Submitted:
06 June 2023
Posted:
25 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
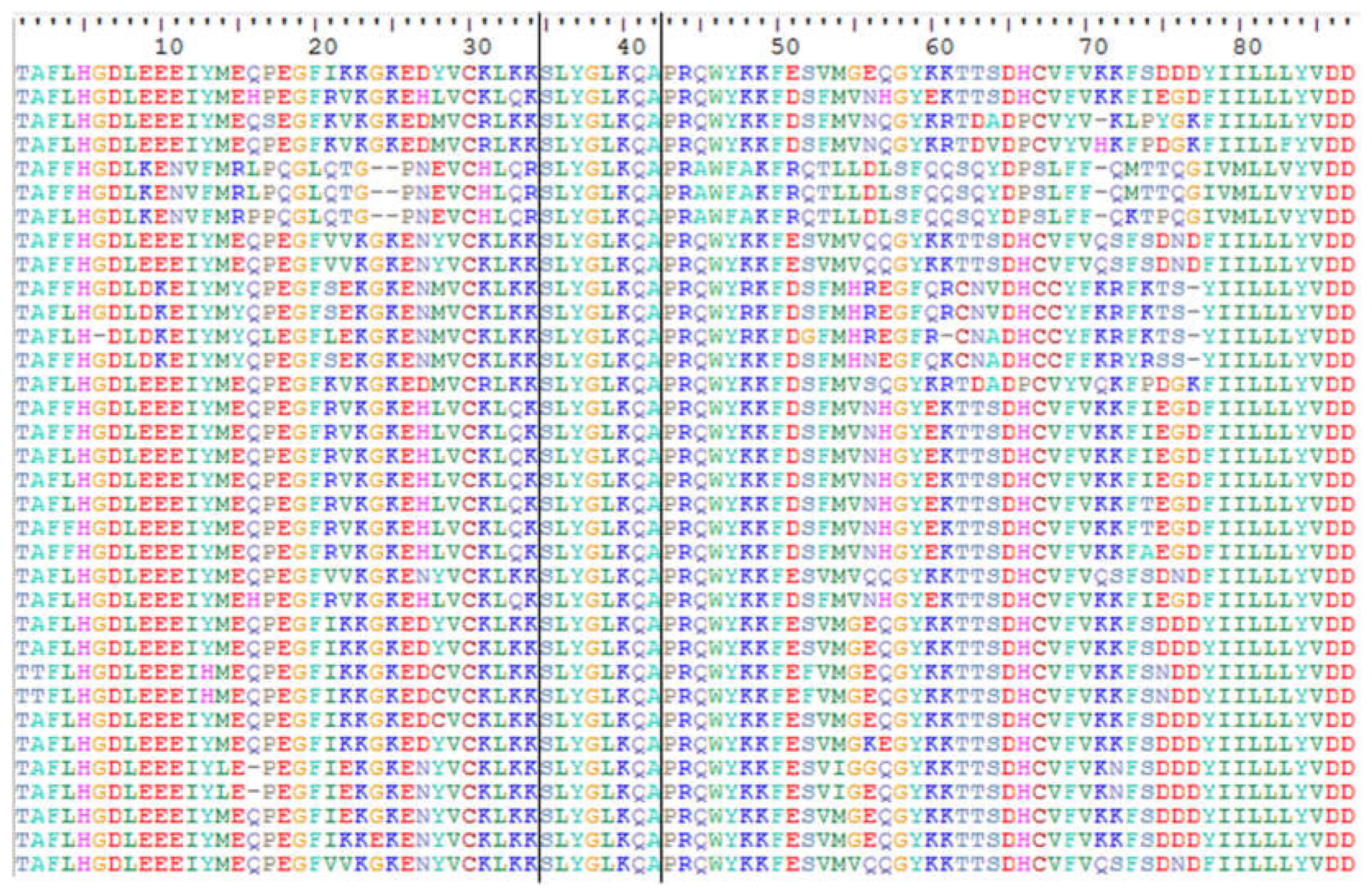
2.1. Characterization of Ty1-copia sequences in the J. curcas genome
2.2. Molecular cytogenetic evaluation of the Ty1-copia in Jatropha based on FISH
3. Materials and Methods
3.1. Isolation and characterization of Ty1-copia retrotransposons in J. curcas
3.2. FISH technique for study of chromosomal distribution of Ty1-copia elements
3.2.1. Chromosome preparation
3.2.2. Probe labeling
3.2.3. Hybridization
4. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ranade, S.A.; Srivastava, A.P.; Rana, T. S.; Srivastava, J.; Tuli, R. Easy assessment of diversity in Jatropha curcas L. plants using two single-primer amplification reaction (SPAR) methods. Biomass Bioenergy 2008, 32, 533–540. [Google Scholar] [CrossRef]
- Maes, W.H.; Trabucco, A.; Achten, W.M.J.; Muys, B. Climatic growing condition of Jatropha curcas. Biomass Bioenergy 2009, 33, 1481–1485. [Google Scholar] [CrossRef]
- Openshaw, H. A. review of Jatropha curcas: an oil plant of unfulfilled promise. Biomass Bioenergy 2000, 19, 1–15. [Google Scholar] [CrossRef]
- Augustus, G.D.P.S.; Jayabalan, M; Seiler, G. J. Evaluation and bioinduction of energy components of Jatropha curcas. Biomass Bioenergy 2002, 23, 161–164. [Google Scholar] [CrossRef]
- Kaushik, N.; Kumar, K.; Kumar, S.; Kaushik, N.; Roy, S. Genetic variability and divergence studies in seed traits and oil content of Jatropha (Jatropha curcas L.) accessions. Biomass Bioenergy 2007, 31, 497–502. [Google Scholar] [CrossRef]
- Sirisomboon, P.; Kitchaiya, P.; Pholpho, T.; Mahuttanyavanitch, W. Physical and mechanical properties of Jatropha curcas L. fruits, nuts and kernels. Biosyst. Eng. 2007, 97, 201–207. [Google Scholar] [CrossRef]
- Sujatha, M. Biotechnological intervention for improving Jatropha and castor for biofuels. In Petrotech, New Delhi, India, 11-15 January 2009. [Google Scholar]
- Finnegan, D.J. Transposable elements and DNA transposition in eukaryotes. Curr. Opin. Cell Biol. 1990, 2, 471–477. [Google Scholar] [CrossRef]
