-1.
Table 6.
Detected resistance genes and resistance patterns of ESBL and KPC harboring E. coli isolates.
Table 6.
Detected resistance genes and resistance patterns of ESBL and KPC harboring E. coli isolates.
| Isolate ID |
Origin |
PHP-cluster |
Resistance Genes |
Resistance Pattern1
|
| DK01EC050 |
water |
M-20 |
blaCTX-M-1
|
AM, CN, CXM, CTX, FEP |
| DR01EC012 |
water |
M-31 |
blaSHV-12, blaTEM-1
|
AM, AMC, CN, CXM, FOX , CAZ, GM, MXF, CIP, NA, SXT, TE, C |
| DR01EC036 |
water |
M-31 |
blaCTX-M-15
|
AM, AMC, CN, CXM, CTX, CAZ, FEP, TE |
| KD01EC006 |
sediment |
Singlt. |
blaCTX-M-14
|
AM, AMC, CN, CXM, CTX, FEP, MXF, CIP , NA, SXT |
| KD01EC110 |
sediment |
Singlt. |
blaCTX-M-1, blaTEM-1, blaKPC-2
|
AM, AMC, CN, CXM, FOX, CTX, TZP, CAZ, FEP, MEM, IPM, MXF, CIP, NA, C |
| KD01EC112 |
sediment |
M-42 |
blaCTX-M-15,blaTEM-1
|
AM, AMC, CN, CXM, CTX, CAZ, FEP, MXF, CIP, NA |
| W04EC016 |
sediment |
M-18 |
blaCTX-M-15
|
AM,CN, CXM, CTX, CAZ, FEP, NA, SXT, TE |
| W04EC018 |
sediment |
M-18 |
blaCTX-M-15,blaTEM-1
|
AM, AMC, CN, CXM, CTX, CAZ, FEP, NA, SXT, TE |
| W04EC029 |
sediment |
M-18 |
blaCTX-M-15
|
AM, CN, FOX, CTX, CAZ, FEP, NA, SXT, TE |
| W04EC057 |
sediment |
Singlt. |
blaCTX-M-1
|
AM, CN, CXM, CTX, CAZ,FB, TE |
| W04EC088 |
water |
M-18 |
blaCTX-M-15,blaTEM-1
|
AM, CN, CXM, CTX, CAZ FEP, NA, SXT, TE |
| W04EC090 |
water |
M-18 |
blaCTX-M-15
|
AM, CN, CXM, CTX, CAZ FEP, NA, SXT, , TE |
| W04EC093 |
water |
M-18 |
blaCTX-M-15
|
AM, CN, CXM, CTX, CAZ FEP, NA, SXT, TE |
3. Discussion
Many studies show that human induced resistances can be found in nearly every kind of environment [
19]. Rivers and lakes seem to have a key role, as the waste waters from cities and hospitals eventually flows into public waters. It was previously described that river sediments seem to play a role in the prolonged persistence of various bacteria and resistance mechanisms, more so than in open river water [
9,
17,
20,
21]. In the current study, it could be shown that three quarters of the tested
E. coli Mur River (water and sediment) isolates were susceptible to all tested antibiotics. In comparison with studies done in other locations, the proportion of resistant and multi-resistant
E. coli isolates in the Drava River and the Mur Riverways are low [
5,
6,
22,
23,
24,
25]. This could be related to the fact that the Mur and Drava flow through only a few major cities, and that waste-water treatment processes are well conducted in Austria.
It was somewhat surprising to discover the presence of resistant and especially multi-drug resistant E. coli in the area downstream of the WWT did not differ from the sections taken from upstream the river near a treatment plant. Only the water from the Drava River had a significantly higher proportion of E. coli resistant to one or two antibiotic classes with the additional city flow through Villach and its WWTP, but no resistance to a specific antibiotic was significantly increased.
The samples were intentionally not taken from the wastewater stream, but were intended to show the general and long-term effects of the discharge from upstream located WWTPs. These results are in line with other studies that shows the influence of wastewater treatment plants tends to have a significant impact typically on rare resistance mechanism (like carbapenemases) only in the area of the direct wastewater plume, unless the river is small and/or largely untouched by human influence prior to wastewater inflow [
5,
6,
7,
26,
27,
28].
