Submitted:
20 June 2023
Posted:
20 June 2023
You are already at the latest version
Abstract

Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, PCR Primers and Growth Conditions
2.2. Recombinant DNA Techniques
2.3. Construction of Plasmids for Production of mCherry in A. vinosum
2.4. Fluoresence Microscopy
2.5. Construction of A. vinosum ΔsgpD::ΩKm
2.6. Characterization of Phenotypes, Detection of Sulfur Species and Protein Determination
3. Results
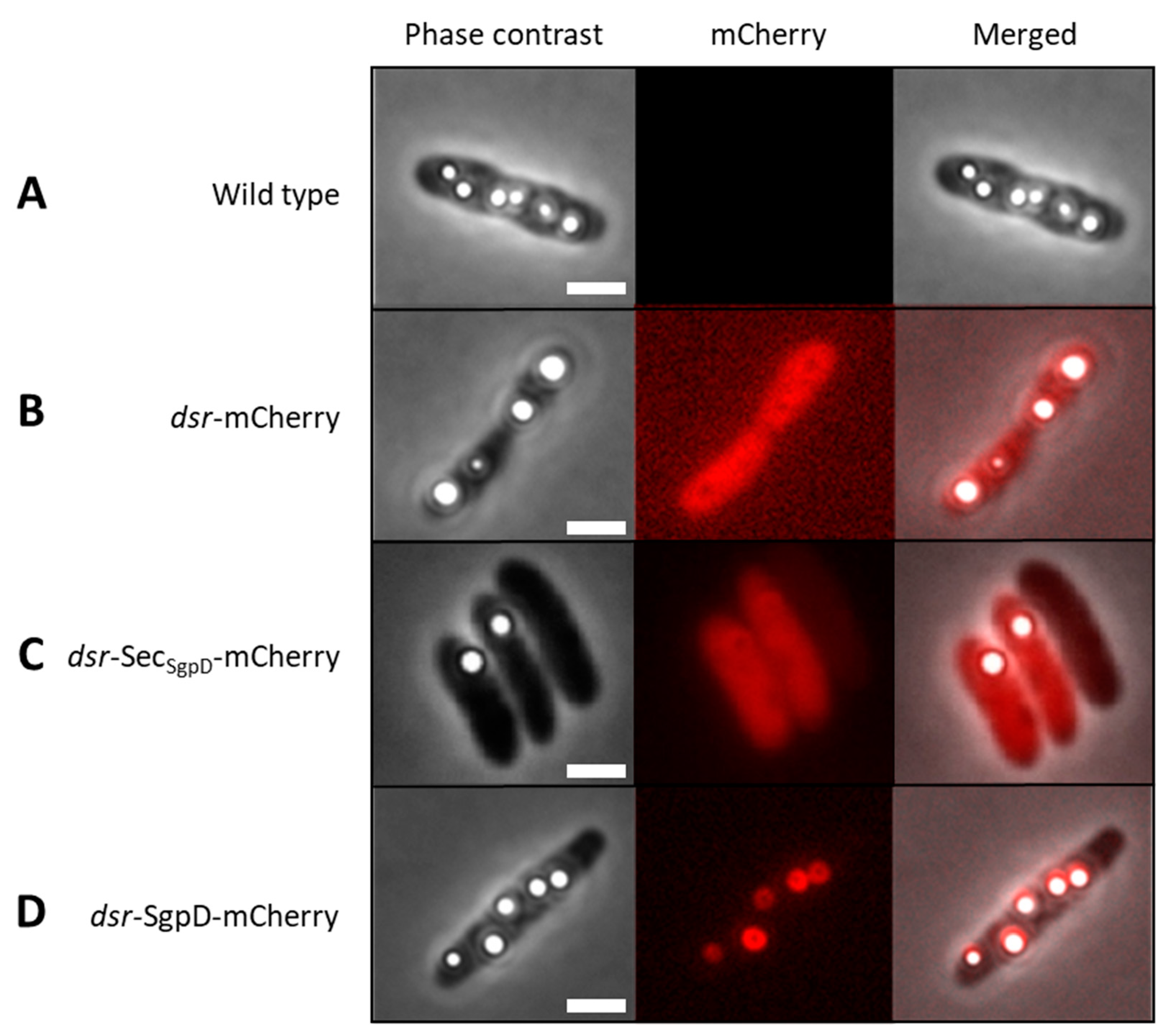
3.1. Suitability of mCherry for the Purple Sulfur Bacterium Allochromatium Vinosum
3.2. Fusions with mCherry Reveals Intracellular Localization of SgpD in A. vinosum
3.3. Stability of mCherry Fluorescence in Allochromatium Vinosum

3.2. In Vivo Role of SgpD
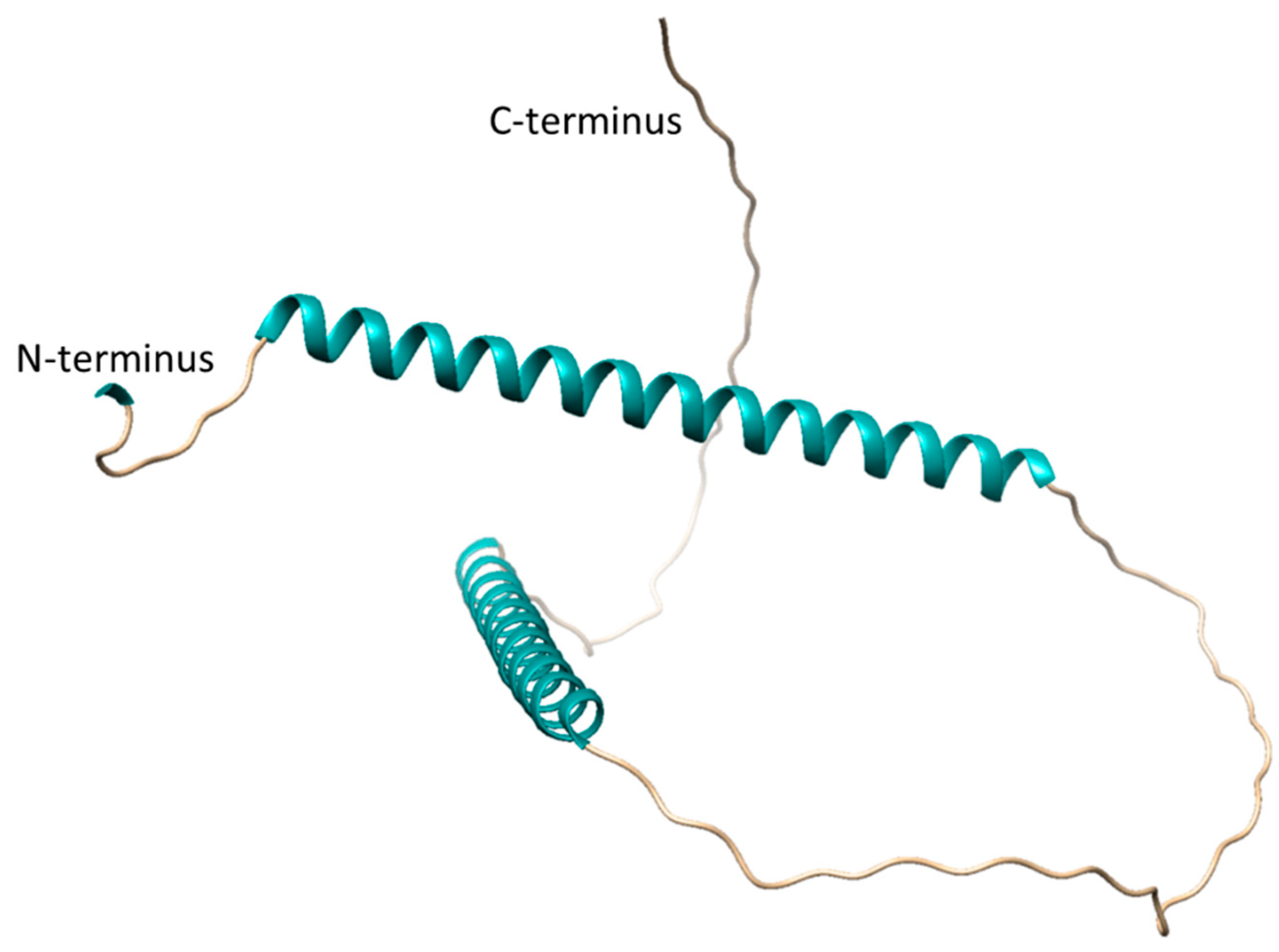

4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dahl, C. Bacterial intracellular sulfur globules. In Bacterial organelles and organelle-like inclusions; Jendrossek, D., Ed.; Springer: Cham, 2020; pp. 19–51. [Google Scholar]
- Marnocha, C.L.; Levy, A.T.; Powell, D.H.; Hanson, T.E.; Chan, C.S. Mechanisms of extracellular S0 globule production and degradation in Chlorobaculum tepidum via dynamic cell-globule interactions. Microbiology 2016, 162, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Marnocha, C.L.; Sabanayagam, C.R.; Modla, S.; Powell, D.H.; Henri, P.A.; Steele, A.S.; Hanson, T.E.; Webb, S.M.; Chan, C.S. Insights into the mineralogy and surface chemistry of extracellular biogenic S0 globules produced by Chlorobaculum tepidum. Front. Microbiol. 2019, 10, 271. [Google Scholar] [CrossRef]
- Dahl, C. A biochemical view on the biological sulfur cycle. In Environmental technologies to treat sulfur pollution: principles and engineering, 2 ed; Lens, P., Ed.; IWA Publishing: London, 2020; pp. 55–96. [Google Scholar]
- Pattaragulwanit, K.; Brune, D.C.; Trüper, H.G.; Dahl, C. Molecular genetic evidence for extracytoplasmic localization of sulfur globules in Chromatium vinosum. Arch. Microbiol. 1998, 169, 434–444. [Google Scholar] [CrossRef]
- Dahl, C.; Prange, A. Bacterial sulfur globules: occurrence, structure and metabolism. In Inclusions in prokaryotes; Shively, J. M., Ed.; Springer: Berlin Heidelberg, 2006; Vol. 1, pp. 21–51. [Google Scholar]