- Kumar, A.; Bennetzen, J.L. Plant retrotransposons. Annu. Rev. Genet. 1999, 33, 479–532. [Google Scholar] [CrossRef]
- Habibollahi, H.; Noormohammadi, Z.; Sheidai, M.; Farahani, F.; Talebi, S.M.; Torabizadeh, E. Assessments of genetic diversity in Iranian flax populations using retrotransposon microsatellite amplification polymorphisms (REMAP) markers. The Nucleus 2017, 61, 55–60. [Google Scholar] [CrossRef]
- Ohmido, N.; Fukui, K.; Kinoshita, T. Recent advances in rice genome and Chromosome structure research by fluorescence in situ hybridization (FISH). Proc. Jpn. Acad. B. 2010, 86, 103–116. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.; Brandes, A.; Taketa, S.; Schmidt, T.; Vershinin, A.V.; Alkhimova, E.G.; Kamm, A.; Doudrick, R.L.; Schwarzacher, T.; Katsiotis, A.; Kubis, S.; Kumar, A.; Pearce, S.R.; Flavell, A.J.; Harrison, G.E. The chromosomal distribution of Ty1-copia group retrotransposable elements in higher plants and their implication for genome evolution. Genetica. 1997, 100, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Flavell, A.J.; Dunbar. E.; Anderson, R.; Pearce, S.R.; Hartley, R.; Kumar, A. Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 1992, 20, 3639–3644. [Google Scholar] [CrossRef]
- Voytas, D.F.; Cummings, M.P.; Konieczy, A.; Ausubel, F.M. Copia-like retrotransposons are ubiquitous among plants. Proc. Natl. Acad. Sci. USA. 1992, 89, 7124–7128. [Google Scholar] [CrossRef] [PubMed]
- Hirochika, H; Hirochika, R. Ty1-copia group retrotransposons as ubiquitous components of plant genomes. Jpn. J. Genet. 1993, 68, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Ambrozova, K.; Mandakova, T.; Bures, P.; Neumann, P.; Leitch, I.J.; Kobizkova, I.; Macas, J.; Lysak, M.A. Diverse retrotransposon families and AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Ann. Bot. 2011, 107, 255–268. [Google Scholar] [CrossRef]
- Sato, S.; Hirakawa, H.; Isobe, S.; Fukai, E.; Watanabe, A.; Kato, M.; Kawashima, K.; Minami, C.; Muraki, A.; Nakazaki, N.; Takahashi, C. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011, 18, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.B.; Li, L.F.; Li, Y.; Wu, G.J.; Ge, X.J. SSR and AFLP Markers Reveal Low Genetic Diversity in the Biofuel Plant Jatropha curcas in China. Crop Sci. 2008, 48, 1865–1871. [Google Scholar] [CrossRef]