However, no significant differences in the resistance patterns in sediment and river water samples could be found. This could be due to the contamination of the sediment samples by river water, which unfortunately cannot be avoided during sampling. Also, it is possible that many of the E. coli populations in the open water derive from the sediment and biofilm and only a small proportion of them represent freshly introduced E. coli (as could be seen from the comparison of before and after the treatment plants). These two factors may be reasons why the sediment samples have shown a slightly higher variability in phenotypic clustering as well. Overall, the phenotypic relationship analysis shows, apart from one case, that there is no strong clustering of a clone in either sample material. This high variability strengthens the statements about the resistance data obtained.
One observation is still worth mentioning, especially since it was not the result of the study planning, but of the given condition of the course of the two rivers: the E. coli sediment samples before and after Villach did not differ significantly in their antibiotic resistance patterns. In the Mur, however, the isolates from the sediment samples downstream of Graz and its WWTP were significantly more susceptible to the antibiotics tested. This sediment sample from the Mur River was the only one that was collected from an area without a power station in the immediate area, so the river is not affected by its flow velocity. The sinking, or rather the lack of sinking, of particles from the river due to a damming (and thus reduced introduction of toxic substances) could be the cause of this finding. However, due to the design of the study and the number of sampling points, this is pure speculation.
In this study, ESBL and KPC harboring
E. coli isolates were detected without using any kind of selective media with added antibiotics in culture. The KPC-2 harboring isolate found was the first
E. coli with these resistance mechanisms to be isolated from the environment in Austria and the first KPC expressing isolate ever found in a river in Austria [
29]. In previous studies of river water and sediment isolates, selective media were always used in order to detect these kinds of resistance genes in different bacteria. Furthermore, in parallel isolation without antibiotics, the same studies failed to isolate carbapenem-resistant
E. coli [
5,
21,
30,
31,
32]. This is particularly interesting, as clinical studies for Austria also show that carbapenemase-producing
E. coli are extremely rare. However, it is possible that KPC-forming Enterobacteriaceae are better able to persist in sediment (or in the biofilm found there) than was previously suspected, or that the colonization of the normal healthy population is much higher than the clinical data suggests [
33,
34].
4. Conclusions
The study did not reveal a significant impact of the sample material, whether water or sediment, don the observed outcomes. The influence of the sampling location showed some discernible differences, albeit not as substantial as initially expected. It is worth noting that the relatively low proportion of (multi-) resistant E. coli isolates, in comparison to previous studies, was overshadowed by the identification of a KPC-2-producing isolate, which raises concerns regarding the presence of highly resistant strains.
These findings provide valuable insights into the dissemination of resistant isolates within the environment and highlight the potential risk of their long-term establishment as permanent inhabitants of aquatic ecosystems. The data contributes to our understanding of the spread of antibiotic resistance and emphasizes the need for continued vigilance and preventive measures to address this issue effectively.
Materials and Methods
Water samples were collected in 500 ml sterile plastic flasks (VWR International, Austria) 30 cm below the river surface and 50 cm from the bank. Sediment samples were lifted from the riverbed using a paddle and packed into sterile homogenizing bags (BagLight® HD PolySilk®, Interscience, France). Further samples were scraped from stones and packed in sterile bags.
On November 24, 2016, three water and three sediment samples were taken near the Weinzödlbrücke (W0X) upstream of Graz and the municipal wastewater treatment plant. On December 12, 2016, three water and three sediment samples were taken in Kalsdorf (KD01) downstream of Graz and the WWTP. On April 3, 2017, three water and three sediment samples were taken from the Drava River at Rennsteinerstraße (DR01) upstream of Villach and the WWTPthere. On April 3, 2017, three water and three sediment samples were taken from the Drava River at Klampfererweg (DK01) downstream of Villach and its WWTP (Table 5).
Table 5.
List of sampling sites including sampling date, site name, geographic name and coordinates.
Table 5.
List of sampling sites including sampling date, site name, geographic name and coordinates.
