Submitted:
20 June 2023
Posted:
21 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Search strategy and selection criteria
2.2. Data extraction
2.3. Statistical analysis
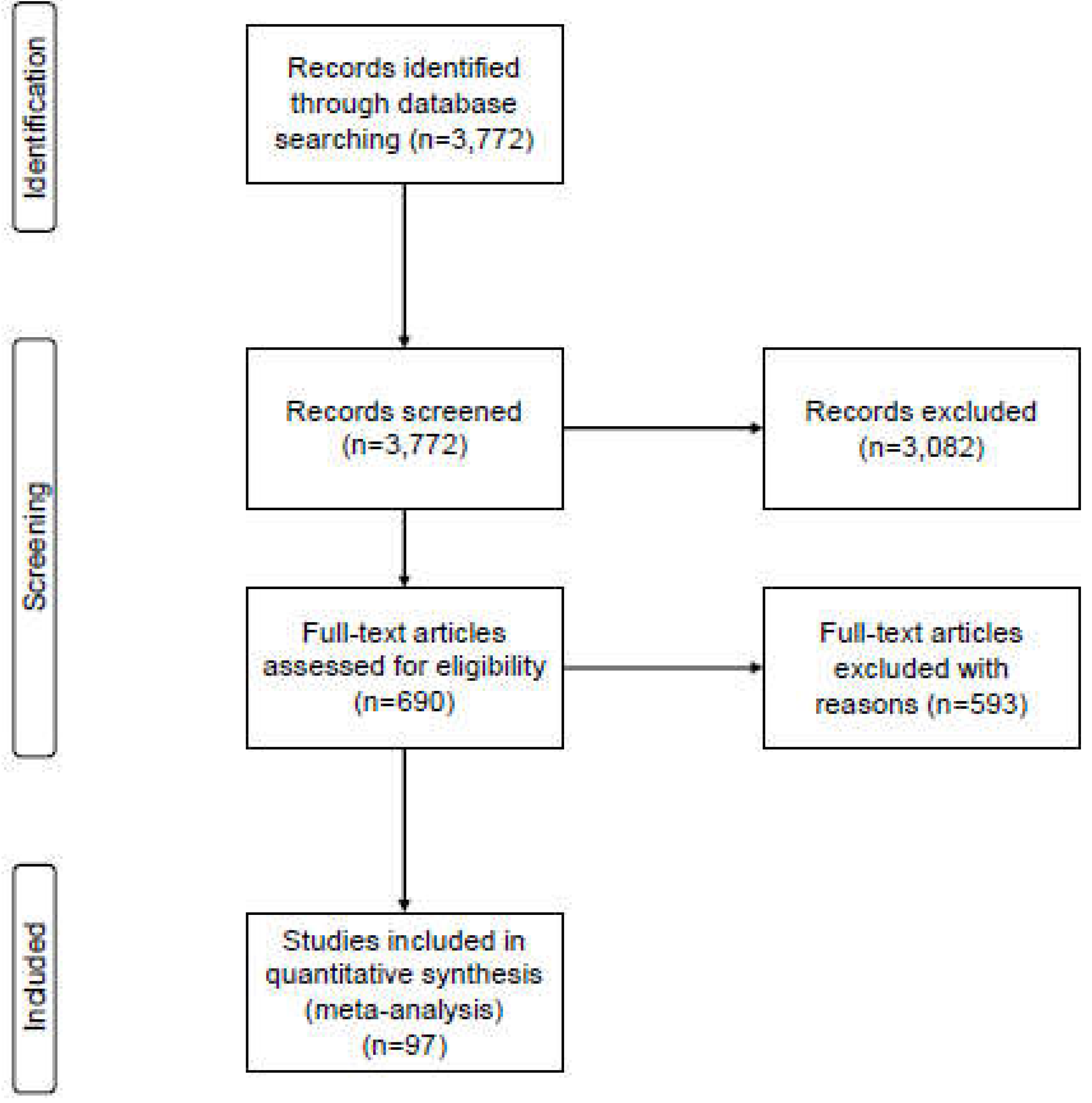
3. Results
3.1. Studies included in our analysis
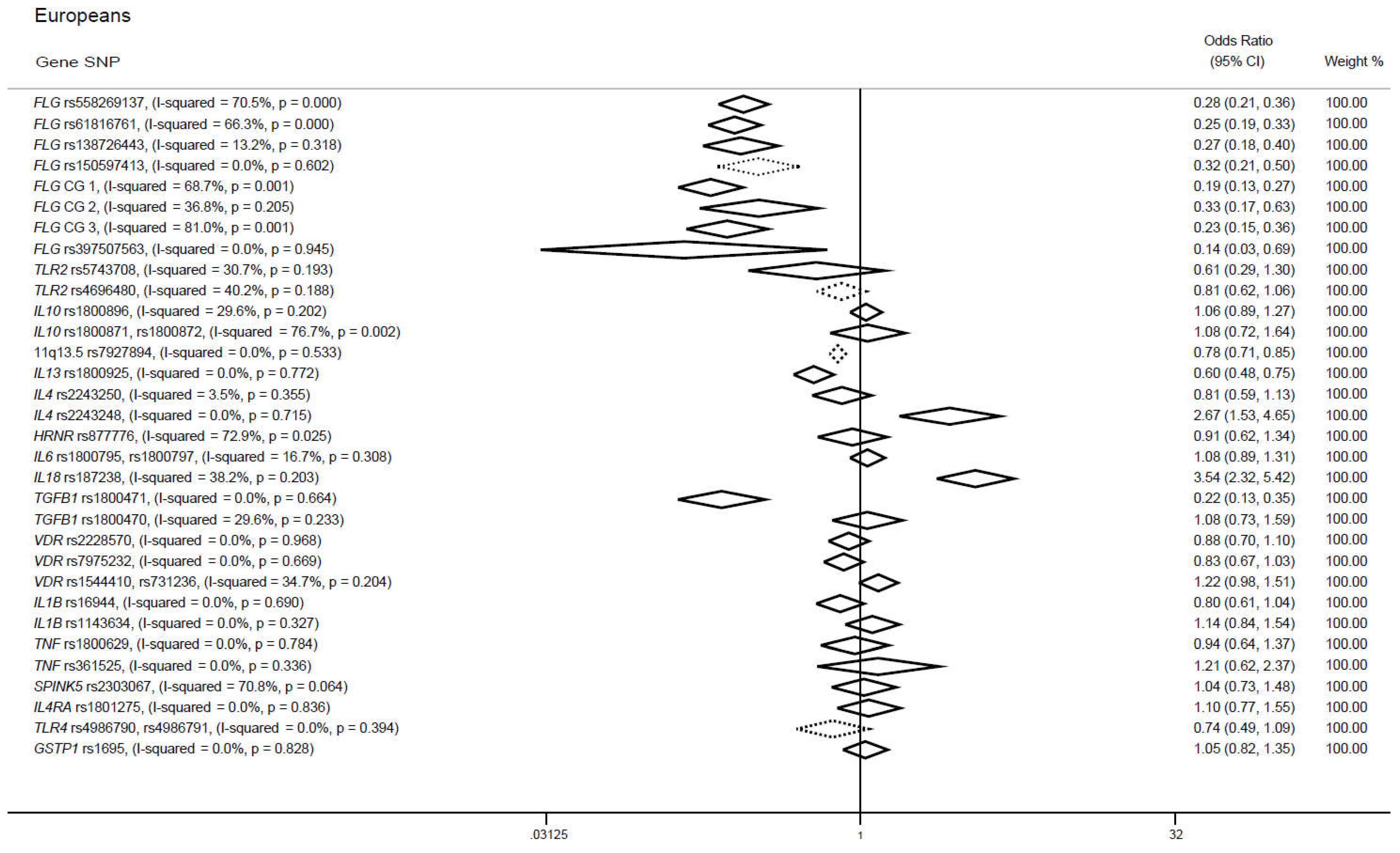
3.2. Candidate gene approaches in cases of European ancestry
3.2.1. FLG gene
3.2.2. TLR2 gene
3.2.3. IL10 gene
3.2.4. 11. q13.5 locus
3.2.5. IL13 gene
3.2.6. SNPs in IL4, IL18 and TGFB1 genes
3.2.7. SNPs in IL6, HRNR, VDR, IL1B, TNF, SPINK5, IL4RA, TLR4 and GSTP1 genes
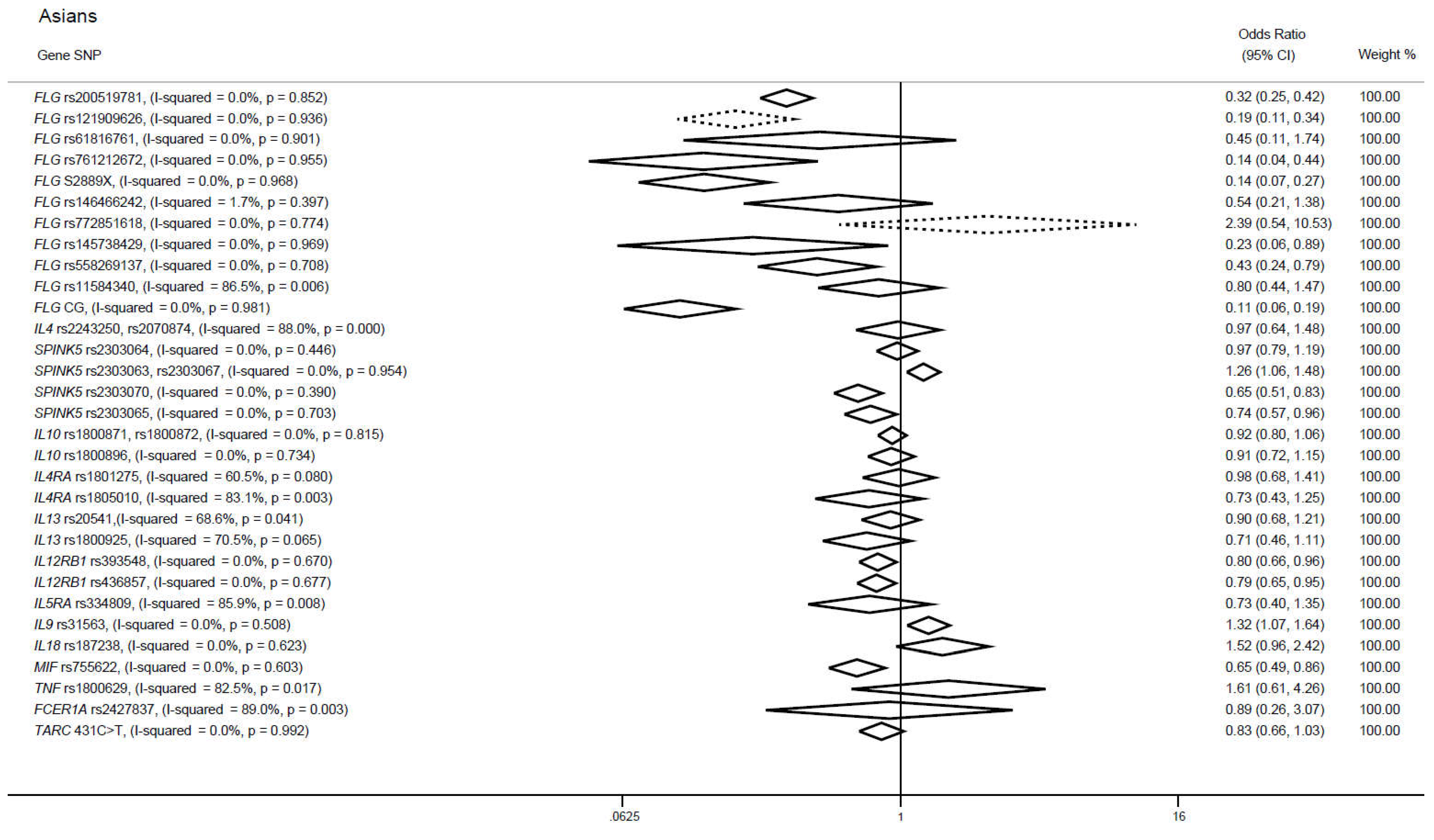
3.3. Candidate gene approaches in cases of Asian ancestry
3.3.1. FLG gene
3.3.2. IL4 gene
3.3.3. SPINK5 gene
3.3.4. SNPs in IL10, IL4RA and IL13 genes
3.3.5. SNPs in IL12RB1, IL9 and MIF genes
3.3.6. SNPs in IL5RA, IL18, TNF, FCER1A and TARC genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Langan, S.M.; Irvine, A.D.; Weidinger, S. Atopic Dermatitis. Lancet 2020, 396, 345–360. [Google Scholar] [CrossRef]
- Thomsen, S.F.; Ulrik, C.S.; Kyvik, K.O.; Hjelmborg, J. v B.; Skadhauge, L.R.; Steffensen, I.; Backer, V. Importance of Genetic Factors in the Etiology of Atopic Dermatitis: A Twin Study. Allergy Asthma Proc 2007, 28, 535–539. [CrossRef]
- Paternoster, L.; Standl, M.; Waage, J.; Baurecht, H.; Hotze, M.; Strachan, D.P.; Curtin, J.A.; Bønnelykke, K.; Tian, C.; Takahashi, A.; et al. Multi-Ancestry Genome-Wide Association Study of 21,000 Cases and 95,000 Controls Identifies New Risk Loci for Atopic Dermatitis. Nat Genet 2015, 47, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Mucha, S.; Baurecht, H.; Novak, N.; Rodríguez, E.; Bej, S.; Mayr, G.; Emmert, H.; Stölzl, D.; Gerdes, S.; Jung, E.S.; et al. J Allergy Clin Immunol 2020, 145, 1208–1218. [CrossRef] [PubMed]
- Ferreira, M.A.R.; Vonk, J.M.; Baurecht, H.; Marenholz, I.; Tian, C.; Hoffman, J.D.; Helmer, Q.; Tillander, A.; Ullemar, V.; Lu, Y.; et al. Eleven Loci with New Reproducible Genetic Associations with Allergic Disease Risk. J Allergy Clin Immunol 2019, 143, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Hoober, J.K.; Eggink, L.L. The Discovery and Function of Filaggrin. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Kypriotou, M.; Huber, M.; Hohl, D. The Human Epidermal Differentiation Complex: Cornified Envelope Precursors, S100 Proteins and the “fused Genes” Family. Exp Dermatol 2012, 21, 643–649. [Google Scholar] [CrossRef]
- Wynn, T.A. Type 2 Cytokines: Mechanisms and Therapeutic Strategies. Nat Rev Immunol 2015, 15, 271–282. [Google Scholar] [CrossRef]
- Amirzargar, A.A.; Movahedi, M.; Rezaei, N.; Moradi, B.; Dorkhosh, S.; Mahloji, M.; Mahdaviani, S.A. Polymorphisms in IL4 and ILARA Confer Susceptibility to Asthma. J Investig Allergol Clin Immunol 2009, 19, 433–438. [Google Scholar]
- Cameron, L.; Webster, R.B.; Strempel, J.M.; Kiesler, P.; Kabesch, M.; Ramachandran, H.; Yu, L.; Stern, D.A.; Graves, P.E.; Lohman, I.C.; et al. Th2 Cell-Selective Enhancement of Human IL13 Transcription by IL13-1112C>T, a Polymorphism Associated with Allergic Inflammation. J Immunol 2006, 177, 8633–8642. [Google Scholar] [CrossRef]
- Gooderham, M.J.; Hong, H.C.-H.; Eshtiaghi, P.; Papp, K.A. Dupilumab: A Review of Its Use in the Treatment of Atopic Dermatitis. J Am Acad Dermatol 2018, 78, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Koido, M.; Suzuki, A.; Otomo, N.; Suetsugu, H.; Kochi, Y.; Tomizuka, K.; Momozawa, Y.; Kamatani, Y.; Ikegawa, S.; et al. Eight Novel Susceptibility Loci and Putative Causal Variants in Atopic Dermatitis. J Allergy Clin Immunol 2021, 148, 1293–1306. [Google Scholar] [CrossRef]
- Brown, S.J. What Have We Learned from GWAS for Atopic Dermatitis? J Invest Dermatol 2021, 141, 19–22. [Google Scholar] [CrossRef]
- Vardar Acar, N.; Cavkaytar, Ö.; Arik Yilmaz, E.; Büyüktiryaki, A.B.; Uysal Soyer, Ö.; Şahiner, Ü.M.; Şekerel, B.E.; Karaaslan, I.Ç.; Saçkesen, C. Rare Occurrence of Common Filaggrin Mutations in Turkish Children with Food Allergy and Atopic Dermatitis. Turk J Med Sci 2020, 50, 1865–1871. [Google Scholar] [CrossRef]
- Jurakic Toncic, R.; Kezic, S.; Jakasa, I.; Ljubojevic Hadzavdic, S.; Balic, A.; Petkovic, M.; Pavicic, B.; Zuzul, K.; Marinovic, B. Filaggrin Loss-of-Function Mutations and Levels of Filaggrin Degradation Products in Adult Patients with Atopic Dermatitis in Croatia. J Eur Acad Dermatol Venereol 2020, 34, 1789–1794. [Google Scholar] [CrossRef]
- González-Tarancón, R.; Sanmartín, R.; Lorente, F.; Salvador-Rupérez, E.; Hernández-Martín, A.; Rello, L.; Puzo, J.; Gilaberte, Y. Prevalence of FLG Loss-of-Function Mutations R501X, 2282del4, and R2447X in Spanish Children with Atopic Dermatitis. Pediatr Dermatol 2020, 37, 98–102. [Google Scholar] [CrossRef]