- Brune, D.C. Isolation and characterization of sulfur globule proteins from Chromatium vinosum and Thiocapsa roseopersicina. Arch. Microbiol. 1995, 163, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Frigaard, N.U.; Dahl, C. Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 2009, 54, 103–200. [Google Scholar] [PubMed]
- Prange, A.; Engelhardt, H.; Trüper, H.G.; Dahl, C. The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT PCR. Arch. Microbiol. 2004, 182, 165–174. [Google Scholar] [CrossRef]
- Pott, A.S.; Dahl, C. Sirohaem-sulfite reductase and other proteins encoded in the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 1998, 144, 1881–1894. [Google Scholar] [CrossRef]
- Dahl, C.; Engels, S.; Pott-Sperling, A.S.; Schulte, A.; Sander, J.; Lübbe, Y.; Deuster, O.; Brune, D.C. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 2005, 187, 1392–1404. [Google Scholar] [CrossRef]
- Sander, J.; Engels-Schwarzlose, S.; Dahl, C. Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch. Microbiol. 2006, 186, 357–366. [Google Scholar] [CrossRef]
- Overmann, J. Mahoney Lake: A case study of the ecological significance of phototrophic sulfur bacteria. Adv. Microb. Ecol. 1997, 15, 251–288. [Google Scholar]
- van Gemerden, H. Growth measurements of Chromatium cultures. Arch. Mikrobiol. 1968, 64, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Weissgerber, T.; Sylvester, M.; Kröninger, L.; Dahl, C. A comparative quantitative proteome study identifies new proteins relevant for sulfur oxidation in the purple sulfur bacterium Allochromatium vinosum. Appl. Environ. Microbiol. 2014, 80, 2279–2292. [Google Scholar] [CrossRef] [PubMed]
- Weissgerber, T.; Dobler, N.; Polen, T.; Latus, J.; Stockdreher, Y.; Dahl, C. Genome-wide transcriptional profiling of the purple sulfur bacterium Allochromatium vinosum DSM 180T during growth on different reduced sulfur compounds. J. Bacteriol. 2013, 195, 4231–4245. [Google Scholar] [CrossRef] [PubMed]
- Simm, D.; Hatje, K.; Waack, S.; Kollmar, M. Critical assessment of coiled-coil predictions based on protein structure data. Sci. Rep. 2021, 11, 12439. [Google Scholar] [CrossRef]
- Fogg, P.C.; Westbye, A.B.; Beatty, J.T. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One 2012, 7, e43772. [Google Scholar] [CrossRef]
- Jiang, M.; Zeng, Y.; Cui, L.; Wang, M.; Zheng, Y. A red fluorescent protein reporter system developed for measuring gene expression in photosynthetic bacteria under anaerobic conditions. Microorganisms 2022, 10. [Google Scholar] [CrossRef]