| Sampling date |
Sample name |
Location |
Coordinates |
| 2016-11-24 |
W0X |
Graz, Weinzödlbrücke |
47°06'30.3"N 15°23'25.4"E |
| 2016-12-21 |
KD01 |
Kalsdorf |
46°58'01.7"N 15°29'24.6"E |
| 2017-03-04 |
DR01 |
Villach, Rennsteinerstraße |
46°38'30.7"N 13°48'21.1"E |
| 2017-03-04 |
DK01 |
Villach, Klampfererweg |
46°36'46.0"N 13°55'16.3"E |
The water samples were filtered using a pump (EZ-Stream™ Pump, Merck KGaA, Germany) and sterile membrane filters (EZ-Pak® mixed cellulose ester filters, 47 mm, 0.45 µm, Millipore S.A.S 67120, Austria). 100 ml from each sample were filtered into two portions of 50 ml each and placed on COL plates (Chromo Cult Coliform Agar, MERCK, Austria). Additionally, 5 ml in 500 µl portions of the water samples were plated directly onto ten COL plates. For each of the three samples from each sampling point, this procedure was performed separately. The COL plates were incubated at 42°C for 24 hours (h). All presumptive E. coli colonies (according to the manufacture’s manual) were picked with sterile inoculating loops, transferred onto blood agar (COL-S, BD BBL Stacker Plates, Germany) and incubated for 24 hours at 37°C. Confirmed E. coli were stored at -70°C in bacterial storage flasks (mWE medical wire, Viabank®, England).
Sediment and stone samples were diluted 1:10 (1 g sediment + 9 ml Ringer Tween solution 0.3%) in 50 ml sterile plastic tubes (Greiner, bio-One, Austria) with 0.3% Ringer Tween (TWEEN 80, Amresco, VWR Austria) solution and incubated at current river temperatures for one hour on a roll mixer (CATRM5, servoLAB, Austria). 500 µl of the diluted samples were then plated on COL agar plates. After incubation, the colonies were isolated and stored using the same procedure used for the water samples.
Species identification of the isolates was performed by MALDI-TOF (Vitek® MS, bioMérieux Austria GmbH, Austria).
Susceptibility testing was performed for all
E. coli isolates as recommended by the European Committee on Antimicrobial Susceptibility testing (EUCAST) for 21 antibiotics [
35]. Tetracycline, chloramphenicol and nalidixic acid tests were carried out according to the Clinical Laboratory Standards Institute (CLSI) [
36]. Interpretation of zone diameters was done according to EUCAST or CLSI. To determine (clinical) resistance to colistin, protocols by Gales et al. and Boyen et al. were used [
37,
38].
The susceptibility testing was performed with ampicillin (10 µg), amoxicillin/clavulanic acid (20/10 µg), cefalexin (30 µg), cefuroxime (30 µg), cefoxitin (30 µg), cefotaxime (5 µg), piperacillin/tazobactam (30/6 µg), ceftazidime (10 µg), cefepime (30 µg), imipenem (10 µg), meropenem (10 µg), moxifloxacin (5 µg), ciprofloxacin (5 µg), nalidixic acid (30 µg), tetracycline (30 µg), tigecycline (15 µg), gentamicin (10 µg), amikacin (30 µg), trimethoprim/sulfamethoxazole (1,25/23,75 µg), colistin (10 µg) and chloramphenicol (30 µg) and BD BBLTM Sensi-DiscTM paper discs (Becton Dickinson, Austria).
E. coli ATCC 25922 was used as control strain in all performed tests.
E. coli susceptible to all antibiotics tested were classified as wildtype (WT), isolates with resistance to one or two classes of antibiotics were labeled as resistant, and isolates resistant to three or more classes of antibiotics were considered multi-resistant.
The minimum inhibitory concentrations (MICs) for imipenem and meropenem were determined using Etest® (bioMérieux Austria GmbH, Vienna, Austria) for all isolates showing resistance to at least one of the tested carbapenems. The expression of carbapenemases was confirmed with the modified Hodge Test.
Presumptive extended spectrum beta-lactamases (ESBL) were confirmed using the double disc tests according to CLSI (30 μg ceftazidime, 30 μg cefepime, 30/10 μg ceftazidime-clavulanic acid and 30/10 μg cefepime-clavulanic acid; bioMérieux Austria GmbH, Vienna, Austria). KPC/MBL and OXA-48 Confirm Kit: Carbapenemases (Rosco Diagnostica, Taastrup, Denmark) were used to determinate the type of carbapenemases. Isolates that revealed an ESBL and/or carbapenemase phenotype were screened for antimicrobial resistance genes.
PCR detection and gene identification were performed for different β-lactamase gene families,
blaCTX-M-1-group, blaCTX-M-2 -group, blaCTX-M-9-group,
blaGES,
blaSHV,
blaTEM and
blaVEB. PCR and sequencing procedures were performed as previously described and carried out for all isolates that showed an ESBL-positive phenotype [
39,
40,
41,
42]. For confirmation of
blaKPC, PCR and sequencing protocols were used as previously described [
43].
