Submitted:
17 June 2023
Posted:
19 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Environmental Nutrient Sensors Determine Metabolisms in Cryptococcus with Appropriate Release of Transcription Factors
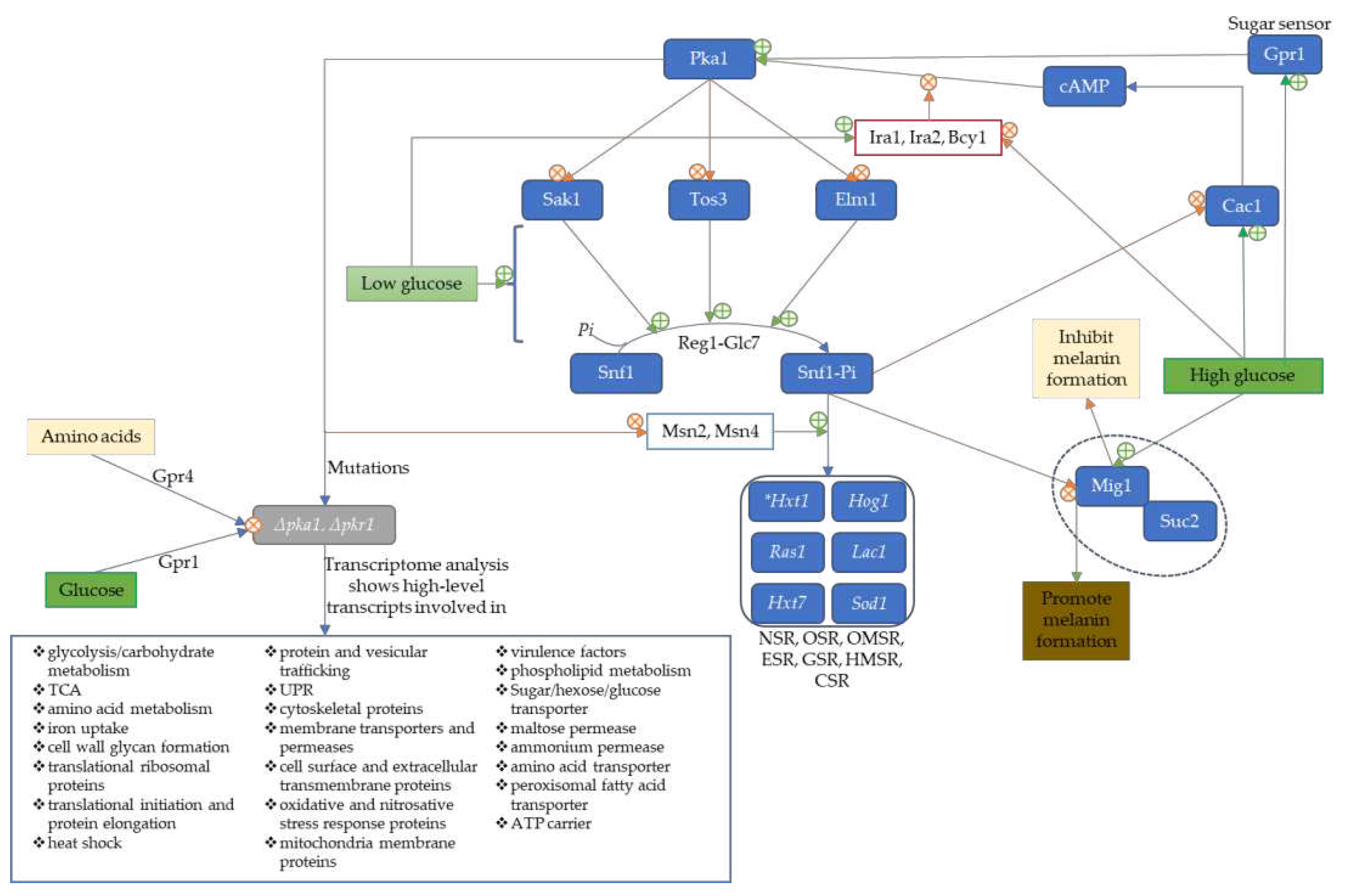
2.1. Metabolism in C. neoformans is Influenced by cAMP/Pka
2.2. Involvement of NADPH in Cryptococcus Metabolism
2.3. Iron Regulators Induced Iron-Dependent Metabolic Enzymes
3. Essential Enzymes of Metabolism and Membrane Transporters/Permeases are Controlled by Transcription Factors in cryptococcal cells for Virulence, Survival, Resistance, and Adaptation
3.1. Deleting Phosphoglucose Isomerase Impairs the Hog1 Pathway, Capsule Production, Membrane Integrity, and Alternative Pathways
3.2. Low Glucose Level Activates Serine/Threonine Protein Kinase 1 Complex to Induce genes with SRE-Promoter Sequence for Stress Response
3.3. Sugar/Hexose transporters (Hxt1 and Hxt7) Are Unexclusively Dependent on the Induction of Serine/Threonine Protein Kinase I Complex (Snf1)
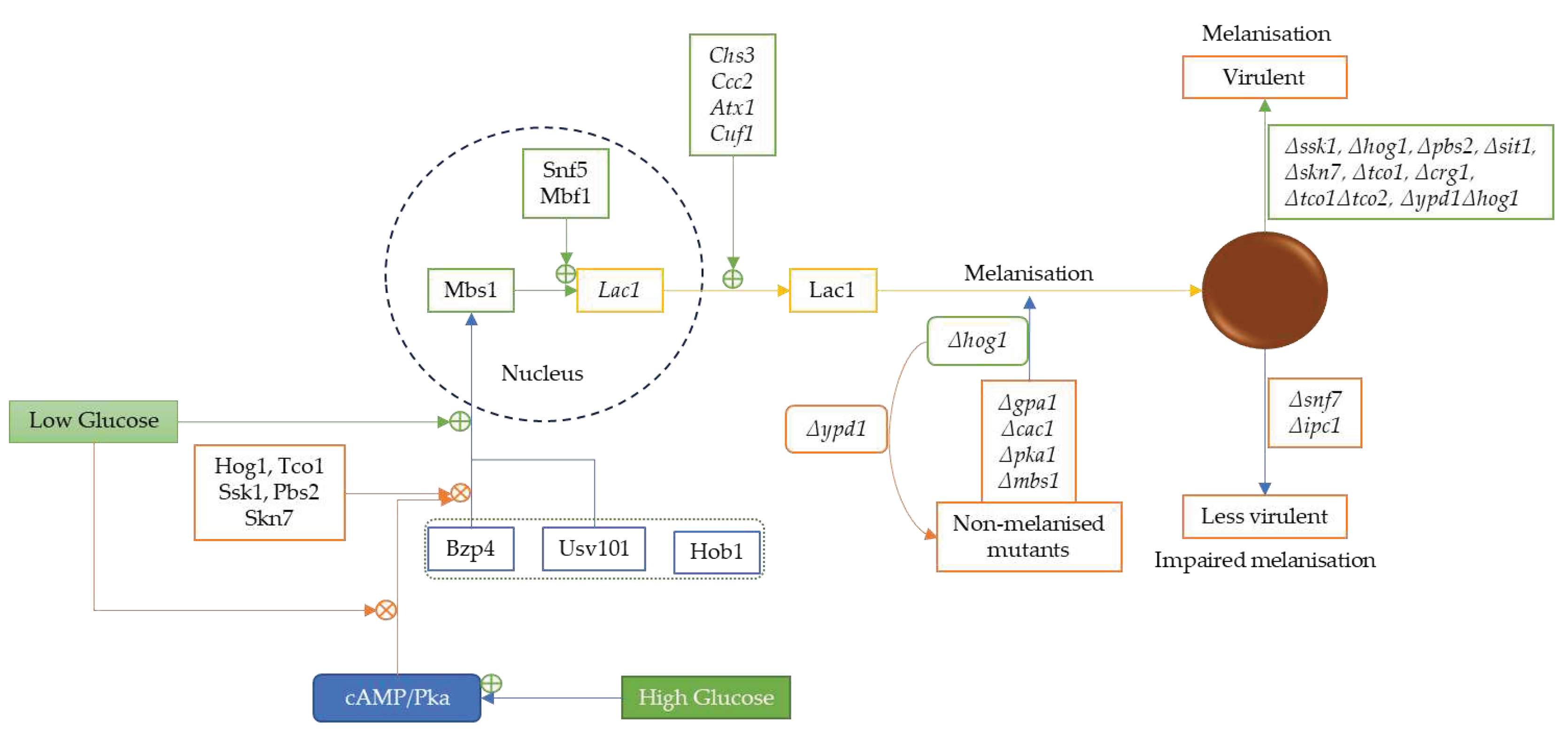
3.4. Low Glucose Level Promotes Melanin and Antioxidative Protein Expression via Serine/Threonine Protein Kinase 1 Complex
3.5. Deleting Acetyl-CoA Forming Enzymes in C. neoformans Impairs Alternative Use of Carbon Sources and Attenuates the Virulence
3.6. Phospholipases B is a Crucial Enzyme to Cellular Tight-Junction Penetration by C. neoformans
3.7. Deleting Membrane-Associated Antioxidative Enzymes Severely Reduced PLB, Urease, and Laccase Expression in C. neoformans
3.8. Phospholipase C orchestrates the Release of PLB, DAG, and IP3 during C. neoformans Infection
3.9. Glycerol-3-Phosphate Phosphatase Controls S-Containing Amino Acid Synthesis via Cystathionine-γ-lyase and Calcineurin Pathways
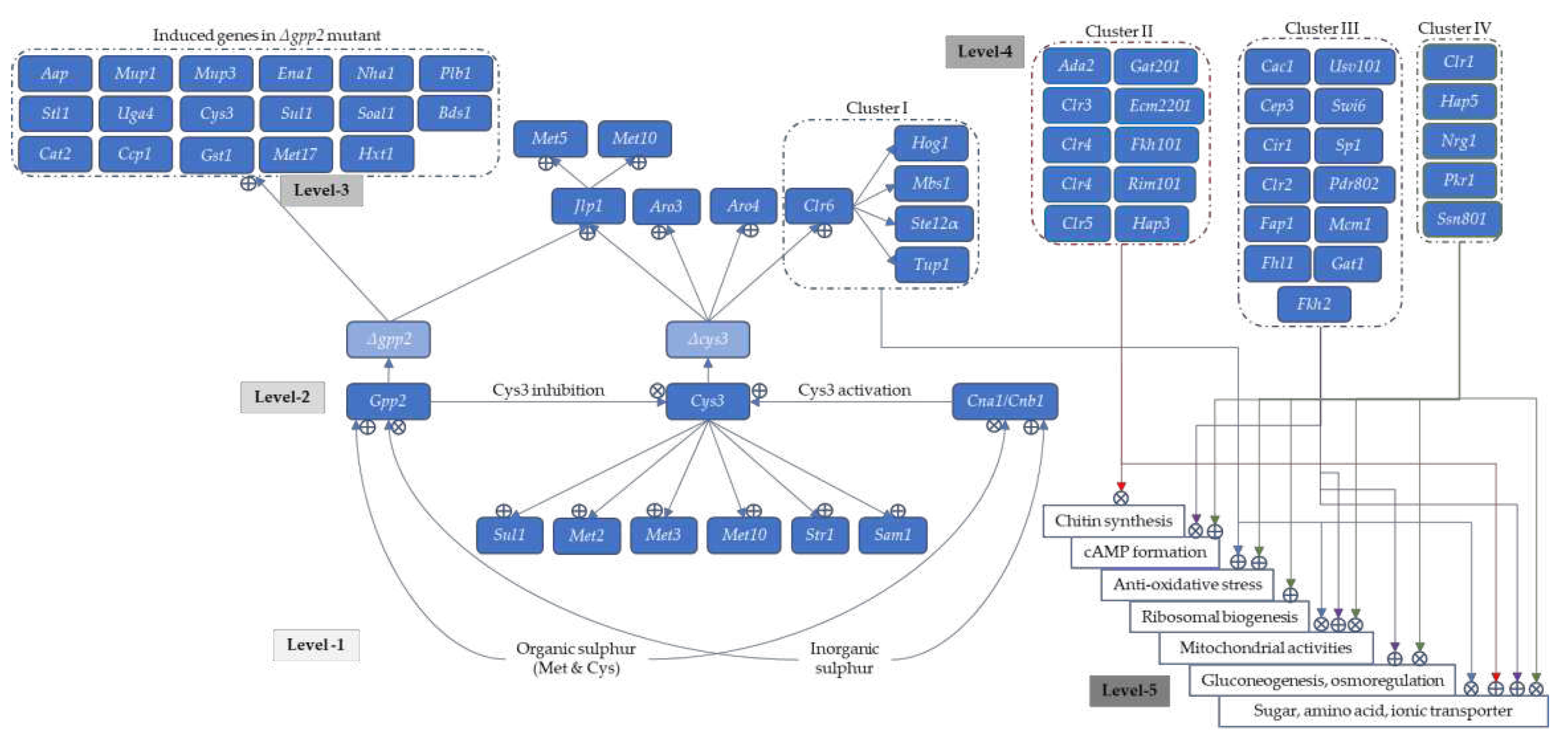
3.10. Transcriptional Activation Factor Controls the Lipid and Sterol Biosynthesis in Cryptococcal Cells for Membrane Integrity and Thermotolerance
3.11. Cryptococcus Virulence Is under Nitrogen Catabolite Repression Regulated by GATA-Sequence Binding Factors
3.12. A Limited Number of Amino Acid Permeases/Transporters Are Crucial for Cryptococcus Virulence
- ❖
- are not affected in rich media (YPD) at high temperatures except Δaap4Δaap5,
- ❖
- are not affected in AAM at 37oC and in capsule formation except Δaap1Δaap2, Δaap4Δaap5, and Δmup1Δmup3,
- ❖
- are not sensitive to any stress agent except Δaap4Δaap5,
- ❖
- are virulent in G. mellonella except for Δaap1Δaap2 and Δaap8 (hypovirulent), Δaap4Δaap5 (avirulent as well as in murine model),
- ❖
- shared appreciable level of sequence homology (Aap1 vs Aap2, 80.9%; Aap1 vs Aap2 vs Aap3, 49%; Aap4 vs Aap5, 89.5%; and Aap6 vs Aap7, 41.4%), and each of these Aaps in the doubly mutated yeast appeared redundant in their functions,
- ❖
- the Aap3 and Aap7 expressions are below detection in YPD or SD; however, Lys-containing medium induced Aap2 and Aap3 expressions under alleviated NCR,
- ❖
- irrespective of the nitrogen sources, the expression of Aap6 remained relatively unchanged,
- ❖
- the Aap8 responds to amino acid-supplemented media only, but the highest expression is usually found in Aap2, Aap4, and Aap5,
- ❖
- Mup1 and Mup3 expressions are under the NCR regulation as Aap2 and Aap5 but can be induced by His, Trp, and Met when NRC is shut down,
- ❖
- only Mup1 can be induced by S-containing amino acids,
- ❖
- galactose induces the expression of all the Aap and Mup genes,
- ❖
- expression of Aap6 and Aap8 is temperature-independent,
- ❖
- Aap4, Aap5, and Mup1 induction increased from 30 to 37oC in SD medium,
- ❖
- Aap2, Aap4, Aap5, and Mup3 expressions are further repressed from 30 to 37oC except for Mup1, which is further induced,
- ❖
- irrespective of the growing media, there is no significant change in the growth of Δaap2, Δaap4, Δaap5, Δmup1, Δmup3 and Δmup1Δmup3 mutants at 30 or 37oC,
- ❖
- the significant growth defect of Δaap4Δaap5 mutants at especially 37oC in YPD or SD showed that the two permeases (or at least one of them) are important for thermotolerance,
- ❖
- the use of amino acids as nitrogen sources impaired the growth in Δaap4Δaap5 mutants,
- ❖
- relative to NH4+ at 30oC, Val, Ile, and Met-containing SD media poorly support the growth of C. neoformans, but Leu, Ser, Lys, and Phe are better nitrogen sources,
- ❖
- Gly, Asp, Asn, Glu, Gln, Arg, Trp, and Pro are highly competitive with NH4+ in culturable AAM,
- ❖
- at 37oC, Val and Met poorly support the growth of C. neoformans in SD media, but Gly, Leu, Ile, Ser, Trp, and Phe are good nitrogen sources, while Asp, Asn, Glu, Gln, Arg, Lys, and Pro are better nitrogen sources,
- ❖
- stereospecifically, var. gattii metabolise D-amino acids because of the more active expression of Dao1, Dao2, and Dao3 genes (encoding D-amino acid oxidase) but less metabolisable for var. neoformans, which prefers L-amino acids as nitrogen sources due to the inefficient evolutionary expression of Dao genes,
- ❖
- in all, growth is denser in L-amino acids containing media than the corresponding D-amino acids media,
- ❖
- pathologically, Δdao mutants of C. neoformans are virulent, but Δdao mutants of C. gattii are attenuated,
- ❖
- positive correlation of Gat1 expression that represses NCR is confirmed to aid the expression of Aap when the preferred nitrogen source is absent/limited but uncertain with Dao expression,
- ❖
- L-Tyr failed to dissolve at permissive pH for culturing C. neoformans, hence not suitable to be tested,
- ❖
- Aap2, Aap3, Mup1, and Mup3 may be global amino acid permeases/transporters because of the most significant growth defect of their corresponding mutants, especially in their double mutant states, from 30 to 37oC and their ability to be induced by various amino acids,
- ❖