- Woźniak, M.; Kaczmarek-Skamira, E.; Romańska-Gocka, K.; Czajkowski, R.; Kałużna, L.; Zegarska, B. The Prevalence of Mutations in the Gene Encoding Filaggrin in the Population of Polish Patients with Atopic Dermatitis. Postepy Dermatol Alergol 2016, 33, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Trzeciak, M.; Gleń, J.; Rębała, K.; Bandurski, T.; Sikorska, M.; Nowicki, R. Coexistence of 2282del4 FLG Gene Mutation and IL-18 -137G/C Gene Polymorphism Enhances the Risk of Atopic Dermatitis. Postepy Dermatol Alergol 2016, 33, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Trzeciak, M.; Wesserling, M.; Bandurski, T.; Glen, J.; Nowicki, R.; Pawelczyk, T. Association of a Single Nucleotide Polymorphism in a Late Cornified Envelope-like Proline-Rich 1 Gene (LELP1) with Atopic Dermatitis. Acta Derm Venereol 2016, 96, 459–463. [Google Scholar] [CrossRef]
- Ballardini, N.; Kull, I.; Söderhäll, C.; Lilja, G.; Wickman, M.; Wahlgren, C.F. Eczema Severity in Preadolescent Children and Its Relation to Sex, Filaggrin Mutations, Asthma, Rhinitis, Aggravating Factors and Topical Treatment: A Report from the BAMSE Birth Cohort. Br J Dermatol 2013, 168, 588–594. [Google Scholar] [CrossRef]
- Ercan, H.; Ispir, T.; Kirac, D.; Baris, S.; Ozen, A.; Oztezcan, S.; Cengizlier, M.R. Predictors of Atopic Dermatitis Phenotypes and Severity: Roles of Serum Immunoglobulins and Filaggrin Gene Mutation R501X. Allergol Immunopathol (Madr) 2013, 41, 86–93. [Google Scholar] [CrossRef]
- Mlitz, V.; Latreille, J.; Gardinier, S.; Jdid, R.; Drouault, Y.; Hufnagl, P.; Eckhart, L.; Guinot, C.; Tschachler, E. Impact of Filaggrin Mutations on Raman Spectra and Biophysical Properties of the Stratum Corneum in Mild to Moderate Atopic Dermatitis. J Eur Acad Dermatol Venereol 2012, 26, 983–990. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, G.M.; Campbell, L.E.; Cordell, H.J.; Irvine, A.D.; McLean, W.H.I.; Brown, S.J. Chromosome 11q13.5 Variant Associated with Childhood Eczema: An Effect Supplementary to Filaggrin Mutations. J Allergy Clin Immunol 2010, 125, 170–174.e1-2. [Google Scholar] [CrossRef] [PubMed]
- Greisenegger, E.; Novak, N.; Maintz, L.; Bieber, T.; Zimprich, F.; Haubenberger, D.; Gleiss, A.; Stingl, G.; Kopp, T.; Zimprich, A. Analysis of Four Prevalent Filaggrin Mutations (R501X, 2282del4, R2447X and S3247X) in Austrian and German Patients with Atopic Dermatitis. J Eur Acad Dermatol Venereol 2010, 24, 607–610. [Google Scholar] [CrossRef]
- Gao, P.-S.; Rafaels, N.M.; Hand, T.; Murray, T.; Boguniewicz, M.; Hata, T.; Schneider, L.; Hanifin, J.M.; Gallo, R.L.; Gao, L.; et al. Filaggrin Mutations That Confer Risk of Atopic Dermatitis Confer Greater Risk for Eczema Herpeticum. J Allergy Clin Immunol 2009, 124, 507–513, 513.e1-7. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Relton, C.L.; Liao, H.; Zhao, Y.; Sandilands, A.; Wilson, I.J.; Burn, J.; Reynolds, N.J.; McLean, W.H.I.; Cordell, H.J. Filaggrin Null Mutations and Childhood Atopic Eczema: A Population-Based Case-Control Study. J Allergy Clin Immunol 2008, 121, 940–946.e3. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Sandilands, A.; Zhao, Y.; Liao, H.; Relton, C.L.; Meggitt, S.J.; Trembath, R.C.; Barker, J.N.W.N.; Reynolds, N.J.; Cordell, H.J.; et al. Prevalent and Low-Frequency Null Mutations in the Filaggrin Gene Are Associated with Early-Onset and Persistent Atopic Eczema. J Invest Dermatol 2008, 128, 1591–1594. [Google Scholar] [CrossRef]
- Giardina, E.; Paolillo, N.; Sinibaldi, C.; Novelli, G. R501X and 2282del4 Filaggrin Mutations Do Not Confer Susceptibility to Psoriasis and Atopic Dermatitis in Italian Patients. Dermatology 2008, 216, 83–84. [Google Scholar] [CrossRef]
- Weidinger, S.; O’Sullivan, M.; Illig, T.; Baurecht, H.; Depner, M.; Rodriguez, E.; Ruether, A.; Klopp, N.; Vogelberg, C.; Weiland, S.K.; et al. Filaggrin Mutations, Atopic Eczema, Hay Fever, and Asthma in Children. J Allergy Clin Immunol 2008, 121, 1203–1209.e1. [Google Scholar] [CrossRef]
- Rogers, A.J.; Celedón, J.C.; Lasky-Su, J.A.; Weiss, S.T.; Raby, B.A. Filaggrin Mutations Confer Susceptibility to Atopic Dermatitis but Not to Asthma. J Allergy Clin Immunol 2007, 120, 1332–1337. [Google Scholar] [CrossRef]
- Lerbaek, A.; Bisgaard, H.; Agner, T.; Ohm Kyvik, K.; Palmer, C.N.A.; Menné, T. Filaggrin Null Alleles Are Not Associated with Hand Eczema or Contact Allergy. Br J Dermatol 2007, 157, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Sandilands, A.; Terron-Kwiatkowski, A.; Hull, P.R.; O’Regan, G.M.; Clayton, T.H.; Watson, R.M.; Carrick, T.; Evans, A.T.; Liao, H.; Zhao, Y.; et al. Comprehensive Analysis of the Gene Encoding Filaggrin Uncovers Prevalent and Rare Mutations in Ichthyosis Vulgaris and Atopic Eczema. Nat Genet 2007, 39, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, S.; Rodríguez, E.; Stahl, C.; Wagenpfeil, S.; Klopp, N.; Illig, T.; Novak, N. Filaggrin Mutations Strongly Predispose to Early-Onset and Extrinsic Atopic Dermatitis. J Invest Dermatol 2007, 127, 724–726. [Google Scholar] [CrossRef] [PubMed]
- Marenholz, I.; Nickel, R.; Rüschendorf, F.; Schulz, F.; Esparza-Gordillo, J.; Kerscher, T.; Grüber, C.; Lau, S.; Worm, M.; Keil, T.; et al. Filaggrin Loss-of-Function Mutations Predispose to Phenotypes Involved in the Atopic March. J Allergy Clin Immunol 2006, 118, 866–871. [Google Scholar] [CrossRef]
- Stemmler, S.; Parwez, Q.; Petrasch-Parwez, E.; Epplen, J.T.; Hoffjan, S. Two Common Loss-of-Function Mutations within the Filaggrin Gene Predispose for Early Onset of Atopic Dermatitis. J Invest Dermatol 2007, 127, 722–724. [Google Scholar] [CrossRef]