To reveal the relationship between E. coli isolates, a biochemical fingerprint method (PhenePlate AB, Sweden) was used according to the manufactural protocol. The OD620 was measured after 16 hours and 24 hours (Zenyth 3100 Multimode Detector, Anthos Mikrosysteme GmbH, Germany). Analyses were performed using the PhenePlate software (PhPlate AB). Isolates with an identification level of 97.5% or higher were grouped in PHP-clusters and considered as clones. To calculate the diversity level, the number of isolates in one sample set was divided by the number of clusters (singletons count as one cluster with one member).
Statistical analyses were conducted with IBM SPSS Statistics 27.0.1.0. To determine the p-values, Chi-square-tests (Fisher`s exact test) were performed. P-values less than 0.05 were considered statistically significant.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: List of all Isolates; Table S2: Antibiotic resistances of E. coli Mur River water and sediment isolates in comparison between upstream and downstream of the WWTP.; Table S3: Antibiotic resistances of E. coli Drava River water and sediment isolates in comparison between upstream and downstream of the WWTP.; Table S4: Antibiotic resistances of E. coli Mur River isolates from upstream and downstream of the WWTP in comparison between water and sediment; Table S5 Antibiotic resistances of E. coli Drava River isolates from upstream and downstream of the WWTP in comparison between water and sediment.
Author Contributions
Conceptualization, C.K. and G.Z.; methodology, S.K., M.K., R.B., C.K. and G.Z.; software, S.K., M.K., R.B., J.H., F.T. and G.Z..; validation, S.K., M.K. and G.Z.; formal analysis, S.K., M.K., C.K. and G.Z..; investigation, S.K., M.K., R.B., J.H., F.T., C.K. and G.Z.; resources, F.T., C.K. and G.Z.; data curation, S.K., M.K., F.T. and G.Z.; writing—original draft preparation, S.K., M.K. and G.Z.; writing—review and editing, F.T., C.K. and G.Z.; visualization, S.K., M.K. and F.T..; supervision, C.K. and G.Z.; project administration, G.Z.; funding acquisition, G.Z.. All authors have read and agreed to the published version of the manuscript. S.K., M.K., R.B., J.H., F.T., C.K. and G.Z.
Funding
This work was funded by grant P32464 from the Austrian Science Fund (FWF).
Institutional Review Board Statement
“Not applicable” for studies not involving humans or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data can be obtained from the corresponding author upon reasonable request.
Acknowledgments
We would like to thank Sara Parenteaus for the text proofreading.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Bürgmann, H. Increased Levels of Multiresistant Bacteria and Resistance Genes after Wastewater Treatment and Their Dissemination into Lake Geneva, Switzerland. Front. Microbio. 2012, 3, doi:10.3389/fmicb.2012.00106.
- Kummerer, K. Resistance in the Environment. Journal of Antimicrobial Chemotherapy 2004, 54, 311–320, doi:10.1093/jac/dkh325.
- Singh, Ts.; Tsering, D.; Poonia, S. Antibiotic Susceptibility Profile of Bacteria Isolated from Natural Sources of Water from Rural Areas of East Sikkim. Indian J Community Med 2014, 39, 156, doi:10.4103/0970-0218.137152.
- Zou, H.-Y.; He, L.-Y.; Gao, F.-Z.; Zhang, M.; Chen, S.; Wu, D.-L.; Liu, Y.-S.; He, L.-X.; Bai, H.; Ying, G.-G. Antibiotic Resistance Genes in Surface Water and Groundwater from Mining Affected Environments. Science of The Total Environment 2021, 772, 145516, doi:10.1016/j.scitotenv.2021.145516.
- Kittinger, C.; Lipp, M.; Folli, B.; Kirschner, A.; Baumert, R.; Galler, H.; Grisold, A.J.; Luxner, J.; Weissenbacher, M.; Farnleitner, A.H.; et al. Enterobacteriaceae Isolated from the River Danube: Antibiotic Resistances, with a Focus on the Presence of ESBL and Carbapenemases. PLoS ONE 2016, 11, e0165820, doi:10.1371/journal.pone.0165820.
- Paulshus, E.; Kühn, I.; Möllby, R.; Colque, P.; O’Sullivan, K.; Midtvedt, T.; Lingaas, E.; Holmstad, R.; Sørum, H. Diversity and Antibiotic Resistance among Escherichia Coli Populations in Hospital and Community Wastewater Compared to Wastewater at the Receiving Urban Treatment Plant. Water Research 2019, 161, 232–241, doi:10.1016/j.watres.2019.05.102.