- in addition to being a global permease and redundant transcription factors, Aap4 and Aap5 promote thermotolerance and response to oxidative stress, and the growth of the double mutant is significantly impacted from 30 to 37oC in a single amino acid medium or the presence of ≥5 mM H2O2,
- ❖
- unlike the Δaap4 and Δaap5 mutants, the growth defect of Δaap4Δaap5 mutants at 37oC appeared to be restored as pH increased gradually into the alkaline state or when supplemented with 0.75 M NaCl (this condition generates H+ via Na+/H+ antiporter that drives other amino acid permeases to compensate for the deletion of Aap4 and Aap5),
- ❖
3.13. Mutating the Ras1 Gene Attenuates Cryptococcus Thermotolerance, Capsule Formation, and a More Significant Proportion of Amino Acid Permeases/Transporters
4. Transcriptional Factors Regulate Virulence-Associated Membrane-Anchored Proteins, Extracellular Enzymes, and Vesicular Secretions in Cryptococcal Cells for Morphology, Growth, and Virulence
4.1. Proteases (Proteinases/Peptidases)
4.2. Urease
4.3. Laccase
4.4. Inositol-Phosphorylceramide Synthase
4.5. Chitin Synthase
4.6. Dnases
4.7. Phosphatase
4.8. Multifunctional Hydrolytic Enzymes in C. neoformans
4.9. Extracellular Vesicles
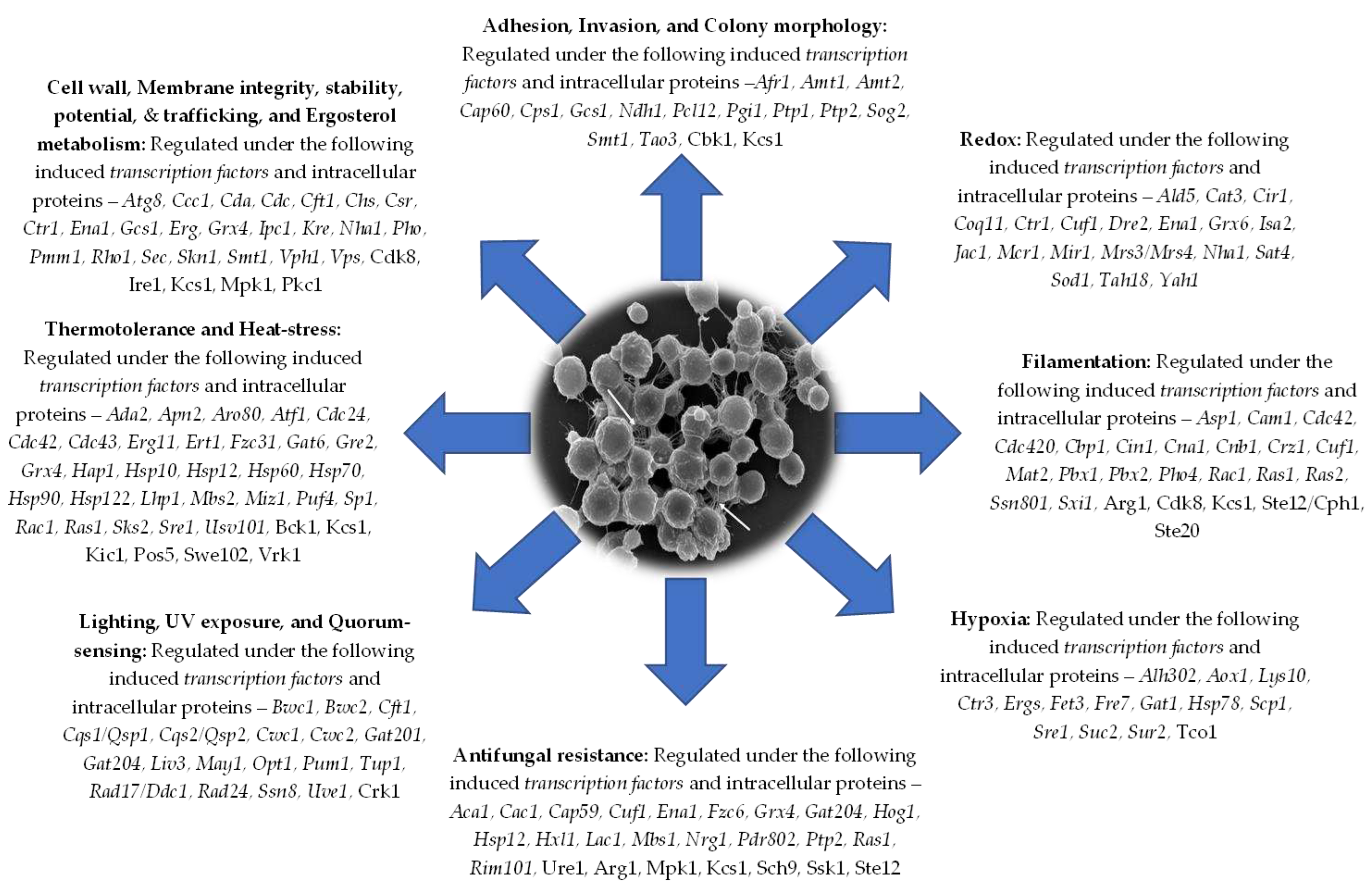
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elhariri:, M.; Hamza, D.; Elhelw, R.; Refai, M. Eucalyptus Tree: A Potential Source of Cryptococcus neoformans in Egyptian Environment. Int J Microbiol 2016, 2016, 4080725. [Google Scholar] [CrossRef]
- Cogliati, M.; D'Amicis, R.; Zani, A.; Montagna, M.T.; Caggiano, G.; De Giglio, O.; Balbino, S.; De Donno, A.; Serio, F.; Susever, S. Environmental distribution of Cryptococcus neoformans and C. gattii around the Mediterranean basin. FEMS yeast research 2016, 16. [Google Scholar] [CrossRef]
- Ergin, C.; Sengul, M.; Aksoy, L.; Dogen, A.; Sun, S.; Averette, A.F.; Cuomo, C.A.; Seyedmousavi, S.; Heitman, J.; Ilkit, M. Cryptococcus neoformans Recovered From Olive Trees (Olea europaea) in Turkey Reveal Allopatry With African and South American Lineages. Front Cell Infect Microbiol 2019, 9, 384. [Google Scholar] [CrossRef]
- Feldmesser, M.; Kress, Y.; Novikoff, P.; Casadevall, A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 2000, 68, 4225–4237. [Google Scholar] [CrossRef] [PubMed]
- Kmetzsch, L.; Staats, C.C.; Cupertino, J.B.; Fonseca, F.L.; Rodrigues, M.L.; Schrank, A.; Vainstein, M.H. The calcium transporter Pmc1 provides Ca2+ tolerance and influences the progression of murine cryptococcal infection. FEBS J 2013, 280, 4853–4864. [Google Scholar] [CrossRef]
- Kmetzsch, L.; Staats, C.C.; Simon, E.; Fonseca, F.L.; Oliveira, D.L.; Joffe, L.S.; Rodrigues, J.; Lourenco, R.F.; Gomes, S.L.; Nimrichter, L.; et al. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet Biol 2011, 48, 192–199. [Google Scholar] [CrossRef]
- Aslanyan, L.; Sanchez, D.A.; Valdebenito, S.; Eugenin, E.A.; Ramos, R.L.; Martinez, L.R. The Crucial Role of Biofilms in Cryptococcus neoformans Survival within Macrophages and Colonization of the Central Nervous System. Journal of Fungi 2017, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: a common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Mowat, E.; Jones, B.; Williams, C.; Lopez-Ribot, J. Our current understanding of fungal biofilms. Critical reviews in microbiology 2009, 35, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annual review of microbiology 1995, 49, 711–745. [Google Scholar] [CrossRef]
- Martinez, L.R.; Casadevall, A. Cryptococcus neoformans cells in biofilms are less susceptible than planktonic cells to antimicrobial molecules produced by the innate immune system. Infection and immunity 2006, 74, 6118–6123. [Google Scholar] [CrossRef]
- Martinez, L.R.; Casadevall, A. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrobial agents and chemotherapy 2006, 50, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Casadevall, A. Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Applied and environmental microbiology 2007, 73, 4592–4601. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Casadevall, A. Biofilm Formation by Cryptococcus neoformans. Microbiology Spectrum 2015, 3, 3.3.05. [Google Scholar] [CrossRef]
- Benaducci, T.; Sardi Jde, C.; Lourencetti, N.M.; Scorzoni, L.; Gullo, F.P.; Rossi, S.A.; Derissi, J.B.; de Azevedo Prata, M.C.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J. Virulence of Cryptococcus sp. Biofilms In Vitro and In Vivo using Galleria mellonella as an Alternative Model. Front Microbiol 2016, 7, 290. [Google Scholar] [CrossRef]
- Walsh, T.J.; Schlegel, R.; Moody, M.M.; Costerton, J.W.; Salcman, M. Ventriculoatrial shunt infection due to Cryptococcus neoformans: an ultrastructural and quantitative microbiological study. Neurosurgery 1986, 18, 373–375. [Google Scholar] [CrossRef]
- Bach, M.C.; Tally, P.W.; Godofsky, E.W. Use of cerebrospinal fluid shunts in patients having acquired immunodeficiency syndrome with cryptococcal meningitis and uncontrollable intracranial hypertension. Neurosurgery 1997, 41, 1280–1283. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.K.; Janssen, D.A.; Marcus, J.R.; Kauffman, C.A. Cryptococcal infection of a prosthetic dialysis fistula. American journal of kidney diseases 1994, 24, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, U.; Gupta, K.; Venugopal, P. A case of prosthetic valve endocarditis caused by Cryptococcus neoformans var. neoformans. J Med Vet Mycol 1997, 35, 139–141. [Google Scholar] [CrossRef]
- Santi, L.; Beys-da-Silva, W.O.; Berger, M.; Calzolari, D.; Guimaraes, J.A.; Moresco, J.J.; Yates, J.R. , 3rd. Proteomic profile of Cryptococcus neoformans biofilm reveals changes in metabolic processes. J Proteome Res 2014, 13, 1545–1559. [Google Scholar] [CrossRef]