- Feilmeier, B.J.; Iseminger, G.; Schroeder, D.; Webber, H.; Phillips, G.J. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000, 182, 4068–4076. [Google Scholar] [CrossRef]
- Cormack, B.P.; Valdivia, R.H.; Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38. [Google Scholar] [CrossRef]
- Elsliger, M.A.; Wachter, R.M.; Hanson, G.T.; Kallio, K.; Remington, S.J. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry 1999, 38, 5296–5301. [Google Scholar] [CrossRef]
- Patterson, G.H.; Knobel, S.M.; Sharif, W.D.; Kain, S.R.; Piston, D.W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997, 73, 2782–2790. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Chen, J.C.; Viollier, P.H.; Shapiro, L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 2005, 55, 1085–1103. [Google Scholar] [CrossRef]
- Lewenza, S.; Vidal-Ingigliardi, D.; Pugsley, A.P. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J. Bacteriol. 2006, 188, 3516–3524. [Google Scholar] [CrossRef]
- Carroll, P.; Schreuder, L.J.; Muwanguzi-Karugaba, J.; Wiles, S.; Robertson, B.D.; Ripoll, J.; Ward, T.H.; Bancroft, G.J.; Schaible, U.E.; Parish, T. Sensitive detection of gene expression in mycobacteria under replicating and non-replicating conditions using optimized far-red reporters. PLoS One 2010, 5, e9823. [Google Scholar] [CrossRef]
- Heim, R.; Prasher, D.C.; Tsien, R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 1994, 91, 12501–12504. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, X.H.; Lou, K. Rapid detection of a gfp-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol. Lett. 2005, 249, 211–218. [Google Scholar] [CrossRef]
- Ransom, E.M.; Ellermeier, C.D.; Weiss, D.S. Use of mCherry Red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl. Environ. Microbiol. 2015, 81, 1652–1660. [Google Scholar] [CrossRef] [PubMed]
- Streett, H.; Charubin, K.; Papoutsakis, E.T. Anaerobic fluorescent reporters for cell identification, microbial cell biology and high-throughput screening of microbiota and genomic libraries. Curr. Opin. Biotechnol. 2021, 71, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Plamont, M.A.; Billon-Denis, E.; Maurin, S.; Gauron, C.; Pimenta, F.M.; Specht, C.G.; Shi, J.; Querard, J.; Pan, B.; Rossignol, J.; Moncoq, K.; Morellet, N.; Volovitch, M.; Lescop, E.; Chen, Y.; Triller, A.; Vriz, S.; Le Saux, T.; Jullien, L.; Gautier, A. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Hinz, A.J.; Stenzler, B.; Poulain, A.J. Golden gate assembly of aerobic and anaerobic microbial bioreporters. Appl. Environ. Microbiol. 2022, 88, e0148521. [Google Scholar] [CrossRef]