- Suzuki, Y.; Hashimoto, R.; Xie, H.; Nishimura, E.; Nishiyama, M.; Nukazawa, K.; Ishii, S. Growth and Antibiotic Resistance Acquisition of Escherichia Coli in a River That Receives Treated Sewage Effluent. Science of The Total Environment 2019, 690, 696–704, doi:10.1016/j.scitotenv.2019.07.050.
- Titilawo, Y.; Sibanda, T.; Obi, L.; Okoh, A. Multiple Antibiotic Resistance Indexing of Escherichia Coli to Identify High-Risk Sources of Faecal Contamination of Water. Environ Sci Pollut Res 2015, 22, 10969–10980, doi:10.1007/s11356-014-3887-3.
- Verburg, I.; García-Cobos, S.; Hernández Leal, L.; Waar, K.; Friedrich, A.W.; Schmitt, H. Abundance and Antimicrobial Resistance of Three Bacterial Species along a Complete Wastewater Pathway. Microorganisms 2019, 7, 312, doi:10.3390/microorganisms7090312.
- Böger, B.; Surek, M.; Vilhena, R. de O.; Fachi, M.M.; Junkert, A.M.; Santos, J.M.; Domingos, E.L.; Cobre, A. de F.; Momade, D.R.; Pontarolo, R. Occurrence of Antibiotics and Antibiotic Resistant Bacteria in Subtropical Urban Rivers in Brazil. Journal of Hazardous Materials 2021, 402, 123448, doi:10.1016/j.jhazmat.2020.123448.
- Carattoli, A. Animal Reservoirs for Extended Spectrum Beta-Lactamase Producers. Clin Microbiol Infect 2008, 14 Suppl 1, 117–123, doi:10.1111/j.1469-0691.2007.01851.x.
- Müller, A.; Stephan, R.; Nüesch-Inderbinen, M. Distribution of Virulence Factors in ESBL-Producing Escherichia Coli Isolated from the Environment, Livestock, Food and Humans. Science of The Total Environment 2016, 541, 667–672, doi:10.1016/j.scitotenv.2015.09.135.
- Poirel, L.; Potron, A.; De La Cuesta, C.; Cleary, T.; Nordmann, P.; Munoz-Price, L.S. Wild Coastline Birds as Reservoirs of Broad-Spectrum-β-Lactamase-Producing Enterobacteriaceae in Miami Beach, Florida. Antimicrob Agents Chemother 2012, 56, 2756–2758, doi:10.1128/AAC.05982-11.
- Arias-Andres, M.; Klümper, U.; Rojas-Jimenez, K.; Grossart, H.-P. Microplastic Pollution Increases Gene Exchange in Aquatic Ecosystems. Environmental Pollution 2018, 237, 253–261, doi:10.1016/j.envpol.2018.02.058.
- Stalder, T.; Top, E. Plasmid Transfer in Biofilms: A Perspective on Limitations and Opportunities. npj Biofilms Microbiomes 2016, 2, 16022, doi:10.1038/npjbiofilms.2016.22.
- Ma, Y.; Chen, J.; Fong, K.; Nadya, S.; Allen, K.; Laing, C.; Ziebell, K.; Topp, E.; Carroll, L.M.; Wiedmann, M.; et al. Antibiotic Resistance in Shiga Toxigenic Escherichia Coli Isolates from Surface Waters and Sediments in a Mixed Use Urban Agricultural Landscape. Antibiotics 2021, 10, 237, doi:10.3390/antibiotics10030237.
- VanMensel, D.; Chaganti, S.R.; Droppo, I.G.; Weisener, C.G. Exploring Bacterial Pathogen Community Dynamics in Freshwater Beach Sediments: A Tale of Two Lakes. Environmental Microbiology 2020, 22, 568–583, doi:10.1111/1462-2920.14860.
- Dickinson, A.W.; Power, A.; Hansen, M.G.; Brandt, K.K.; Piliposian, G.; Appleby, P.; O’Neill, P.A.; Jones, R.T.; Sierocinski, P.; Koskella, B.; et al. Heavy Metal Pollution and Co-Selection for Antibiotic Resistance: A Microbial Palaeontology Approach. Environment International 2019, 132, 105117, doi:10.1016/j.envint.2019.105117.