- Barker, J.N.W.N.; Palmer, C.N.A.; Zhao, Y.; Liao, H.; Hull, P.R.; Lee, S.P.; Allen, M.H.; Meggitt, S.J.; Reynolds, N.J.; Trembath, R.C.; et al. Null Mutations in the Filaggrin Gene (FLG) Determine Major Susceptibility to Early-Onset Atopic Dermatitis That Persists into Adulthood. J Invest Dermatol 2007, 127, 564–567. [Google Scholar] [CrossRef]
- Palmer, C.N.A.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.D.; et al. Common Loss-of-Function Variants of the Epidermal Barrier Protein Filaggrin Are a Major Predisposing Factor for Atopic Dermatitis. Nat Genet 2006, 38, 441–446. [Google Scholar] [CrossRef]
- Dêbiñska, A.; Danielewicz, H.; Drabik-Chamerska, A.; Kalita, D.; Boznañski, A. Chromosome 11q13.5 Variant as a Risk Factor for Atopic Dermatitis in Children. Postepy Dermatol Alergol 2020, 37, 103–110. [Google Scholar] [CrossRef]
- Can, C.; Yazıcıoğlu, M.; Gürkan, H.; Tozkır, H.; Görgülü, A.; Süt, N.H. Lack of Association Between Toll-like Receptor 2 Polymorphisms (R753Q and A-16934T) and Atopic Dermatitis in Children from Thrace Region of Turkey. Balkan Med J 2017, 34, 232–238. [Google Scholar] [CrossRef]
- Salpietro, C.; Rigoli, L.; Miraglia Del Giudice, M.; Cuppari, C.; Di Bella, C.; Salpietro, A.; Maiello, N.; La Rosa, M.; Marseglia, G.L.; Leonardi, S.; et al. TLR2 and TLR4 Gene Polymorphisms and Atopic Dermatitis in Italian Children: A Multicenter Study. Int J Immunopathol Pharmacol 2011, 24, 33–40. [Google Scholar] [CrossRef]
- Galli, E.; Ciucci, A.; Cersosimo, S.; Pagnini, C.; Avitabile, S.; Mancino, G.; Delle Fave, G.; Corleto, V.D. Eczema and Food Allergy in an Italian Pediatric Cohort: No Association with TLR-2 and TLR-4 Polymorphisms. Int J Immunopathol Pharmacol 2010, 23, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-Y.; Schumann, R.R.; Hamann, L.; Neumann, K.; Worm, M.; Heine, G. Association of the Toll-like Receptor 2 A-16934T Promoter Polymorphism with Severe Atopic Dermatitis. Allergy 2009, 64, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, M.; Langnickel, J.; Draing, C.; Renz, H.; Kapp, A.; Werfel, T. Dysregulation of Toll-like Receptor-2 (TLR-2)-Induced Effects in Monocytes from Patients with Atopic Dermatitis: Impact of the TLR-2 R753Q Polymorphism. Allergy 2008, 63, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Levchenko, L.Yu.; Izmailova, O.V.; Shlykova, O.A.; Kaidashev, I.P. TLR4 896A/G Gene Polymorphism, Rather than the TLR4 1196C/T and TLR2 2258G/A Gene Polymorphisms, Determines the Severe and Aggravated Course of Atopic Dermatitis in Children. Cytology and Genetics 2013, 47, 167–173. [Google Scholar] [CrossRef]
- Ahmad-Nejad, P.; Mrabet-Dahbi, S.; Breuer, K.; Klotz, M.; Werfel, T.; Herz, U.; Heeg, K.; Neumaier, M.; Renz, H. The Toll-like Receptor 2 R753Q Polymorphism Defines a Subgroup of Patients with Atopic Dermatitis Having Severe Phenotype. J Allergy Clin Immunol 2004, 113, 565–567. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Patria, M.F.; Spena, S.; Codecà, C.; Tagliabue, C.; Zampiero, A.; Lelii, M.; Montinaro, V.; Pelucchi, C.; Principi, N. Impact of Genetic Polymorphisms on Paediatric Atopic Dermatitis. Int J Immunopathol Pharmacol 2015, 28, 286–295. [Google Scholar] [CrossRef]
- Lesiak, A.; Zakrzewski, M.; Przybyłowska, K.; Rogowski-Tylman, M.; Wozniacka, A.; Narbutt, J. Atopic Dermatitis Patients Carrying G Allele in -1082 G/A IL-10 Polymorphism Are Predisposed to Higher Serum Concentration of IL-10. Arch Med Sci 2014, 10, 1239–1243. [Google Scholar] [CrossRef]
- Kayserova, J.; Sismova, K.; Zentsova-Jaresova, I.; Katina, S.; Vernerova, E.; Polouckova, A.; Capkova, S.; Malinova, V.; Striz, I.; Sediva, A. A Prospective Study in Children with a Severe Form of Atopic Dermatitis: Clinical Outcome in Relation to Cytokine Gene Polymorphisms. J Investig Allergol Clin Immunol 2012, 22, 92–101. [Google Scholar]
- Stavric, K.; Peova, S.; Trajkov, D.; Spiroski, M. Gene Polymorphisms of 22 Cytokines in Macedonian Children with Atopic Dermatitis. Iran J Allergy Asthma Immunol 2012, 11, 37–50. [Google Scholar]
- Lesiak, A.; Kuna, P.; Zakrzewski, M.; van Geel, M.; Bladergroen, R.S.; Przybylowska, K.; Stelmach, I.; Majak, P.; Hawro, T.; Sysa-Jedrzejowska, A.; et al. Combined Occurrence of Filaggrin Mutations and IL-10 or IL-13 Polymorphisms Predisposes to Atopic Dermatitis. Exp Dermatol 2011, 20, 491–495. [Google Scholar] [CrossRef]
- Reich, K.; Westphal, G.; König, I.R.; Mössner, R.; Schupp, P.; Gutgesell, C.; Hallier, E.; Ziegler, A.; Neumann, C. Cytokine Gene Polymorphisms in Atopic Dermatitis. Br J Dermatol 2003, 148, 1237–1241. [Google Scholar] [CrossRef]
- Arkwright, P.D.; Chase, J.M.; Babbage, S.; Pravica, V.; David, T.J.; Hutchinson, I.V. Atopic Dermatitis Is Associated with a Low-Producer Transforming Growth Factor Beta(1) Cytokine Genotype. J Allergy Clin Immunol 2001, 108, 281–284. [Google Scholar] [CrossRef]
- Ponińska, J.K.; Samoliński, B.; Tomaszewska, A.; Raciborski, F.; Samel-Kowalik, P.; Walkiewicz, A.; Lipiec, A.; Piekarska, B.; Krzych-Fałta, E.; Namysłowski, A.; et al. Haplotype Dependent Association of Rs7927894 (11q13.5) with Atopic Dermatitis and Chronic Allergic Rhinitis: A Study in ECAP Cohort. PLoS One 2017, 12, e0183922. [Google Scholar] [CrossRef]