- Soonthornyatara, S.; Sripichitt, P.; Kaveeta, R.; Hongtrakul, V. Assessment of genetic diversity of Jatropha curcas L. using AFLP and ISSR markers. Chiang Mai J. Sci. 2015, 42, 614–625. [Google Scholar]
- Woodrow, P.; Pontecorvo, G.; Fantaccione, S.; Fuggi, A.; Kafantaris, I.; Parisi, D.; Carillo, P. Polymorphism of a new Ty1-copia retrotransposon in durum wheat under salt and light stresses. Theor. Appl. Genet. 2010, 121, 311–322. [Google Scholar] [CrossRef]
- Vitte, C.; Bennetzen, J.L. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA. 2006, 103, 17638–17643. [Google Scholar] [CrossRef]
- Wicker, T.; Keller, B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007, 17, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Vitte, C.; Panaud, O.; Quesneville, H. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genom. 2007, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol. 2004, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Devos, K.M.; Bennetzen, J.L. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004, 14, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Baucom, R.S.; Estill, J.C.; Chaparro, C.; Upshaw, N.; Jogi, A.; Deragon, J.M.; Westerman, R.P.; SanMiguel, P.J.; Bennetzen, J.L. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009, 5, e1000732. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Tsuchimoto, S.; Sakai, H.; Nakayama, S.; Fujishiro, T.; Kishida, Y.; Kohara, M.; Watanabe, A.; Yamada, M.; Aizu, T.; Toyoda, A.; Fujiyama, A.; Tabata, S.; Fukui, K.; Sato, T. Upgraded genomic information of Jatropha curcas L. Plant Biotechnol. 2012, 29, 123–130. [Google Scholar] [CrossRef]
- Du, J.; Tian, Z.; Hans, C.S.; Laten, H.M.; Cannon, S.B.; Jackson, S.A.; Shoemaker, R.C.; Ma, J. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 2010, 63, 584–598. [Google Scholar] [CrossRef]
- Tuntipaiboontana, R.; Kuleung, C.; Hongtrakul, V. Diverse Ty1-copia retrotransposons found in waterlilies of the genus Nymphaea. Hort. J. 2018, 87, 524–531. [Google Scholar] [CrossRef]