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol Spectr 2018, 6, 6.2.10, doi:10.1128/microbiolspec.ARBA-0009-2017.
- Hocquet, D.; Muller, A.; Bertrand, X. What Happens in Hospitals Does Not Stay in Hospitals: Antibiotic-Resistant Bacteria in Hospital Wastewater Systems. Journal of Hospital Infection 2016, 93, 395–402, doi:10.1016/j.jhin.2016.01.010.
- Piedra-Carrasco, N.; Fàbrega, A.; Calero-Cáceres, W.; Cornejo-Sánchez, T.; Brown-Jaque, M.; Mir-Cros, A.; Muniesa, M.; González-López, J.J. Carbapenemase-Producing Enterobacteriaceae Recovered from a Spanish River Ecosystem. PLoS ONE 2017, 12, e0175246, doi:10.1371/journal.pone.0175246.
- Dhawde, R.; Macaden, R.; Saranath, D.; Nilgiriwala, K.; Ghadge, A.; Birdi, T. Antibiotic Resistance Characterization of Environmental E. Coli Isolated from River Mula-Mutha, Pune District, India. IJERPH 2018, 15, 1247, doi:10.3390/ijerph15061247.
- Lepper, H.C.; Woolhouse, M.E.J.; van Bunnik, B.A.D. The Role of the Environment in Dynamics of Antibiotic Resistance in Humans and Animals: A Modelling Study. Antibiotics 2022, 11, 1361, doi:10.3390/antibiotics11101361.
- Pantanella, F.; Lekunberri, I.; Gagliardi, A.; Venuto, G.; Sànchez-Melsió, A.; Fabiani, M.; Balcázar, J.L.; Schippa, S.; De Giusti, M.; Borrego, C.; et al. Effect of Urban Wastewater Discharge on the Abundance of Antibiotic Resistance Genes and Antibiotic-Resistant Escherichia Coli in Two Italian Rivers. IJERPH 2020, 17, 6813, doi:10.3390/ijerph17186813.
- Rizzo, L.; Fiorentino, A.; Anselmo, A. Effect of Solar Radiation on Multidrug Resistant E. Coli Strains and Antibiotic Mixture Photodegradation in Wastewater Polluted Stream. Science of The Total Environment 2012, 427–428, 263–268, doi:10.1016/j.scitotenv.2012.03.062.
- Hooban, B.; Fitzhenry, K.; Cahill, N.; Joyce, A.; O’ Connor, L.; Bray, J.E.; Brisse, S.; Passet, V.; Abbas Syed, R.; Cormican, M.; et al. A Point Prevalence Survey of Antibiotic Resistance in the Irish Environment, 2018–2019. Environment International 2021, 152, 106466, doi:10.1016/j.envint.2021.106466.
- Khan, F.A.; Söderquist, B.; Jass, J. Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater. Front. Microbiol. 2019, 10, 688, doi:10.3389/fmicb.2019.00688.
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban Wastewater Treatment Plants as Hotspots for Antibiotic Resistant Bacteria and Genes Spread into the Environment: A Review. Science of The Total Environment 2013, 447, 345–360, doi:10.1016/j.scitotenv.2013.01.032.
- Galler, H.; Feierl, G.; Petternel, C.; Reinthaler, F.; Haas, D.; Habib, J.; Kittinger, C.; Luxner, J.; Zarfel, G. Multiresistant Bacteria Isolated from Activated Sludge in Austria. IJERPH 2018, 15, 479, doi:10.3390/ijerph15030479.
- Bessa, L.J.; Barbosa-Vasconcelos, A.; Mendes, Â.; Vaz-Pires, P.; Martins da Costa, P. High Prevalence of Multidrug-Resistant Escherichia Coli and Enterococcus Spp. in River Water, Upstream and Downstream of a Wastewater Treatment Plant. Journal of Water and Health 2014, 12, 426–435, doi:10.2166/wh.2014.160.
- Bleichenbacher, S.; Stevens, M.J.A.; Zurfluh, K.; Perreten, V.; Endimiani, A.; Stephan, R.; Nüesch-Inderbinen, M. Environmental Dissemination of Carbapenemase-Producing Enterobacteriaceae in Rivers in Switzerland. Environmental Pollution 2020, 265, 115081, doi:10.1016/j.envpol.2020.115081.