- Greisenegger, E.K.; Zimprich, F.; Zimprich, A.; Gleiss, A.; Kopp, T. Association of the Chromosome 11q13.5 Variant with Atopic Dermatitis in Austrian Patients. Eur J Dermatol 2013, 23, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Marenholz, I.; Bauerfeind, A.; Esparza-Gordillo, J.; Kerscher, T.; Granell, R.; Nickel, R.; Lau, S.; Henderson, J.; Lee, Y.-A. The Eczema Risk Variant on Chromosome 11q13 (Rs7927894) in the Population-Based ALSPAC Cohort: A Novel Susceptibility Factor for Asthma and Hay Fever. Hum Mol Genet 2011, 20, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Hummelshoj, T.; Bodtger, U.; Datta, P.; Malling, H.J.; Oturai, A.; Poulsen, L.K.; Ryder, L.P.; Sorensen, P.S.; Svejgaard, E.; Svejgaard, A. Association between an Interleukin-13 Promoter Polymorphism and Atopy. Eur J Immunogenet 2003, 30, 355–359. [Google Scholar] [CrossRef]
- Dębińska, A.; Danielewicz, H.; Sozańska, B. Genetic Variants in Epidermal Differentiation Complex Genes as Predictive Biomarkers for Atopic Eczema, Allergic Sensitization, and Eczema-Associated Asthma in a 6-Year Follow-Up Case-Control Study in Children. J Clin Med 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Trzeciak, M.; Gleń, J.; Roszkiewicz, J.; Nedoszytko, B. Association of Single Nucleotide Polymorphism of Interleukin-18 with Atopic Dermatitis. J Eur Acad Dermatol Venereol 2010, 24, 78–79. [Google Scholar] [CrossRef] [PubMed]
- Kılıç, S.; Sılan, F.; Hız, M.M.; Işık, S.; Ögretmen, Z.; Özdemir, Ö. Vitamin D Receptor Gene BSMI, FOKI, APAI, and TAQI Polymorphisms and the Risk of Atopic Dermatitis. J Investig Allergol Clin Immunol 2016, 26, 106–110. [Google Scholar] [CrossRef]
- Heine, G.; Hoefer, N.; Franke, A.; Nöthling, U.; Schumann, R.R.; Hamann, L.; Worm, M. Association of Vitamin D Receptor Gene Polymorphisms with Severe Atopic Dermatitis in Adults. Br J Dermatol 2013, 168, 855–858. [Google Scholar] [CrossRef]
- Dežman, K.; Korošec, P.; Rupnik, H.; Rijavec, M. SPINK5 Is Associated with Early-Onset and CHI3L1 with Late-Onset Atopic Dermatitis. Int J Immunogenet 2017, 44, 212–218. [Google Scholar] [CrossRef]
- Fölster-Holst, R.; Stoll, M.; Koch, W.A.; Hampe, J.; Christophers, E.; Schreiber, S. Lack of Association of SPINK5 Polymorphisms with Nonsyndromic Atopic Dermatitis in the Population of Northern Germany. Br J Dermatol 2005, 152, 1365–1367. [Google Scholar] [CrossRef]
- Vavilin, V.A.; Safronova, O.G.; Lyapunova, A.A.; Lyakhovich, V.V.; Kaznacheeva, L.F.; Manankin, N.A.; Molokova, A.V. Interaction of GSTM1, GSTT1, and GSTP1 Genotypes in Determination of Predisposition to Atopic Dermatitis. Bull Exp Biol Med 2003, 136, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Safronova, O.G.; Vavilin, V.A.; Lyapunova, A.A.; Makarova, S.I.; Lyakhovich, V.V.; Kaznacheeva, L.F.; Manankin, N.A.; Batychko, O.A.; Gavalov, S.M. Relationship between Glutathione S-Transferase P1 Polymorphism and Bronchial Asthma and Atopic Dermatitis. Bull Exp Biol Med 2003, 136, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Wang, H.-Y.; Lee, H.; Lee, S.-Y.; Hong, S.-J.; Choi, E.H. Clinical Characteristics and Genetic Variations in Early-Onset Atopic Dermatitis Patients. Ann Dermatol 2019, 31, 286–293. [Google Scholar] [CrossRef]
- Sasaki, T.; Furusyo, N.; Shiohama, A.; Takeuchi, S.; Nakahara, T.; Uchi, H.; Hirota, T.; Tamari, M.; Shimizu, N.; Ebihara, T.; et al. Filaggrin Loss-of-Function Mutations Are Not a Predisposing Factor for Atopic Dermatitis in an Ishigaki Island under Subtropical Climate. J Dermatol Sci 2014, 76, 10–15. [Google Scholar] [CrossRef]
- Meng, L.; Wang, L.; Tang, H.; Tang, X.; Jiang, X.; Zhao, J.; Gao, J.; Li, B.; Fu, X.; Chen, Y.; et al. Filaggrin Gene Mutation c.3321delA Is Associated with Various Clinical Features of Atopic Dermatitis in the Chinese Han Population. PLoS One 2014, 9, e98235. [Google Scholar] [CrossRef]
- Lee, D.-E.; Park, S.-Y.; Han, J.-Y.; Ryu, H.-M.; Lee, H.-C.; Han, Y.S. Association between Filaggrin Mutations and Atopic Dermatitis in Korean Pregnant Women. Int J Dermatol 2013, 52, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Common, J.E.A.; Haines, R.L.; Balakrishnan, A.; Brown, S.J.; Goh, C.S.M.; Cordell, H.J.; Sandilands, A.; Campbell, L.E.; Kroboth, K.; et al. Wide Spectrum of Filaggrin-Null Mutations in Atopic Dermatitis Highlights Differences between Singaporean Chinese and European Populations. Br J Dermatol 2011, 165, 106–114. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, Y.; Wang, W.; Shi, M.; Chen, X.; Yao, Z. Mutations in the Filaggrin Gene in Han Chinese Patients with Atopic Dermatitis. Allergy 2011, 66, 420–427. [Google Scholar] [CrossRef]
- Osawa, R.; Konno, S.; Akiyama, M.; Nemoto-Hasebe, I.; Nomura, T.; Nomura, Y.; Abe, R.; Sandilands, A.; McLean, W.H.I.; Hizawa, N.; et al. Japanese-Specific Filaggrin Gene Mutations in Japanese Patients Suffering from Atopic Eczema and Asthma. J Invest Dermatol 2010, 130, 2834–2836. [Google Scholar] [CrossRef]