- Charubin, K.; Streett, H.; Papoutsakis, E.T. Development of strong anaerobic fluorescent reporters for Clostridium acetobutylicum and Clostridium ljungdahlii using HaloTag and SNAP-tag proteins. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Karunatilaka, K.S.; Cameron, E.A.; Martens, E.C.; Koropatkin, N.M.; Biteen, J.S. Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts. mBio 2014, 5, e02172. [Google Scholar] [CrossRef] [PubMed]
- Weaver, P.F.; Wall, J.D.; Gest, H. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 1975, 105, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Pfennig, N.; Trüper, H.G. The family Chromatiaceae. In The Prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications; Balows, A., Trüper, H. G., Dworkin, M., Harder, W., Schleifer, K. H., Eds.; Springer-Verlag: New York, 1992; Vol. 2, pp. 3200–3221. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: a laboratory manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, N.Y., 1989. [Google Scholar]
- Bazaral, M.; Helinski, D.R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli J. Mol. Biol. 1968, 36, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Dahl, C. Insertional gene inactivation in a phototrophic sulphur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology 1996, 142, 3363–3372. [Google Scholar] [CrossRef] [PubMed]
- Lübbe, Y.J.; Youn, H.S.; Timkovich, R.; Dahl, C. Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c diamide synthase for sulphur oxidation. FEMS Microbiol. Lett. 2006, 261, 194–202. [Google Scholar] [CrossRef]
- Horton, R.M. PCR mediated recombination and mutagenesis: SOEing together tailor-made genes. Mol. Biotechnol. 1995, 3, 93–99. [Google Scholar] [CrossRef]
- Pattaragulwanit, K.; Dahl, C. Development of a genetic system for a purple sulfur bacterium: conjugative plasmid transfer in Chromatium vinosum. Arch. Microbiol. 1995, 164, 217–222. [Google Scholar] [CrossRef]