- Martinez, L.R.; Casadevall, A. Specific Antibody Can Prevent Fungal Biofilm Formation and This Effect Correlates with Protective Efficacy. Infection and Immunity 2005, 73, 6350–6362. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rodas, R.; Zaragoza, O. Catch me if you can: phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS Immunol Med Microbiol 2012, 64, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, J.N.; Shuman, H.A.; Casadevall, A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proceedings of the National Academy of Sciences 2001, 98, 15245–15250. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, J.N.; Nosanchuk, J.D.; Malliaris, S.D.; Casadevall, A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infection and immunity 2003, 71, 4862–4872. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.S.; Liporagi-Lopes, L.C.; Dos Santos, S.R.J.; Tenor, J.L.; Perfect, J.R.; Cuomo, C.A.; Casadevall, A. Amoeba Predation of Cryptococcus neoformans Results in Pleiotropic Changes to Traits Associated with Virulence. mBio 2021, 12. [Google Scholar] [CrossRef]
- Neilson, J.; Ivey, M.; Bulmer, G. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infection and Immunity 1978, 20, 262–266. [Google Scholar] [CrossRef]
- Paul, C.; Emeka, N. Pathogenicity of Cryptococcus neoformans VNI (ST 32) recovered from environmental and clinical isolates in Nigeria. Comparative Clinical Pathology 2019, 28, 1013–1024. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell 2008, 7, 58–67. [Google Scholar] [CrossRef]
- Maliehe, M.; Ntoi, M.A.; Lahiri, S.; Folorunso, O.S.; Ogundeji, A.O.; Pohl, C.H.; Sebolai, O.M. Environmental Factors That Contribute to the Maintenance of Cryptococcus neoformans Pathogenesis. Microorganisms 2020, 8, 180. [Google Scholar] [CrossRef]
- Derengowski Lda, S.; Paes, H.C.; Albuquerque, P.; Tavares, A.H.; Fernandes, L.; Silva-Pereira, I.; Casadevall, A. The transcriptional response of Cryptococcus neoformans to ingestion by Acanthamoeba castellanii and macrophages provides insights into the evolutionary adaptation to the mammalian host. Eukaryot Cell 2013, 12, 761–774. [Google Scholar] [CrossRef]
- Wang, Z.A.; Li, L.X.; Doering, T.L. Unraveling synthesis of the cryptococcal cell wall and capsule. Glycobiology 2018, 28, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Panepinto, J.; Liu, L.; Ramos, J.; Zhu, X.; Valyi-Nagy, T.; Eksi, S.; Fu, J.; Jaffe, H.A.; Wickes, B.; Williamson, P.R. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J Clin Invest 2005, 115, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Kraus, P.R.; Boily, M.J.; Heitman, J. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell 2005, 4, 1420–1433. [Google Scholar] [CrossRef]
- Price, M.S.; Betancourt-Quiroz, M.; Price, J.L.; Toffaletti, D.L.; Vora, H.; Hu, G.; Kronstad, J.W.; Perfect, J.R. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2011, 2, e00103–00111. [Google Scholar] [CrossRef]
- Griffiths, E.J.; Hu, G.; Fries, B.; Caza, M.; Wang, J.; Gsponer, J.; Gates-Hollingsworth, M.A.; Kozel, T.R.; De Repentigny, L.; Kronstad, J.W. A defect in ATP-citrate lyase links acetyl-CoA production, virulence factor elaboration and virulence in Cryptococcus neoformans. Molecular microbiology 2012, 86, 1404–1423. [Google Scholar] [CrossRef]
- Fan, W.; Kraus, P.R.; Boily, M.-J.; Heitman, J. Cryptococcus neoformans Gene Expression during Murine Macrophage Infection. Eukaryotic Cell 2005, 4, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Bahn, Y.S.; Cox, G.M.; Heitman, J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol Biol Cell 2006, 17, 667–679. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Hicks, J.K.; Giles, S.S.; Cox, G.M.; Heitman, J. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell 2004, 3, 1476–1491. [Google Scholar] [CrossRef]
- Hu, G.; Steen, B.R.; Lian, T.; Sham, A.P.; Tam, N.; Tangen, K.L.; Kronstad, J.W. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog 2007, 3, e42. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Bender, J.A.; Fink, G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 2004, 3, 1076–1087. [Google Scholar] [CrossRef]
- Rude, T.H.; Toffaletti, D.L.; Cox, G.M.; Perfect, J.R. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun 2002, 70, 5684–5694. [Google Scholar] [CrossRef] [PubMed]
- McClelland, E.E.; Ramagopal, U.A.; Rivera, J.; Cox, J.; Nakouzi, A.; Prabu, M.M.; Almo, S.C.; Casadevall, A. A Small Protein Associated with Fungal Energy Metabolism Affects the Virulence of Cryptococcus neoformans in Mammals. PLoS Pathog 2016, 12, e1005849. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.; Simmer, M.I.; D'Souza, C.A.; Steen, B.R.; Zuyderduyn, S.D.; Jones, S.J.; Marra, M.A.; Kronstad, J.W. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 2005, 55, 1452–1472. [Google Scholar] [CrossRef]
- Jung, W.H.; Saikia, S.; Hu, G.; Wang, J.; Fung, C.K.; D'Souza, C.; White, R.; Kronstad, J.W. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog 2010, 6, e1001209. [Google Scholar] [CrossRef]
- Eisenstein, R.S. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr 2000, 20, 627–662. [Google Scholar] [CrossRef]
- Ihrig, J.; Hausmann, A.; Hain, A.; Richter, N.; Hamza, I.; Lill, R.; Mühlenhoff, U. Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae. Eukaryotic cell 2010, 9, 460–471. [Google Scholar] [CrossRef]
- Ramos-Alonso, L.; Romero, A.M.; Martinez-Pastor, M.T.; Puig, S. Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front Microbiol 2020, 11, 582830. [Google Scholar] [CrossRef]
- Garcia-Santamarina, S.; Uzarska, M.A.; Festa, R.A.; Lill, R.; Thiele, D.J. Cryptococcus neoformans Iron-Sulfur Protein Biogenesis Machinery Is a Novel Layer of Protection against Cu Stress. mBio 2017, 8, e01742–01717. [Google Scholar] [CrossRef] [PubMed]