- Nomura, Y.; Akiyama, M.; Nomura, T.; Nemoto-Hasebe, I.; Abe, R.; McLean, W.H.I.; Shimizu, H. Chromosome 11q13.5 Variant: No Association with Atopic Eczema in the Japanese Population. J Dermatol Sci 2010, 59, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, L.; Di, Z.-H.; Zhao, L.-P.; Lu, Y.-N.; Xu, J.; Chen, H.-D.; Gao, X.-H. Association Analysis of Filaggrin Gene Mutations and Atopic Dermatitis in Northern China. Br J Dermatol 2010, 162, 225–227. [Google Scholar] [CrossRef] [PubMed]
- Nemoto-Hasebe, I.; Akiyama, M.; Nomura, T.; Sandilands, A.; McLean, W.H.I.; Shimizu, H. FLG Mutation p.Lys4021X in the C-Terminal Imperfect Filaggrin Repeat in Japanese Patients with Atopic Eczema. Br J Dermatol 2009, 161, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Akiyama, M.; Sandilands, A.; Nemoto-Hasebe, I.; Sakai, K.; Nagasaki, A.; Palmer, C.N.A.; Smith, F.J.D.; McLean, W.H.I.; Shimizu, H. Prevalent and Rare Mutations in the Gene Encoding Filaggrin in Japanese Patients with Ichthyosis Vulgaris and Atopic Dermatitis. J Invest Dermatol 2009, 129, 1302–1305. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Akiyama, M.; Sandilands, A.; Nemoto-Hasebe, I.; Sakai, K.; Nagasaki, A.; Ota, M.; Hata, H.; Evans, A.T.; Palmer, C.N.A.; et al. Specific Filaggrin Mutations Cause Ichthyosis Vulgaris and Are Significantly Associated with Atopic Dermatitis in Japan. J Invest Dermatol 2008, 128, 1436–1441. [Google Scholar] [CrossRef]
- Enomoto, H.; Hirata, K.; Otsuka, K.; Kawai, T.; Takahashi, T.; Hirota, T.; Suzuki, Y.; Tamari, M.; Otsuka, F.; Fujieda, S.; et al. Filaggrin Null Mutations Are Associated with Atopic Dermatitis and Elevated Levels of IgE in the Japanese Population: A Family and Case-Control Study. J Hum Genet 2008, 53, 615. [Google Scholar] [CrossRef]
- Nomura, T.; Sandilands, A.; Akiyama, M.; Liao, H.; Evans, A.T.; Sakai, K.; Ota, M.; Sugiura, H.; Yamamoto, K.; Sato, H.; et al. Unique Mutations in the Filaggrin Gene in Japanese Patients with Ichthyosis Vulgaris and Atopic Dermatitis. J Allergy Clin Immunol 2007, 119, 434–440. [Google Scholar] [CrossRef]
- Ching, G.K.; Hon, K.-L.; Ng, P.-C.; Leung, T.-F. Filaggrin Null Mutations in Childhood Atopic Dermatitis among the Chinese. Int J Immunogenet 2009, 36, 251–254. [Google Scholar] [CrossRef]
- Zhong, W.-L.; Wu, X.; Yu, B.; Zhang, J.; Zhang, W.; Xu, N.; Zhou, J.; Zheng, J.-C.; Chen, X.-F.; Dou, X. Filaggrin Gene Mutation c.3321delA Is Associated with Dry Phenotypes of Atopic Dermatitis in the Chinese Han Population. Chin Med J (Engl) 2016, 129, 1498–1500. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Yang, S.W.; Kim, H.-L.; Kim, S.-H.; Kim, S.J.; Park, S.-M.; Son, M.; Ryu, S.; Pyo, Y.-S.; Lee, J.-S.; et al. Association between P478S Polymorphism of the Filaggrin Gene & Atopic Dermatitis. Indian J Med Res 2013, 138, 922–927. [Google Scholar] [PubMed]
- Wang, I.J.; Lin, T.J.; Kuo, C.F.; Lin, S.L.; Lee, Y.L.; Chen, P.C. Filaggrin Polymorphism P478S, IgE Level, and Atopic Phenotypes. Br J Dermatol 2011, 164, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Chen, Q.; Liu, Q.-H.; Guo, Y.-F.; Cheng, R.-H.; Zhang, J.; Zhang, J.-H.; Wang, L.-W.; Ji, R.-X. Interaction between Antibiotic Use and MS4A2 Gene Polymorphism on Childhood Eczema: A Prospective Birth Cohort Study. BMC Pediatr 2021, 21, 314. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Cao, X.-L.; Wan, Y.-J.; Meng, J.; Guo, L.-H. IL-4 Gene Polymorphism May Contribute to an Increased Risk of Atopic Dermatitis in Children. Dis Markers 2016, 2016, 1021942. [Google Scholar] [CrossRef]
- Hussein, Y.M.; Alzahrani, S.S.; Alharthi, A.A.; Alhazmi, A.S.; Ghonaim, M.M.; Alghamdy, A.A.N.; El Askary, A. Gene Polymorphism of Interleukin-4, Interleukin-4 Receptor and STAT6 in Children with Atopic Dermatitis in Taif, Saudi Arabia. Immunol Invest 2016, 45, 223–234. [Google Scholar] [CrossRef]
- Gharagozlou, M.; Behniafard, N.; Amirzargar, A.A.; Hosseinverdi, S.; Sotoudeh, S.; Farhadi, E.; Khaledi, M.; Aryan, Z.; Moghaddam, Z.G.; Mahmoudi, M.; et al. Association between Single Nucleotide Polymorphisms of the Interleukin-4 Gene and Atopic Dermatitis. Acta Dermatovenerol Croat 2015, 23, 96–100. [Google Scholar]
- Hussein, Y.M.; Shalaby, S.M.; Nassar, A.; Alzahrani, S.S.; Alharbi, A.S.; Nouh, M. Association between Genes Encoding Components of the IL-4/IL-4 Receptor Pathway and Dermatitis in Children. Gene 2014, 545, 276–281. [Google Scholar] [CrossRef]
- Tanaka, K.; Sugiura, H.; Uehara, M.; Hashimoto, Y.; Donnelly, C.; Montgomery, D.S. Lack of Association between Atopic Eczema and the Genetic Variants of Interleukin-4 and the Interleukin-4 Receptor Alpha Chain Gene: Heterogeneity of Genetic Backgrounds on Immunoglobulin E Production in Atopic Eczema Patients. Clin Exp Allergy 2001, 31, 1522–1527. [Google Scholar] [CrossRef]
- Kawashima, T.; Noguchi, E.; Arinami, T.; Yamakawa-Kobayashi, K.; Nakagawa, H.; Otsuka, F.; Hamaguchi, H. Linkage and Association of an Interleukin 4 Gene Polymorphism with Atopic Dermatitis in Japanese Families. J Med Genet 1998, 35, 502–504. [Google Scholar] [CrossRef]
- Morizane, S.; Ouchida, M.; Sunagawa, K.; Sugimoto, S.; Kobashi, M.; Sugihara, S.; Nomura, H.; Tsuji, K.; Sato, A.; Miura, Y.; et al. Analysis of All 34 Exons of the SPINK5 Gene in Japanese Atopic Dermatitis Patients. Acta Med Okayama 2018, 72, 275–282. [Google Scholar] [CrossRef]