- Rethmeier, J.; Rabenstein, A.; Langer, M.; Fischer, U. Detection of traces of oxidized and reduced sulfur compounds in small samples by combination of different high- performance liquid chromatography methods. J.Chromatogr.A 1997, 760, 295–302. [Google Scholar] [CrossRef]
- Shu, X.; Shaner, N.C.; Yarbrough, C.A.; Tsien, R.Y.; Remington, S.J. Novel chromophores and buried charges control color in mFruits. Biochemistry 2006, 45, 9639–9647. [Google Scholar] [CrossRef]
- Heger, A.; Holm, L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins 2000, 41, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; Bridgland, A.; Meyer, C.; Kohl, S.A.A.; Ballard, A.J.; Cowie, A.; Romera-Paredes, B.; Nikolov, S.; Jain, R.; Adler, J.; Back, T.; Petersen, S.; Reiman, D.; Clancy, E.; Zielinski, M.; Steinegger, M.; Pacholska, M.; Berghammer, T.; Bodenstein, S.; Silver, D.; Vinyals, O.; Senior, A.W.; Kavukcuoglu, K.; Kohli, P.; Hassabis, D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Fellay, R.; Frey, J.; Krisch, H.M. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 1987, 52, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Salamov, A. Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and its applications in agriculture, biomedicine and environmental studies; Li, R. W., Ed.; Nova Science Publishers: Hauppage, N.Y, 2010; pp. 71–78. [Google Scholar]
- Reynolds, R.; Bermudez-Cruz, R.M.; Chamberlin, M.J. Parameters affecting transcription termination by Escherichia coli RNA. I. Analysis of 13 rho-independent terminators. J. Mol. Biol. 1992, 224, 31–51. [Google Scholar] [CrossRef]
- Fodor, B.; Rákhely, G.; Kovács, A.T.; Kovács, K.L. Transposon mutagenesis in purple sulfur photosynthetic bacteria: identification of hypF, encoding a protein capable of processing [NiFe] hydrogenases in α, β and γ subdivisions of the proteobacteria. Appl. Environ. Microbiol. 2001, 67, 2476–2483. [Google Scholar] [CrossRef]
- Dahl, C. Sulfur metabolism in phototrophic bacteria. In Modern topics in the phototrophic prokaryotes: Metabolism, bioenergetics and omics; Hallenbeck, P. C., Ed.; Springer International Publishing: Cham, 2017; pp. 27–66. [Google Scholar]
- Regmi, C.K.; Bhandari, Y.R.; Gerstman, B.S.; Chapagain, P.P. Exploring the diffusion of molecular oxygen in the red fluorescent protein mCherry using explicit oxygen molecular dynamics simulations. J. Phys. Chem. B 2013, 117, 2247–2253. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