- Do, E.; Hu, G.; Caza, M.; Oliveira, D.; Kronstad, J.W.; Jung, W.H. Leu1 plays a role in iron metabolism and is required for virulence in Cryptococcus neoformans. Fungal Genet Biol 2015, 75, 11–19. [Google Scholar] [CrossRef]
- Nazi, I.; Scott, A.; Sham, A.; Rossi, L.; Williamson, P.R.; Kronstad, J.W.; Wright, G.D. Role of homoserine transacetylase as a new target for antifungal agents. Antimicrob Agents Chemother 2007, 51, 1731–1736. [Google Scholar] [CrossRef]
- Pascon, R.C.; Ganous, T.M.; Kingsbury, J.M.; Cox, G.M.; McCusker, J.H. Cryptococcus neoformans methionine synthase: expression analysis and requirement for virulence. Microbiology (Reading) 2004, 150, 3013–3023. [Google Scholar] [CrossRef] [PubMed]
- Do, E.; Park, M.; Hu, G.; Caza, M.; Kronstad, J.W.; Jung, W.H. The lysine biosynthetic enzyme Lys4 influences iron metabolism, mitochondrial function and virulence in Cryptococcus neoformans. Biochem Biophys Res Commun 2016, 477, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wei, D.; Li, Z.; Sun, Z.; Pan, J.; Zhu, X. Cryptococcal phosphoglucose isomerase is required for virulence factor production, cell wall integrity and stress resistance. FEMS Yeast Res 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.; Orlova, M.; Maziarz, M.; Kuchin, S. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 2012, 11, 119–128. [Google Scholar] [CrossRef]
- Hong, S.-P.; Leiper, F.C.; Woods, A.; Carling, D.; Carlson, M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proceedings of the National Academy of Sciences 2003, 100, 8839–8843. [Google Scholar] [CrossRef]
- McCartney, R.R.; Rubenstein, E.M.; Schmidt, M.C. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Current genetics 2005, 47, 335–344. [Google Scholar] [CrossRef]
- McCartney, R.R.; Schmidt, M.C. Regulation of Snf1 kinase: activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. Journal of Biological Chemistry 2001, 276, 36460–36466. [Google Scholar] [CrossRef]
- Tu, J.; Carlson, M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. The EMBO Journal 1995, 14, 5939–5946. [Google Scholar] [CrossRef]
- Nehlin, J.O.; Ronne, H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J 1990, 9, 2891–2898. [Google Scholar] [CrossRef]
- Nicastro, R.; Tripodi, F.; Gaggini, M.; Castoldi, A.; Reghellin, V.; Nonnis, S.; Tedeschi, G.; Coccetti, P. Snf1 Phosphorylates Adenylate Cyclase and Negatively Regulates Protein Kinase A-dependent Transcription in Saccharomyces cerevisiae. J Biol Chem 2015, 290, 24715–24726. [Google Scholar] [CrossRef]
- Hu, G.; Cheng, P.Y.; Sham, A.; Perfect, J.R.; Kronstad, J.W. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol 2008, 69, 1456–1475. [Google Scholar] [CrossRef] [PubMed]
- Pukkila-Worley, R.; Gerrald, Q.D.; Kraus, P.R.; Boily, M.J.; Davis, M.J.; Giles, S.S.; Cox, G.M.; Heitman, J.; Alspaugh, J.A. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot Cell 2005, 4, 190–201. [Google Scholar] [CrossRef]
- Zhu, X.; Williamson, P.R. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res 2004, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Alden, K.M.; Jezewski, A.J.; Beattie, S.R.; Fox, D., 3rd; Krysan, D.J. Genetic Interaction Analysis Reveals that Cryptococcus neoformans Utilizes Multiple Acetyl-CoA-Generating Pathways during Infection. mBio 2022, 13, e0127922. [Google Scholar] [CrossRef]
- Saito, K.; Sugatani, J.; Okumura, T. Phospholipase B from Penicillium notatum. In Methods in enzymology; Elsevier, 1991; Volume 197, pp. 446–456. [Google Scholar]
- Lee, K.S.; Patton, J.L.; Fido, M.; Hines, L.K.; Kohlwein, S.D.; Paltauf, F.; Henry, S.A.; Levin, D.E. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J Biol Chem 1994, 269, 19725–19730. [Google Scholar] [CrossRef]
- Leidich, S.D.; Ibrahim, A.S.; Fu, Y.; Koul, A.; Jessup, C.; Vitullo, J.; Fonzi, W.; Mirbod, F.; Nakashima, S.; Nozawa, Y.; et al. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J Biol Chem 1998, 273, 26078–26086. [Google Scholar] [CrossRef] [PubMed]
- Siafakas, A.R.; Sorrell, T.C.; Wright, L.C.; Wilson, C.; Larsen, M.; Boadle, R.; Williamson, P.R.; Djordjevic, J.T. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J Biol Chem 2007, 282, 37508–37514. [Google Scholar] [CrossRef]
- Chen, S.C.; Muller, M.; Zhou, J.Z.; Wright, L.C.; Sorrell, T.C. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J Infect Dis 1997, 175, 414–420. [Google Scholar] [CrossRef]
- Chayakulkeeree, M.; Sorrell, T.C.; Siafakas, A.R.; Wilson, C.F.; Pantarat, N.; Gerik, K.J.; Boadle, R.; Djordjevic, J.T. Role and mechanism of phosphatidylinositol-specific phospholipase C in survival and virulence of Cryptococcus neoformans. Mol Microbiol 2008, 69, 809–826. [Google Scholar] [CrossRef]
- Lev, S.; Desmarini, D.; Li, C.; Chayakulkeeree, M.; Traven, A.; Sorrell, T.C.; Djordjevic, J.T. Phospholipase C of Cryptococcus neoformans regulates homeostasis and virulence by providing inositol trisphosphate as a substrate for Arg1 kinase. Infect Immun 2013, 81, 1245–1255. [Google Scholar] [CrossRef]
- Cox, G.M.; McDade, H.C.; Chen, S.C.; Tucker, S.C.; Gottfredsson, M.; Wright, L.C.; Sorrell, T.C.; Leidich, S.D.; Casadevall, A.; Ghannoum, M.A.; et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol 2001, 39, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.S.; Mirbod, F.; Filler, S.G.; Banno, Y.; Cole, G.T.; Kitajima, Y.; Edwards, J.E., Jr.; Nozawa, Y.; Ghannoum, M.A. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect Immun 1995, 63, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Wickes, B.L.; Miller, G.F.; Penoyer, L.A.; Kwon-Chung, K.J. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J Exp Med 2000, 191, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Narasipura, S.D.; Chaturvedi, V.; Chaturvedi, S. Characterisation of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol Microbiol 2005, 55, 1782–1800. [Google Scholar] [CrossRef]