- Novovic, K.; Filipic, B.; Veljovic, K.; Begovic, J.; Mirkovic, N.; Jovcic, B. Environmental Waters and BlaNDM-1 in Belgrade, Serbia: Endemicity Questioned. Science of The Total Environment 2015, 511, 393–398, doi:10.1016/j.scitotenv.2014.12.072.
- Paulitsch-Fuchs, A.H.; Melchior, N.; Haitzmann, T.; Fingerhut, T.; Feierl, G.; Baumert, R.; Kittinger, C.; Zarfel, G. Analysis of Extended Spectrum Beta Lactamase (ESBL) Genes of Non-Invasive ESBL Enterobacterales in Southeast Austria in 2017. Antibiotics 2022, 12, 1, doi:10.3390/antibiotics12010001.
- Zarfel, G.; Hoenigl, M.; Leitner, E.; Salzer, H.J.F.; Feierl, G.; Masoud, L.; Valentin, T.; Krause, R.; Grisold, A.J. Emergence of New Delhi Metallo-β-Lactamase, Austria. Emerg. Infect. Dis. 2011, 17, 129–130, doi:10.3201/eid1701.101331.
- EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.1, 2017. Http://Www.Eucast.Org. The European Committee on Antimicrobial Susceptibility Testing. 2017.
- CLSI M100 Performance Standards for Antimicrobial Susceptibility Testing. Clinical Microbiology and Infection 2008.
- Boyen, F.; Vangroenweghe, F.; Butaye, P.; De Graef, E.; Castryck, F.; Heylen, P.; Vanrobaeys, M.; Haesebrouck, F. Disk Prediffusion Is a Reliable Method for Testing Colistin Susceptibility in Porcine E. Coli Strains. Veterinary Microbiology 2010, 144, 359–362, doi:10.1016/j.vetmic.2010.01.010.
- Gales, A.C.; Reis, A.O.; Jones, R.N. Contemporary Assessment of Antimicrobial Susceptibility Testing Methods for Polymyxin B and Colistin: Review of Available Interpretative Criteria and Quality Control Guidelines. J Clin Microbiol 2001, 39, 183–190, doi:10.1128/JCM.39.1.183-190.2001.
- Eckert, C.; Gautier, V.; Saladin-Allard, M.; Hidri, N.; Verdet, C.; Ould-Hocine, Z.; Barnaud, G.; Delisle, F.; Rossier, A.; Lambert, T.; et al. Dissemination of CTX-M-Type Beta-Lactamases among Clinical Isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother 2004, 48, 1249–1255, doi:10.1128/AAC.48.4.1249-1255.2004.
- Kiratisin, P.; Apisarnthanarak, A.; Laesripa, C.; Saifon, P. Molecular Characterization and Epidemiology of Extended-Spectrum- β-Lactamase-Producing Escherichia Coli and Klebsiella Pneumoniae Isolates Causing Health Care-Associated Infection in Thailand, Where the CTX-M Family Is Endemic. Antimicrob Agents Chemother 2008, 52, 2818–2824, doi:10.1128/AAC.00171-08.
- Asai, T.; Masani, K.; Sato, C.; Hiki, M.; Usui, M.; Baba, K.; Ozawa, M.; Harada, K.; Aoki, H.; Sawada, T. Phylogenetic Groups and Cephalosporin Resistance Genes of Escherichia Coli from Diseased Food-Producing Animals in Japan. Acta Vet Scand 2011, 53, 52, doi:10.1186/1751-0147-53-52.
- Zarfel, G.; Galler, H.; Feierl, G.; Haas, D.; Kittinger, C.; Leitner, E.; Grisold, A.J.; Mascher, F.; Posch, J.; Pertschy, B.; et al. Comparison of Extended-Spectrum-β-Lactamase (ESBL) Carrying Escherichia Coli from Sewage Sludge and Human Urinary Tract Infection. Environmental Pollution 2013, 173, 192–199, doi:10.1016/j.envpol.2012.09.019.
- Gröbner, S.; Linke, D.; Schütz, W.; Fladerer, C.; Madlung, J.; Autenrieth, I.B.; Witte, W.; Pfeifer, Y. Emergence of Carbapenem-Non-Susceptible Extended-Spectrum β-Lactamase-Producing Klebsiella Pneumoniae Isolates at the University Hospital of Tübingen, Germany. Journal of Medical Microbiology 2009, 58, 912–922, doi:10.1099/jmm.0.005850-0.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).