- Zhao, L.P.; Di, Z.; Zhang, L.; Wang, L.; Ma, L.; Lv, Y.; Hong, Y.; Wei, H.; Chen, H.D.; Gao, X.H. Association of SPINK5 Gene Polymorphisms with Atopic Dermatitis in Northeast China. J Eur Acad Dermatol Venereol 2012, 26, 572–577. [Google Scholar] [CrossRef]
- Kato, A.; Fukai, K.; Oiso, N.; Hosomi, N.; Murakami, T.; Ishii, M. Association of SPINK5 Gene Polymorphisms with Atopic Dermatitis in the Japanese Population. Br J Dermatol 2003, 148, 665–669. [Google Scholar] [CrossRef]
- Bin Huraib, G.; Al Harthi, F.; Arfin, M.; Al-Sugheyr, M.; Rizvi, S.; Al-Asmari, A. Cytokine Gene Polymorphisms in Saudi Patients With Atopic Dermatitis: A Case-Control Study. Biomark Insights 2018, 13, 1177271918777760. [Google Scholar] [CrossRef] [PubMed]
- Behniafard, N.; Amirzargar, A.A.; Gharagozlou, M.; Delavari, F.; Hosseinverdi, S.; Sotoudeh, S.; Farhadi, E.; Mahmoudi, M.; Khaledi, M.; Moghaddam, Z.G.; et al. Single Nucleotide Polymorphisms of the Genes Encoding IL-10 and TGF-Β1 in Iranian Children with Atopic Dermatitis. Allergol Immunopathol (Madr) 2018, 46, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Sohn, M.H.; Song, J.S.; Kim, K.-W.; Kim, E.-S.; Kim, K.-E.; Lee, J.M. Association of Interleukin-10 Gene Promoter Polymorphism in Children with Atopic Dermatitis. J Pediatr 2007, 150, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Tanaka, K.; Arakawa, M. Case-Control Study of Eczema in Relation to IL4Rα Genetic Polymorphisms in Japanese Women: The Kyushu Okinawa Maternal and Child Health Study. Scand J Immunol 2013, 77, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Tanaka, K.; Arakawa, M. IL13 Genetic Polymorphisms, Smoking, and Eczema in Women: A Case-Control Study in Japan. BMC Med Genet 2011, 12, 142. [Google Scholar] [CrossRef]
- Miyake, Y.; Kiyohara, C.; Koyanagi, M.; Fujimoto, T.; Shirasawa, S.; Tanaka, K.; Sasaki, S.; Hirota, Y. Case-Control Study of Eczema Associated with IL13 Genetic Polymorphisms in Japanese Children. Int Arch Allergy Immunol 2011, 154, 328–335. [Google Scholar] [CrossRef]
- Takahashi, N.; Akahoshi, M.; Matsuda, A.; Ebe, K.; Inomata, N.; Obara, K.; Hirota, T.; Nakashima, K.; Shimizu, M.; Tamari, M.; et al. Association of the IL12RB1 Promoter Polymorphisms with Increased Risk of Atopic Dermatitis and Other Allergic Phenotypes. Hum Mol Genet 2005, 14, 3149–3159. [Google Scholar] [CrossRef]
- Namkung, J.-H.; Lee, J.-E.; Kim, E.; Park, G.T.; Yang, H.S.; Jang, H.Y.; Shin, E.-S.; Cho, E.-Y.; Yang, J.-M. An Association between IL-9 and IL-9 Receptor Gene Polymorphisms and Atopic Dermatitis in a Korean Population. J Dermatol Sci 2011, 62, 16–21. [Google Scholar] [CrossRef]
- Kim, J.S.; Choi, J.; Hahn, H.-J.; Lee, Y.-B.; Yu, D.-S.; Kim, J.-W. Association of Macrophage Migration Inhibitory Factor Polymorphisms with Total Plasma IgE Levels in Patients with Atopic Dermatitis in Korea. PLoS One 2016, 11, e0162477. [Google Scholar] [CrossRef]
- Ma, L.; Xue, H.-B.; Guan, X.-H.; Qi, R.-Q.; Liu, Y.-B. Macrophage Migration Inhibitory Factor Promoter 173G/C Polymorphism Is Associated with Atopic Dermatitis Risk. Int J Dermatol 2014, 53, e75–77. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Tanaka, K.; Arakawa, M. IL5RA Polymorphisms, Smoking and Eczema in Japanese Women: The Kyushu Okinawa Maternal and Child Health Study. Int J Immunogenet 2015, 42, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Tsunemi, Y.; Saeki, H.; Shibata, S.; Sekiya, T.; Nakamura, K.; Kakinuma, T.; Kagami, S.; Fujita, H.; Tada, Y.; et al. Interferon-18 Gene Polymorphism -137 G/C Is Associated with Susceptibility to Psoriasis Vulgaris but Not with Atopic Dermatitis in Japanese Patients. J Dermatol Sci 2009, 53, 162–163. [Google Scholar] [CrossRef] [PubMed]
- Osawa, K.; Etoh, T.; Ariyoshi, N.; Ishii, I.; Ohtani, M.; Kariya, S.; Uchino, K.; Kitada, M. Relationship between Kaposi’s Varicelliform Eruption in Japanese Patients with Atopic Dermatitis Treated with Tacrolimus Ointment and Genetic Polymorphisms in the IL-18 Gene Promoter Region. J Dermatol 2007, 34, 531–536. [Google Scholar] [CrossRef]
- Behniafard, N.; Gharagozlou, M.; Farhadi, E.; Khaledi, M.; Sotoudeh, S.; Darabi, B.; Fathi, S.M.; Gholizadeh Moghaddam, Z.; Mahmoudi, M.; Aghamohammadi, A.; et al. TNF-Alpha Single Nucleotide Polymorphisms in Atopic Dermatitis. Eur Cytokine Netw 2012, 23, 163–165. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, Y.; Lin, L.; Wang, J.; Peng, X.; Li, J.; Li, L. Association of Polymorphisms in the Promoter Region of FCER1A Gene with Atopic Dermatitis, Chronic Uticaria, Asthma, and Serum Immunoglobulin E Levels in a Han Chinese Population. Hum Immunol 2012, 73, 301–305. [Google Scholar] [CrossRef]