- Narasipura, S.D.; Ault, J.G.; Behr, M.J.; Chaturvedi, V.; Chaturvedi, S. Characterisation of Cu,Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: role in biology and virulence. Mol Microbiol 2003, 47, 1681–1694. [Google Scholar] [CrossRef]
- Chaturvedi, V.; Wong, B.; Newman, S.L. Oxidative killing of Cryptococcus neoformans by human neutrophils - Evidence that fungal mannitol protects by scavenging reactive oxygen intermediates. Journal of Immunology 1996, 156, 3836–3840. [Google Scholar] [CrossRef]
- Narasipura, S.D.; Chaturvedi, V.; Chaturvedi, S. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Molecular microbiology 2005, 55, 1782–1800. [Google Scholar] [CrossRef]
- Garcia-Martos, P.; Marin, P.; Hernandez-Molina, J.M.; Garcia-Agudo, L.; Aoufi, S.; Mira, J. Extracellular enzymatic activity in 11 Cryptococcus species. Mycopathologia 2001, 150, 1–4. [Google Scholar] [CrossRef]
- Park, M.; Do, E.; Jung, W.H. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 2013, 41, 67–72. [Google Scholar] [CrossRef]
- Maeng, S.; Ko, Y.J.; Kim, G.B.; Jung, K.W.; Floyd, A.; Heitman, J.; Bahn, Y.S. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot Cell 2010, 9, 360–378. [Google Scholar] [CrossRef]
- Martho, K.F.; Brustolini, O.J.B.; Vasconcelos, A.T.; Vallim, M.A.; Pascon, R.C. The Glycerol Phosphatase Gpp2: A Link to Osmotic Stress, Sulfur Assimilation and Virulence in Cryptococcus neoformans. Front Microbiol 2019, 10, 2728. [Google Scholar] [CrossRef]
- de Melo, A.T.; Martho, K.F.; Roberto, T.N.; Nishiduka, E.S.; Machado, J., Jr.; Brustolini, O.J.B.; Tashima, A.K.; Vasconcelos, A.T.; Vallim, M.A.; Pascon, R.C. The regulation of the sulfur amino acid biosynthetic pathway in Cryptococcus neoformans: the relationship of Cys3, Calcineurin, and Gpp2 phosphatases. Sci Rep 2019, 9, 11923. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.J.; Yu, Y.M.; Kim, G.B.; Lee, G.W.; Maeng, P.J.; Kim, S.; Floyd, A.; Heitman, J.; Bahn, Y.S. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 2009, 8, 1197–1217. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.I.; Bonaduce, M.J.; Klar, A.J. Histone deacetylase homologs regulate the epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 1998, 150, 563–576. [Google Scholar] [CrossRef]
- Maier, E.J.; Haynes, B.C.; Gish, S.R.; Wang, Z.A.; Skowyra, M.L.; Marulli, A.L.; Doering, T.L.; Brent, M.R. Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome Res 2015, 25, 690–700. [Google Scholar] [CrossRef]
- Kraus, P.R.; Boily, M.J.; Giles, S.S.; Stajich, J.E.; Allen, A.; Cox, G.M.; Dietrich, F.S.; Perfect, J.R.; Heitman, J. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot Cell 2004, 3, 1249–1260. [Google Scholar] [CrossRef]
- Chellappa, R.; Kandasamy, P.; Oh, C.S.; Jiang, Y.; Vemula, M.; Martin, C.E. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J Biol Chem 2001, 276, 43548–43556. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.E.; Oh, C.S.; Jiang, Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta 2007, 1771, 271–285. [Google Scholar] [CrossRef]
- Lee, I.R.; Chow, E.W.; Morrow, C.A.; Djordjevic, J.T.; Fraser, J.A. Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics 2011, 188, 309–323. [Google Scholar] [CrossRef]
- Ries, L.N.A.; Beattie, S.; Cramer, R.A.; Goldman, G.H. Overview of carbon and nitrogen catabolite metabolism in the virulence of human pathogenic fungi. Mol Microbiol 2018, 107, 277–297. [Google Scholar] [CrossRef]
- Fernandes, J.D.; Martho, K.; Tofik, V.; Vallim, M.A.; Pascon, R.C. The Role of Amino Acid Permeases and Tryptophan Biosynthesis in Cryptococcus neoformans Survival. PLoS One 2015, 10, e0132369. [Google Scholar] [CrossRef] [PubMed]
- Kingsbury, J.M.; McCusker, J.H. Threonine biosynthetic genes are essential in Cryptococcus neoformans. Microbiology (Reading) 2008, 154, 2767–2775. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.H.; Chen, J.S.; Reddy, V.; Day, J.L.; Shlykov, M.A.; Wakabayashi, S.T.; Saier, M.H., Jr. The amino acid-polyamine-organocation superfamily. J Mol Microbiol Biotechnol 2012, 22, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Mikros, E.; Diallinas, G. Tales of tails in transporters. Open Biol 2019, 9, 190083. [Google Scholar] [CrossRef]
- Wyman, C.L. Analysis Of Membrane Transporter Systems Expressed During Symbiotic Nitrogen Fixation In The Model Legume Medicago Truncatula; West Virginia University, 2018. [Google Scholar]
- Calvete, C.L.; Martho, K.F.; Felizardo, G.; Paes, A.; Nunes, J.M.; Ferreira, C.O.; Vallim, M.A.; Pascon, R.C. Amino acid permeases in Cryptococcus neoformans are required for high temperature growth and virulence; and are regulated by Ras signalling. PLoS One 2019, 14, e0211393. [Google Scholar] [CrossRef] [PubMed]
- Martho, K.F.; de Melo, A.T.; Takahashi, J.P.; Guerra, J.M.; Santos, D.C.; Purisco, S.U.; Melhem, M.S.; Fazioli, R.D.; Phanord, C.; Sartorelli, P.; et al. Amino Acid Permeases and Virulence in Cryptococcus neoformans. PLoS One 2016, 11, e0163919. [Google Scholar] [CrossRef]
- Garbe, E.; Vylkova, S. Role of Amino Acid Metabolism in the Virulence of Human Pathogenic Fungi. Current Clinical Microbiology Reports 2019, 6, 108–119. [Google Scholar] [CrossRef]
- Chang, Y.C.; Khanal Lamichhane, A.; Bradley, J.; Rodgers, L.; Ngamskulrungroj, P.; Kwon-Chung, K.J. Differences between Cryptococcus neoformans and Cryptococcus gattii in the Molecular Mechanisms Governing Utilization of D-Amino Acids as the Sole Nitrogen Source. PLoS One 2015, 10, e0131865. [Google Scholar] [CrossRef]
- Alspaugh, J.A.; Cavallo, L.M.; Perfect, J.R.; Heitman, J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol 2000, 36, 352–365. [Google Scholar] [CrossRef]
- Nichols, C.B.; Perfect, Z.H.; Alspaugh, J.A. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 2007, 63, 1118–1130. [Google Scholar] [CrossRef]