- Brandes, A.; Heslop-Harrison, J.S.; Kamm, A.; Kubis, S.; Doudrick, R.L.; Schmidt, T. Comparative analysis of the chromosomal and genomic organization of Ty1-copia-like retrotransposons in pteridophytes, gymnosperms and angiosperms. Plant Mol. Biol. 1997, 33, 11–21. [Google Scholar] [CrossRef]
- Kikuchi, S.; Tsujimoto, H.; Sassa, H.; Koba, T. JcSat1, a novel subtelomeric repeat of Jatropha curcas L. and its use in karyotyping. Chromosome Sci. 2011, 13, 11–16. [Google Scholar]
- Herrera, J.C.; Camayo, G.; Torre, G.D.L.T.; Galeano, N.; Salcedo, E.; Rivera, L.F.; Duran, A. Identification and chromosomal distribution of copia-like retrotransposon sequences in the coffee (Coffea L.) genome. Agron. Colomb. 2013, 31, 269–278. [Google Scholar]
- Kolano, B.; Bednara, E.; Weiss-Schneeweiss, H. Isolation and characterization of reverse transcriptase fragments of LTR retrotransposons from the genome of Chenopodium quinoa (Amaranthaceae). Plant Cell Rep. 2013, 32, 1575–1588. [Google Scholar] [PubMed]
- Felice, B.D.; Wilson, R.R.; Argenziano, C.; Kafantaris, I; Conicella, C. A transcriptionally active copia-like retroelement in citrus limon. Cell. Mol. Biol. Lett. 2009, 14, 289–304. [Google Scholar]
- Kamm, A.; Doudrick, R.L.; Heslop-Harrison, J.S.; Schmidt, T. The genomic and physical organization of Ty1-copia-like sequence as a component of large genomes in Pinus elliottii var. elliottii and other gymnosperms. Proc. Natl. Acad. Sci. USA. 1996, 93, 2708–2713. [Google Scholar]
- Linares, C.; Loarce, Y.; Serna, A.; Fominaya, A. Isolation and characterization of two novel retrotransposons of the Ty1-copia group in oat genomes. Chromosoma. 2001, 110, 25. [Google Scholar]
- Ito, H. Environmental stress and transposons in plants. Genes Genet. Syst. 2022, 97, 169–175. [Google Scholar] [CrossRef]
- Wessler, S.R. Plant retrotransposons: turned on by stress. Curr. Biol. 1996, 6, 959–961. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Williams, C.G. Retroelements contribute to the excess low-copy-number DNA in pine. Mol. Gen. Genet. 2000, 264, 47–55. [Google Scholar] [CrossRef]