- Park, K.Y.; Park, M.K.; Kim, E.J.; Lee, M.-K.; Seo, S.J. FCεRI Gene Promoter Polymorphisms and Total IgE Levels in Susceptibility to Atopic Dermatitis in Korea. J Korean Med Sci 2011, 26, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Tsunemi, Y.; Komine, M.; Sekiya, T.; Saeki, H.; Nakamura, K.; Hirai, K.; Kakinuma, T.; Kagami, S.; Fujita, H.; Asano, N.; et al. The -431C>T Polymorphism of Thymus and Activation-Regulated Chemokine Increases the Promoter Activity but Is Not Associated with Susceptibility to Atopic Dermatitis in Japanese Patients. Exp Dermatol 2004, 13, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Tsunemi, Y.; Miyamasu, M.; Ohta, K.; Morita, A.; Saeki, H.; Matsushima, K.; Yoshie, O.; Tsuchiya, N.; Yamaguchi, M.; et al. Variations in the Human Th2-Specific Chemokine TARC Gene. Immunogenetics 2003, 54, 742–745. [Google Scholar] [CrossRef] [PubMed]
- Grafanaki, K.; Antonatos, C.; Maniatis, A.; Petropoulou, A.; Vryzaki, E.; Vasilopoulos, Y.; Georgiou, S.; Gregoriou, S. Intrinsic Effects of Exposome in Atopic Dermatitis: Genomics, Epigenomics and Regulatory Layers. Journal of Clinical Medicine 2023, 12. [Google Scholar] [CrossRef]
- McAleer, M.A.; Irvine, A.D. The Multifunctional Role of Filaggrin in Allergic Skin Disease. J Allergy Clin Immunol 2013, 131, 280–291. [Google Scholar] [CrossRef]
- Rodríguez, E.; Baurecht, H.; Herberich, E.; Wagenpfeil, S.; Brown, S.J.; Cordell, H.J.; Irvine, A.D.; Weidinger, S. Meta-Analysis of Filaggrin Polymorphisms in Eczema and Asthma: Robust Risk Factors in Atopic Disease. J Allergy Clin Immunol 2009, 123, 1361–1370.e7. [Google Scholar] [CrossRef] [PubMed]
- Nedoszytko, B.; Olszewska, B.; Roszkiewicz, J.; Glen, J.; Zabłotna, M.; Ługowska-Umer, H.; Nowicki, R.; Sokołowska-Wojdyło, M. The Role of Polymorphism of Interleukin-2, -10, -13 and TNF-α Genes in Cutaneous T-Cell Lymphoma Pathogenesis. Postepy Dermatol Alergol 2016, 33, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Wong, L.; Lang, A.; Kraus, C.; Anderson, N.; Elsensohn, A. Cutaneous T-Cell Lymphoma Following Dupilumab Use: A Systematic Review. Int J Dermatol 2022. [Google Scholar] [CrossRef] [PubMed]
- Imboden, M.; Nieters, A.; Bircher, A.J.; Brutsche, M.; Becker, N.; Wjst, M.; Ackermann-Liebrich, U.; Berger, W.; Probst-Hensch, N.M. Cytokine Gene Polymorphisms and Atopic Disease in Two European Cohorts. (ECRHS-Basel and SAPALDIA). Clin Mol Allergy 2006, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Weissler, K.A.; Frischmeyer-Guerrerio, P.A. Genetic Evidence for the Role of Transforming Growth Factor-β in Atopic Phenotypes. Curr Opin Immunol 2019, 60, 54–62. [Google Scholar] [CrossRef]
- Bozza, M.T.; Lintomen, L.; Kitoko, J.Z.; Paiva, C.N.; Olsen, P.C. The Role of MIF on Eosinophil Biology and Eosinophilic Inflammation. Clin Rev Allergy Immunol 2020, 58, 15–24. [Google Scholar] [CrossRef]
- Egger, M.; Smith, G.D. Bias in Location and Selection of Studies. BMJ 1998, 316, 61–66. [Google Scholar] [CrossRef]
- Lou, C.; Mitra, N.; Wubbenhorst, B.; D’Andrea, K.; Hoffstad, O.; Kim, B.S.; Yan, A.; Zaenglein, A.L.; Fuxench, Z.C.; Nathanson, K.L.; et al. Association between Fine Mapping Thymic Stromal Lymphopoietin and Atopic Dermatitis Onset and Persistence. Ann Allergy Asthma Immunol 2019, 123, 595–601.e1. [Google Scholar] [CrossRef]
| Study, Year [REF] | Rs ID | Sample Size |
|---|---|---|
| Vardar Acar N et al., 2020 [14] | rs558269137, rs61816761, rs138726443, rs150597413 | 189 |
| Jurakic Toncic R et al., 2020 [15] | rs558269137, rs61816761, rs138726443 | 150 |
| González-Tarancón R et al., 2020 [16] | rs558269137, rs61816761, rs138726443, CG2 | 214 |
| Woźniak M et al., 2016 [17] | rs558269137, rs61816761, CG1 | 121 |
| Trzeciak M et al. (a), 2016 [18] | rs558269137, rs61816761, rs1800925, rs187238 | 275 |
| Trzeciak M et al. (b), 2016 [19] | rs558269137 | 256 |
| Ballardini N et al., 2013 [20] | rs558269137, rs61816761, rs138726443, CG2 | 1,854 |
| Ercan H et al., 2013 [21] | rs61816761 | 99 |
| Mlitz V et al., 2012 [22] | rs558269137, rs61816761, rs138726443, CG2 | 196 |
| O'Regan GM et al., 2010 [23] | rs558269137, rs61816761, rs138726443, rs150597413, CG3, rs7927894, rs877776 | 1,511 |
| Greisenegger E et al., 2010 [24] | rs558269137, rs61816761, rs138726443, rs150597413, CG3 | 864 |
| Gao PS et al., 2009 [25] | rs558269137, rs61816761, CG1 | 435 |
| Brown SJ et al. (a), 2008 [26] | rs558269137, rs61816761, rs138726443, rs150597413, rs397507563 | 811 |
| Brown SJ et al. (b), 2008 [27] | rs558269137, rs61816761, rs138726443, rs150597413, rs397507563 | 1,221 |
| Giardina E et al., 2008 [28] | rs558269137, rs61816761 | 388 |
| Weidinger S et al., 2008 [29] | rs558269137, rs61816761, rs138726443, rs150597413, CG3 | 3,099 |
| Rogers AJ et al., 2007 [30] | rs558269137, rs61816761, CG1 | 646 |
| Lerbaek A et al., 2007 [31] | rs558269137, rs61816761, CG1 | 215 |
| Sandilands A et al., 2007 [32] | rs558269137, rs61816761, rs138726443, rs150597413, rs397507563 | 924 |
| Weidinger S et al., 2007 [33] | rs558269137, rs61816761, CG1 | 526 |
| Marenholz I et al., 2006 [34] | rs558269137, rs61816761, CG1 | 507 |
| Stemmler S et al., 2007 [35] | rs558269137, rs61816761, CG1 | 1,078 |
| Barker JN et al., 2007 [36] | rs558269137, rs61816761, CG1 | 1,626 |
| Palmer CN et al., 2006 [37] | rs558269137, rs61816761, CG1 | 241 |
| Dêbiñska A et al., 2020 [38] | CG3, rs7927894 | 188 |
| Can C et al., 2017 [39] | rs5743708, rs4696480 | 139 |
| Salpietro C et al., 2011 [40] | rs5743708, rs4696480, rs4986790, rs4986791 | 337 |