- Vallim, M.A.; Nichols, C.B.; Fernandes, L.; Cramer, K.L.; Alspaugh, J.A. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell 2005, 4, 1066–1078. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.D.S.; Martho, K.; Tofik, V.; Vallim, M.A.; Pascon, R.C. The Role of Amino Acid Permeases and Tryptophan Biosynthesis in Cryptococcus neoformans Survival. PLOS ONE 2015, 10, e0132369. [Google Scholar] [CrossRef] [PubMed]
- Pentland, D.R.; Piper-Brown, E.; Muhlschlegel, F.A.; Gourlay, C.W. Ras signalling in pathogenic yeasts. Microb Cell 2017, 5, 63–73. [Google Scholar] [CrossRef]
- Eigenheer, R.A.; Jin Lee, Y.; Blumwald, E.; Phinney, B.S.; Gelli, A. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res 2007, 7, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Novick, P.; Ferro, S.; Schekman, R. Order of events in the yeast secretory pathway. Cell 1981, 25, 461–469. [Google Scholar] [CrossRef]
- Vu, K.; Tham, R.; Uhrig, J.P.; Thompson, G.R., 3rd; Na Pombejra, S.; Jamklang, M.; Bautos, J.M.; Gelli, A. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. mBio 2014, 5, e01101–01114. [Google Scholar] [CrossRef]
- Homer, C.M.; Summers, D.K.; Goranov, A.I.; Clarke, S.C.; Wiesner, D.L.; Diedrich, J.K.; Moresco, J.J.; Toffaletti, D.; Upadhya, R.; Caradonna, I.; et al. Intracellular Action of a Secreted Peptide Required for Fungal Virulence. Cell Host Microbe 2016, 19, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Brueske, C.H. Proteolytic activity of a clinical isolate of Cryptococcus neoformans. J Clin Microbiol 1986, 23, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Yoo Ji, J.; Lee, Y.S.; Song, C.Y.; Kim, B.S. Purification and characterisation of a 43-kilodalton extracellular serine proteinase from Cryptococcus neoformans. J Clin Microbiol 2004, 42, 722–726. [Google Scholar] [CrossRef]
- Chen, L.C.; Blank, E.S.; Casadevall, A. Extracellular proteinase activity of Cryptococcus neoformans. Clin Diagn Lab Immunol 1996, 3, 570–574. [Google Scholar] [CrossRef]
- Ruma-Haynes, P.; Brownlee, A.G.; Sorrell, T.C. A rapid method for detecting extracellular proteinase activity in Cryptococcus neoformans and a survey of 63 isolates. J Med Microbiol 2000, 49, 733–737. [Google Scholar] [CrossRef]
- Leone, R.; Cabeli, P.; Sinicco, A.; Ito-Kuwa, S.; Aoki, S.; Vidotto, V. Relationship between protease production and capsule size in Cryptococcus neoformans. Journal De Mycologie Medicale 1999, 9, 42–44. [Google Scholar]
- Clarke, S.C.; Dumesic, P.A.; Homer, C.M.; O'Donoghue, A.J.; La Greca, F.; Pallova, L.; Majer, P.; Madhani, H.D.; Craik, C.S. Integrated Activity and Genetic Profiling of Secreted Peptidases in Cryptococcus neoformans Reveals an Aspartyl Peptidase Required for Low pH Survival and Virulence. PLoS Pathog 2016, 12, e1006051. [Google Scholar] [CrossRef]
- Monari, C.; Pericolini, E.; Bistoni, G.; Cenci, E.; Bistoni, F.; Vecchiarelli, A. Influence of indinavir on virulence and growth of Cryptococcus neoformans. J Infect Dis 2005, 191, 307–311. [Google Scholar] [CrossRef]
- Sidrim, J.J.; Perdigao-Neto, L.V.; Cordeiro, R.A.; Brilhante, R.S.; Leite, J.J.; Teixeira, C.E.; Monteiro, A.J.; Freitas, R.M.; Ribeiro, J.F.; Mesquita, J.R.; et al. Viral protease inhibitors affect the production of virulence factors in Cryptococcus neoformans. Can J Microbiol 2012, 58, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Pinti, M.; Orsi, C.F.; Gibellini, L.; Esposito, R.; Cossarizza, A.; Blasi, E.; Peppoloni, S.; Mussini, C. Identification and characterisation of an aspartyl protease from Cryptococcus neoformans. FEBS Lett 2007, 581, 3882–3886. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Panting, R.J.; Varma, A.; Saijo, T.; Waldron, K.J.; Jong, A.; Ngamskulrungroj, P.; Chang, Y.C.; Rutherford, J.C.; Kwon-Chung, K.J. Factors required for activation of urease as a virulence determinant in Cryptococcus neoformans. mBio 2013, 4, e00220-00213. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.M.; Mukherjee, J.; Cole, G.T.; Casadevall, A.; Perfect, J.R. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 2000, 68, 443–448. [Google Scholar] [CrossRef]
- Olszewski, M.A.; Noverr, M.C.; Chen, G.H.; Toews, G.B.; Cox, G.M.; Perfect, J.R.; Huffnagle, G.B. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am J Pathol 2004, 164, 1761–1771. [Google Scholar] [CrossRef]
- Bava, A.J.; Negroni, R.; Bianchi, M. Cryptococcosis produced by a urease negative strain of Cryptococcus neoformans. J Med Vet Mycol 1993, 31, 87–89. [Google Scholar] [CrossRef]
- Ruane, P.J.; Walker, L.J.; George, W.L. Disseminated infection caused by urease-negative Cryptococcus neoformans. J Clin Microbiol 1988, 26, 2224–2225. [Google Scholar] [CrossRef]
- Almeida, F.; Wolf, J.M.; Casadevall, A. Virulence-Associated Enzymes of Cryptococcus neoformans. Eukaryot Cell 2015, 14, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Tangen, K.L.; Jung, W.H.; Sham, A.P.; Lian, T.; Kronstad, J.W. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilisation, melanisation and cell wall structure in Cryptococcus neoformans. Microbiology (Reading) 2007, 153, 29–41. [Google Scholar] [CrossRef]
- Lee, D.; Jang, E.H.; Lee, M.; Kim, S.W.; Lee, Y.; Lee, K.T.; Bahn, Y.S. Unraveling Melanin Biosynthesis and Signalling Networks in Cryptococcus neoformans. mBio 2019, 10, e02267–02219. [Google Scholar] [CrossRef]
- Wang, Y.; Aisen, P.; Casadevall, A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun 1995, 63, 3131–3136. [Google Scholar] [CrossRef]
- Durrell, L.W. The Composition and Structure of Walls of Dark Fungus Spores. Mycopathol Mycol Appl 1964, 23, 337–345. [Google Scholar] [CrossRef]