- McClintock, B. The significance of responses of the genome to challenge. Science. 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Kidwell, M.G.; Lisch, D.R. Perspective: transposable elements, parasitic DNA and genome evolution. Evol. 2001, 2001. 55, 1–24. [Google Scholar]
- Kidwell, M.G.; Lisch, D. Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA. 1997, 94, 7704–7711. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus. 1990, 12, 13–15. [Google Scholar]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; Grosdidier, A.; Hernandez, C.; Ioannidis, V.; Kuznetsov, D.; Liechti, R.; Moretti, S.; Mostaguir, K.; Redaschi, N.; Rossier, G.; Xenarios, I.; Stockinger, H. ExPASy: SIB Bioinformatics Resource Potal. Nucleic Acids Res. 2012, 40, 597–603. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from EMBL-EBI. Nucleic Acids Res. 2013, 14, W597–W600. [Google Scholar] [CrossRef]
- Ohmido, N.; Fukui, K. Visual verification of close disposition between a rice A genome-specific DNA sequence (TrsA) and the telomere sequence. Plant Mol. Biol. 1997, 35, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Ohmido, N.; Akiyama, Y.; Fukui, K. Physical mapping of unique nucleotide sequences on identified rice chromosomes. Plant Mol. Biol. 1998, 38, 1041–1052. [Google Scholar] [CrossRef]
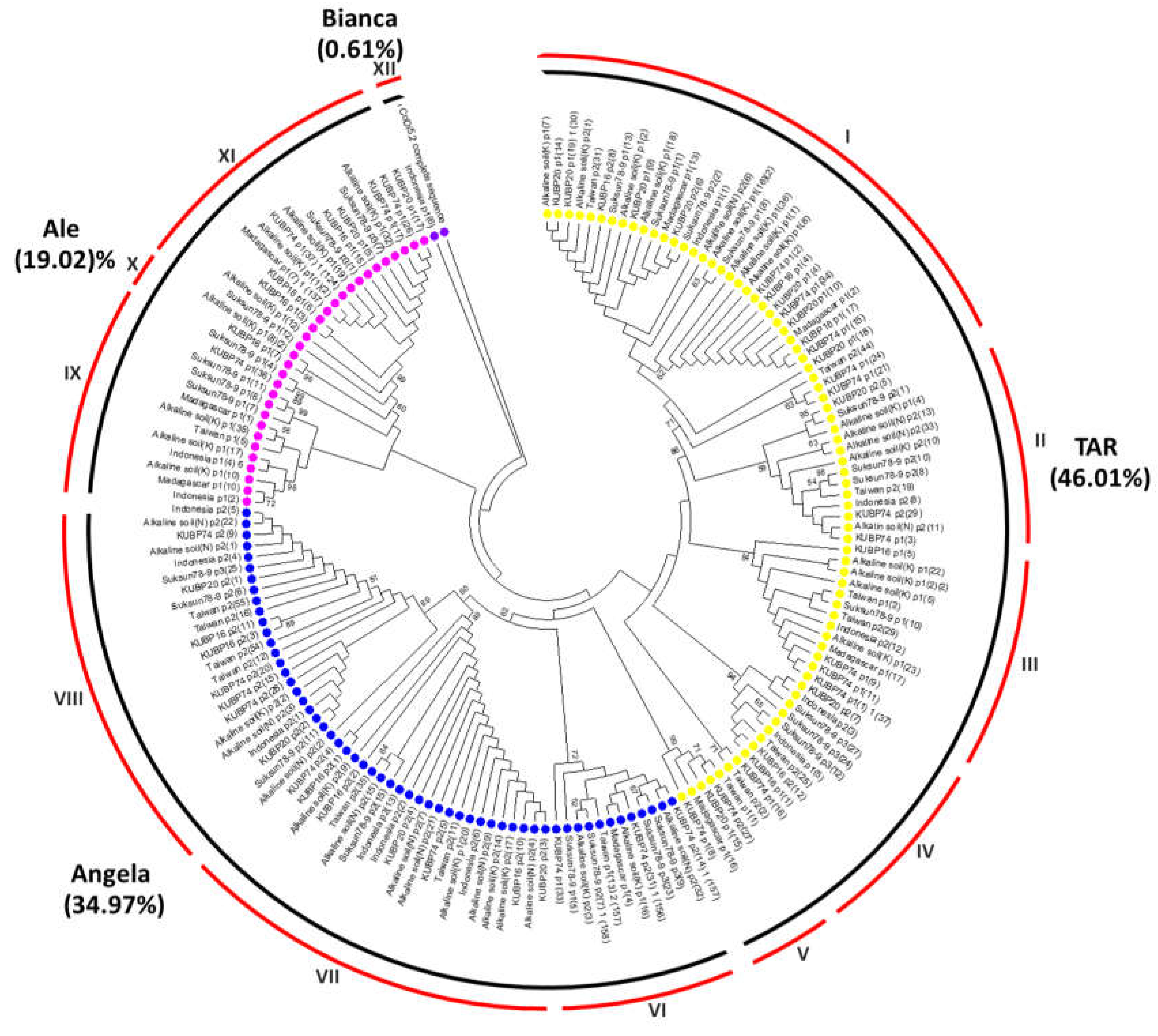


| J. curcas plant samples | Origin | TAR | Angela | Ale | Bianca | Total |
|---|---|---|---|---|---|---|
| KUBP78-9 | Thailand | 11 | 8 | 7 | - | 26 |
| KUBP74 | Thailand | 13 | 9 | 4 | - | 26 |
| KUBP20 | Thailand | 11 | 4 | 2 | - | 17 |
| KUBP16 | Thailand | 6 | 5 | 4 | - | 15 |
| Salt-tolerant #1 from None-Sung district of Nakhon Ratchasima province | Thailand | 4 | 10 | - | - | 14 |
| Salt-tolerant #2 from Kham-Talay-Sor district of Nakhon Ratchasima province | Thailand | 14 | 7 | 8 | - | 29 |
| Madagascar | Madagascar | 4 | 1 | 3 | - | 8 |
| Taiwan | Taiwan | 7 | 7 | 1 | - | 15 |
| Indonesia | Indonesia | 5 | 6 | 2 | 1 | 14 |
| Total | 75 (46.01%) |
57 (34.97%) |
31 (19.02%) |
1 (0.61%) |
164 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