| Galli E et al., 2010 [41] | rs5743708 | 249 |
| Oh DY et al., 2009 [42] | rs5743708, rs4696480, rs4986790, rs4986791 | 265 |
| Niebuhr M et al., 2008 [43] | rs5743708 | 19 |
| Levchenko L Yu et al., 2013 [44] | rs5743708 | 131 |
| Ahmad-Nejad et al., 2003 [45] | rs5743708 | 117 |
| Esposito S et al., 2015 [46] | rs1800896, rs1800872 | 223 |
| Lesiak A et al., 2014 [47] | rs1800896, rs1800925, rs2243250 | 136 |
| Kayserova J et al., 2012 [48] | rs1800896, rs1800871, rs1800872, rs2243250, rs2243248, rs1800795, rs1800797, rs1801275 | 197 |
| Stavric K et al., 2012 [49] | rs1800896, rs1800871, rs1800872, rs2243250, rs2243248, rs1800471, rs1800470, rs1800795, rs1800797, rs16944, rs1143634, rs1800629, rs361525, rs1801275 | 367 |
| Lesiak A et al., 2011 [50] | rs1800896, rs1800925 | 367 |
| Reich K et al., 2003 [51] | rs1800896, rs1800795, rs16944, rs1143634, rs1800629, rs361525 | 308 |
| Arkwright PD et al., 2001 [52] | rs1800896, rs1800471, rs1800470 | 118 |
| Ponińska JK et al., 2017 [53] | rs7927894 | 810 |
| Greisenegger EK et al., 2013 [54] | rs7927894, rs877776 | 518 |
| Marenholz I et al., 2011 [55] | rs7927894 | 2,485 |
| Hummelshoj T et al., 2003 [56] | rs1800925 | 159 |
| Dêbiñska A et al., 2022 [57] | rs877776 | 188 |
| Trzeciak M et al., 2010 [58] | rs187238 | 113 |
| Kılıç S et al., 2016 [59] | rs2228570, rs7975232, rs1544410, rs731236 | 138 |
| Heine G et al., 2013 [60] | rs2228570, rs7975232, rs1544410, rs731236 | 530 |
| Dežman K et al., 2017 [61] | rs2303067 | 405 |
| Fölster-Holst R et al., 2005 [62] | rs2303067 | 569 |
| Vavilin VA et al., 2003 [63] | rs1695 | 325 |
| Safronova OG et al., 2003 [64] | rs1695 | 274 |
| Abbreviations: CG, Combined Genotype. | ||
| Study, Year [REF] | Rs ID | Sample Size |
|---|---|---|
| Kim BJ et al., 2019 [65] | rs200519781, rs146466242, rs2303064, rs2303070, rs2303065, rs393548, rs436857, rs31563, rs334809 | 325 |
| Sasaki T et al., 2014 [66] | rs200519781, rs121909626, rs761212672, rs145738429, rs61816761, rs146466242, rs772851618 | 721 |
| Meng L et al., 2014 [67] | rs200519781 | 1,988 |
| Lee DE et al., 2013 [68] | rs200519781 | 175 |
| Chen H et al., 2011 [69] | rs200519781, rs61816761 | 865 |
| Zhang H et al., 2011 [70] | rs200519781 | 353 |
| Osawa R et al., 2010 [71] | rs200519781, rs121909626, rs761212672, rs145738429, S2889X, rs61816761, rs146466242, rs772851618, CG | 306 |
| Nomura Y et al., 2010 [72] | rs200519781, rs121909626, rs761212672, rs145738429, S2889X, rs61816761, rs146466242, rs772851618, CG | 307 |
| Ma L et al., 2010 [73] | rs200519781 | 329 |
| Nemoto-Hasebe I et al., 2009 [74] | rs200519781, rs121909626, rs761212672, rs145738429, S2889X, rs61816761, rs146466242, rs772851618, CG | 271 |
| Nomura T et al., 2009 [75] | rs200519781, rs121909626, rs761212672, rs145738429, S2889X, rs61816761, rs772851618 | 252 |
| Nomura T et al., 2008 [76] | rs200519781, rs121909626, rs761212672, S2889X | 235 |
| Enomoto H et al., 2008 [77] | rs200519781, rs121909626 | 1,299 |
| Nomura T et al., 2007 [78] | rs200519781, rs121909626 | 299 |
| Ching GK et al., 2009 [79] | rs121909626, S2889X, rs558269137, rs61816761 | 365 |
| Zhong WL et al., 2016 [80] | rs558269137 | 1,017 |
| Kim SY et al., 2013 [81] | rs11584340 | 527 |
| Wang IJ et al., 2011 [82] | rs11584340 | 328 |
| Hua L et al., 2021 [83] | rs2243250, rs2070874, rs1801275, rs1805010, rs20541 | 597 |
| Shang H et al., 2016 [84] | rs2243250, rs2070874 | 182 |
| Hussein YM et al., 2016 [85] | rs2243250, rs2070874, rs1805010 | 100 |
| Gharagozlou M et al., 2015 [86] | rs2243250, rs2070874, rs1801275 | 228 |
| Hussein YM et al., 2014 [87] | rs2243250, rs2070874 | 206 |
| Tanaka K et al., 2001 [88] | rs2243250, rs2070874, rs1805010 | 424 |
| Kawashima T et al., 1998 [89] | rs2243250, rs2070874 | 425 |
| Morizane S et al., 2018 [90] | rs2303064, rs2303063, rs2303067, rs2303070 | 107 |
| Zhao LP et al., 2012 [91] | rs2303064, rs2303063, rs2303067, rs2303070 | 341 |
| Kato A et al., 2003 [92] | rs2303064, rs2303063, rs2303067, rs2303065 | 234 |
| Bin Huraib G et al., 2018 [93] | rs1800871, rs1800872, rs1800896, rs1800629 | 315 |
| Behniafard N et al., 2018 [94] | rs1800871, rs1800872, rs1800896 | 229 |
| Sohn MH et al., 2007 [95] | rs1800871, rs1800872, rs1800896 | 416 |
| Miyake Y et al., 2013 [96] | rs1801275 | 823 |
| Miyake Y et al. (a), 2011 [97] | rs20541, rs1800925 | 1,270 |
| Miyake Y et al. (b), 2011 [98] | rs20541, rs1800925 | 533 |
| Takahashi N et al., 2005 [99] | rs393548, rs436857 | 1,040 |
| Namkung JH et al., 2011 [100] | rs31563 | 1,090 |
| Kim JS et al., 2016 [101] | rs755622 | 258 |
| Ma L et al., 2013 [102] | rs755622 | 391 |
| Miyake Y et al., 2015 [103] | rs334809 | 1,318 |
| Kato T et al., 2009 [104] | rs187238 | 264 |
| Osawa K et al., 2007 [105] | rs187238 | 121 |
| Behniafard N et al., 2012 [106] | rs1800629 | 226 |
| Zhou J et al, 2012 [107] | rs2427837 | 380 |
| Park KY et al., 2011 [108] | rs2427837 | 231 |
| Tsunemi Y et al., 2004 [109] | 431C>T | 351 |
| Sekiya T et al., 2003 [110] | 431C>T | 306 |
| Abbreviations: CG, Combined Genotype. | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