- Rosas, A.L.; Casadevall, A. Melanization affects susceptibility of Cryptococcus neoformans to heat and cold. FEMS Microbiol Lett 1997, 153, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Aisen, P.; Casadevall, A. Melanin, melanin "ghosts," and melanin composition in Cryptococcus neoformans. Infect Immun 1996, 64, 2420–2424. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rivera, J.; Eisenman, H.C.; Nosanchuk, J.D.; Aisen, P.; Zaragoza, O.; Moadel, T.; Dadachova, E.; Casadevall, A. Comparative analysis of Cryptococcus neoformans acid-resistant particles generated from pigmented cells grown in different laccase substrates. Fungal Genet Biol 2005, 42, 989–998. [Google Scholar] [CrossRef]
- Garcia-Rivera, J.; Casadevall, A. Melanization of Cryptococcus neoformans reduces its susceptibility to the antimicrobial effects of silver nitrate. Med Mycol 2001, 39, 353–357. [Google Scholar] [CrossRef]
- Wang, Y.; Casadevall, A. Growth of Cryptococcus neoformans in the presence of L-dopa decreases its susceptibility to amphotericin B. Antimicrob Agents Chemother 1994, 38, 2648–2650. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Casadevall, A. Susceptibility of melanised and nonmelanised Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun 1994, 62, 3004–3007. [Google Scholar] [CrossRef] [PubMed]
- Rosas, A.L.; Nosanchuk, J.D.; Gomez, B.L.; Edens, W.A.; Henson, J.M.; Casadevall, A. Isolation and serological analyses of fungal melanins. J Immunol Methods 2000, 244, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Sussman, A.S. The Fungal Population: An Advanced Treatise; Elsevier Science, 2013; pp. 447–486. [Google Scholar]
- Godinho, R.M.; Crestani, J.; Kmetzsch, L.; Araujo Gde, S.; Frases, S.; Staats, C.C.; Schrank, A.; Vainstein, M.H.; Rodrigues, M.L. The vacuolar-sorting protein Snf7 is required for the export of virulence determinants in members of the Cryptococcus neoformans complex. Sci Rep 2014, 4, 6198. [Google Scholar] [CrossRef]
- Walton, F.J.; Idnurm, A.; Heitman, J. Novel gene functions required for melanisation of the human pathogen Cryptococcus neoformans. Mol Microbiol 2005, 57, 1381–1396. [Google Scholar] [CrossRef]
- Alspaugh, J.A.; Perfect, J.R.; Heitman, J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev 1997, 11, 3206–3217. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Kojima, K.; Cox, G.M.; Heitman, J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell 2005, 16, 2285–2300. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Kojima, K.; Cox, G.M.; Heitman, J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell 2006, 17, 3122–3135. [Google Scholar] [CrossRef]
- Lee, J.W.; Ko, Y.J.; Kim, S.Y.; Bahn, Y.S. Multiple roles of Ypd1 phosphotransfer protein in viability, stress response, and virulence factor regulation in Cryptococcus neoformans. Eukaryot Cell 2011, 10, 998–1002. [Google Scholar] [CrossRef]
- Song, M.H.; Lee, J.W.; Kim, M.S.; Yoon, J.K.; White, T.C.; Floyd, A.; Heitman, J.; Strain, A.K.; Nielsen, J.N.; Nielsen, K.; et al. A flucytosine-responsive Mbp1/Swi4-like protein, Mbs1, plays pleiotropic roles in antifungal drug resistance, stress response, and virulence of Cryptococcus neoformans. Eukaryot Cell 2012, 11, 53–67. [Google Scholar] [CrossRef]
- D'Souza, C.A.; Alspaugh, J.A.; Yue, C.; Harashima, T.; Cox, G.M.; Perfect, J.R.; Heitman, J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol 2001, 21, 3179–3191. [Google Scholar] [CrossRef]
- Salas, S.D.; Bennett, J.E.; Kwon-Chung, K.J.; Perfect, J.R.; Williamson, P.R. Effect of the laccase gene CNLAC1, on the virulence of Cryptococcus neoformans. J Exp Med 1996, 184, 377–386. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Rosas, A.L.; Lee, S.C.; Casadevall, A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 2000, 355, 2049–2050. [Google Scholar] [CrossRef] [PubMed]
- Rosas, A.L.; Nosanchuk, J.D.; Feldmesser, M.; Cox, G.M.; McDade, H.C.; Casadevall, A. Synthesis of polymerised melanin by Cryptococcus neoformans in infected rodents. Infect Immun 2000, 68, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.C.; Polacheck, I.; Kwon-Chung, K.J. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect Immun 1982, 36, 1175–1184. [Google Scholar] [CrossRef]
- Liu, L.; Wakamatsu, K.; Ito, S.; Williamson, P.R. Catecholamine oxidative products, but not melanin, are produced by Cryptococcus neoformans during neuropathogenesis in mice. Infect Immun 1999, 67, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Wakamatsu, K. Chemical degradation of melanins: application to identification of dopamine-melanin. Pigment Cell Res 1998, 11, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, A.; Pezzella, A.; d'Ischia, M.; Prota, G. New pyrrole acids by oxidative degradation of eumelanins with hydrogen peroxide. Further hints to the mechanism of pigment breakdown. Tetrahedron 1996, 52, 8775–8780. [Google Scholar] [CrossRef]
- Napolitano, A.; Pezzella, A.; Vincensi, M.R.; Prota, G. Oxidative degradation of melanins to pyrrole acids: A model study. Tetrahedron 1995, 51, 5913–5920. [Google Scholar] [CrossRef]
- Wang, P.; Cutler, J.; King, J.; Palmer, D. Mutation of the regulator of G protein signalling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot Cell 2004, 3, 1028–1035. [Google Scholar] [CrossRef]
- Luberto, C.; Toffaletti, D.L.; Wills, E.A.; Tucker, S.C.; Casadevall, A.; Perfect, J.R.; Hannun, Y.A.; Del Poeta, M. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in the pathogenesis of C. neoformans. Genes Dev 2001, 15, 201–212. [Google Scholar] [CrossRef]
- Lee, C.G.; Da Silva, C.A.; Dela Cruz, C.S.; Ahangari, F.; Ma, B.; Kang, M.J.; He, C.H.; Takyar, S.; Elias, J.A. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodelling, and injury. Annu Rev Physiol 2011, 73, 479–501. [Google Scholar] [CrossRef]
- Fries, B.C.; Goldman, D.L.; Cherniak, R.; Ju, R.; Casadevall, A. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect Immun 1999, 67, 6076–6083. [Google Scholar] [CrossRef]
- Banks, I.R.; Specht, C.A.; Donlin, M.J.; Gerik, K.J.; Levitz, S.M.; Lodge, J.K. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell 2005, 4, 1902–1912. [Google Scholar] [CrossRef]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 2007, 6, 855–867. [Google Scholar] [CrossRef]
- Upadhya, R.; Baker, L.G.; Lam, W.C.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Cryptococcus neoformans Cda1 and Its Chitin Deacetylase Activity Are Required for Fungal Pathogenesis. mBio 2018, 9, e02087-02018. [Google Scholar] [CrossRef]
- Baker, L.G.; Specht, C.A.; Lodge, J.K. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot Cell 2011, 10, 1264–1268. [Google Scholar] [CrossRef]
- Upadhya, R.; Lam, W.C.; Maybruck, B.; Specht, C.A.; Levitz, S.M.; Lodge, J.K. Induction of Protective Immunity to Cryptococcal Infection in Mice by a Heat-Killed, Chitosan-Deficient Strain of Cryptococcus neoformans. mBio 2016, 7, e00547–00516. [Google Scholar] [CrossRef]
- Cazin, J., Jr.; Kozel, T.R.; Lupan, D.M.; Burt, W.R. Extracellular deoxyribonuclease production by yeasts. J Bacteriol 1969, 100, 760–762. [Google Scholar] [CrossRef]
- Sanchez, M.; Colom, F. Extracellular DNase activity of Cryptococcus neoformans and Cryptococcus gattii. Rev Iberoam Micol 2010, 27, 10–13. [Google Scholar] [CrossRef]
- Gumasta, R.; Nawange, S.R.; Singh, S.M.; Garg, A.; Sethi, R.; Yadu, R. Extracellular Protease and DNase Activities in Clinical and Environmental Isolates of Cryptococcus neoformans Species Complex from Central India. Journal of Drug Delivery and Therapeutics 2019, 9, 328–333. [Google Scholar] [CrossRef]
- Sumby, P.; Barbian, K.D.; Gardner, D.J.; Whitney, A.R.; Welty, D.M.; Long, R.D.; Bailey, J.R.; Parnell, M.J.; Hoe, N.P.; Adams, G.G. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proceedings of the National Academy of Sciences 2005, 102, 1679–1684. [Google Scholar] [CrossRef]
- Fazekas, G.; Schwarz, J. Histology of experimental murine cryptococcosis. The American Journal of Pathology 1958, 34, 517. [Google Scholar]
- Collopy-Junior, I.; Esteves, F.F.; Nimrichter, L.; Rodrigues, M.L.; Alviano, C.S.; Meyer-Fernandes, J.R. An ectophosphatase activity in Cryptococcus neoformans. FEMS Yeast Res 2006, 6, 1010–1017. [Google Scholar] [CrossRef]
- Mahvi, T.A.; Spicer, S.S.; Wright, N.J. Cytochemistry of acid mucosubstance and acid phosphatase in Cryptococcus neoformans. Canadian Journal of Microbiology 1974, 20, 833–838. [Google Scholar] [CrossRef]
- Lev, S.; Crossett, B.; Cha, S.Y.; Desmarini, D.; Li, C.; Chayakulkeeree, M.; Wilson, C.F.; Williamson, P.R.; Sorrell, T.C.; Djordjevic, J.T. Identification of Aph1, a phosphate-regulated, secreted, and vacuolar acid phosphatase in Cryptococcus neoformans. mBio 2014, 5, e01649–01614. [Google Scholar] [CrossRef]
- Plesner, L. Ecto-ATPases: identities and functions. International review of cytology 1995, 158, 141–214. [Google Scholar]
- Junior, I.C.; Rodrigues, M.L.; Alviano, C.S.; Travassos, L.R.; Meyer-Fernandes, J.R. Characterization of an ecto-ATPase activity in Cryptococcus neoformans. FEMS yeast research 2005, 5, 899–907. [Google Scholar] [CrossRef]
- Chen, L.C.; Pirofski, L.A.; Casadevall, A. Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect Immun 1997, 65, 2599–2605. [Google Scholar] [CrossRef]
- Nicola, A.M.; Frases, S.; Casadevall, A. Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot Cell 2009, 8, 1373–1380. [Google Scholar] [CrossRef]
- Rizzo, J.; Wong, S.S.W.; Gazi, A.D.; Moyrand, F.; Chaze, T.; Commere, P.H.; Novault, S.; Matondo, M.; Pehau-Arnaudet, G.; Reis, F.C.G.; et al. Cryptococcus extracellular vesicle properties and their use as vaccine platforms. J Extracell Vesicles 2021, 10, e12129. [Google Scholar] [CrossRef]
- Reis, R.S.; Bonna, I.C.F.; Antonio, I.; Pereira, S.A.; Nascimento, C.; Ferraris, F.K.; Brito-Santos, F.; Ferreira Gremiao, I.D.; Trilles, L. Cryptococcus neoformans VNII as the Main Cause of Cryptococcosis in Domestic Cats from Rio de Janeiro, Brazil. J Fungi (Basel) 2021, 7, e00125-00121. [Google Scholar] [CrossRef]
- Rizzo, J.; Taheraly, A.; Janbon, G. Structure, composition and biological properties of fungal extracellular vesicles. microLife 2021, 2. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Freire-de-Lima, C.G.; Nosanchuk, J.D.; Casadevall, A.; Rodrigues, M.L.; Nimrichter, L. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect Immun 2010, 78, 1601–1609. [Google Scholar] [CrossRef]
- Bielska, E.; Sisquella, M.A.; Aldeieg, M.; Birch, C.; O'Donoghue, E.J.; May, R.C. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat Commun 2018, 9, 1556. [Google Scholar] [CrossRef]
- Marina, C.L.; Bürgel, P.H.; Agostinho, D.P.; Zamith-Miranda, D.; Las-Casas, L.d.O.; Tavares, A.H.; Nosanchuk, J.D.; Bocca, A.L. Nutritional Conditions Modulate C. neoformans Extracellular Vesicles’ Capacity to Elicit Host Immune Response. Microorganisms 2020, 8, 1815. [Google Scholar] [CrossRef]
- Huang, S.H.; Wu, C.H.; Chang, Y.C.; Kwon-Chung, K.J.; Brown, R.J.; Jong, A. Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS One 2012, 7, e48570. [Google Scholar] [CrossRef]
- Panepinto, J.; Komperda, K.; Frases, S.; Park, Y.D.; Djordjevic, J.T.; Casadevall, A.; Williamson, P.R. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol Microbiol 2009, 71, 1165–1176. [Google Scholar] [CrossRef]
- Moyrand, F.; Janbon, G. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37 degrees C and for capsule biosynthesis. Eukaryot Cell 2004, 3, 1601–1608. [Google Scholar] [CrossRef]
- Reis, F.C.G.; Costa, J.H.; Honorato, L.; Nimrichter, L.; Fill, T.P.; Rodrigues, M.L. Small Molecule Analysis of Extracellular Vesicles Produced by Cryptococcus gattii: Identification of a Tripeptide Controlling Cryptococcal Infection in an Invertebrate Host Model. Front Immunol 2021, 12, 654574. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




