Submitted:
15 June 2023
Posted:
15 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
- ➢
- WBCs classification is achieved without the requirement for any preprocessing or convolutional processes.
- ➢
- The effectiveness of using ViT for WBC classification, which can potentially outperform traditional CNN architectures.
- ➢
- The proposed ViT-based method achieves high accuracy rates in both multi-class and binary classification of WBCs, which is crucial for accurate disease diagnosis and treatment.
- ➢
- An explainable method for WBCs classification, which increases the transparency and trustworthiness of the model's decision-making process.
- ➢
- Visiualization of the pixel areas upon which the model focuses its predictions, which can facilitate the adoption of the proposed method in clinical settings.
2. Materials and Methods
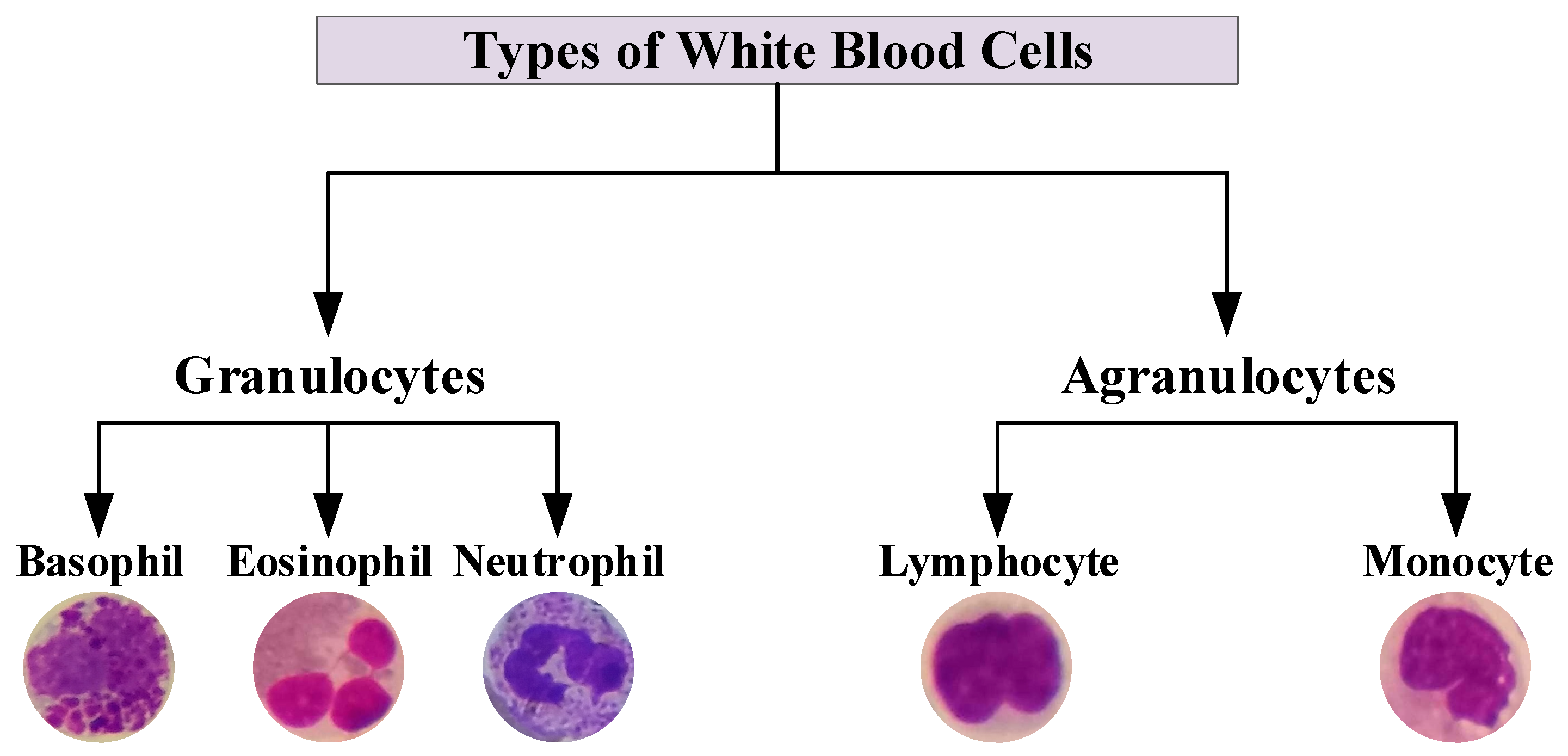
2.1. WBCs Dataset
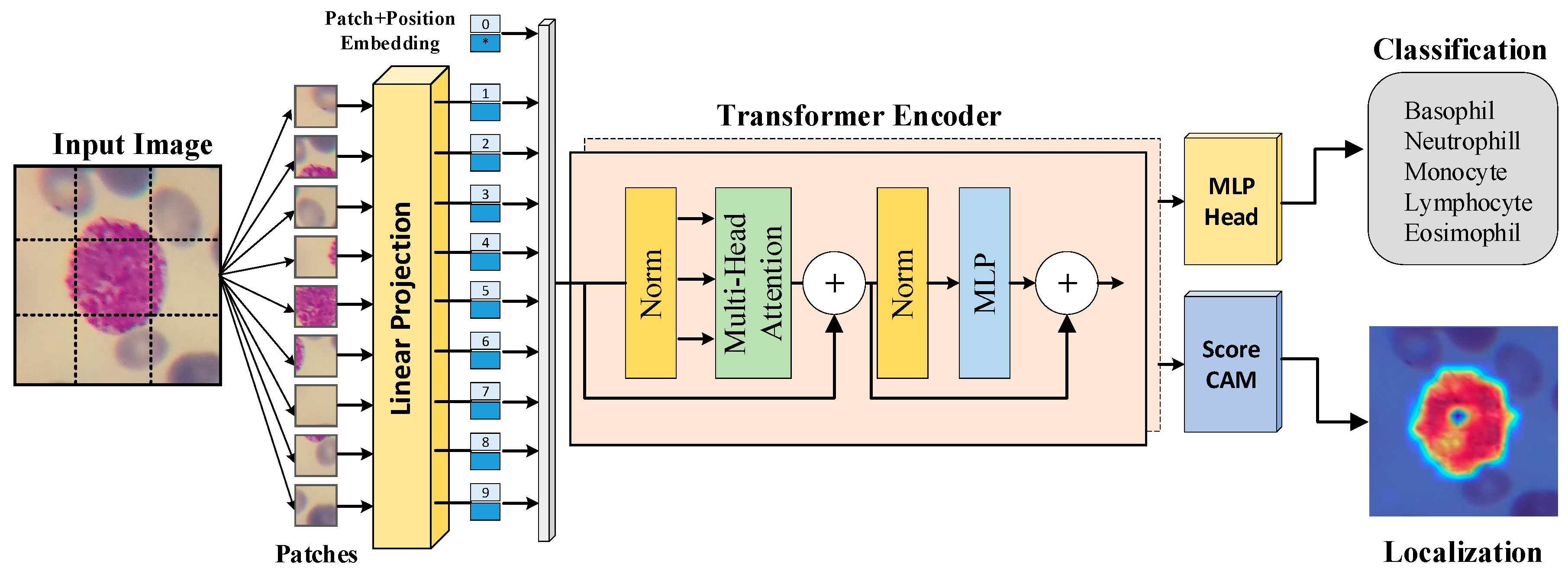
2.2. Transformer-based Image Classification Method
3. Experiments
3.1. Experimental Setup
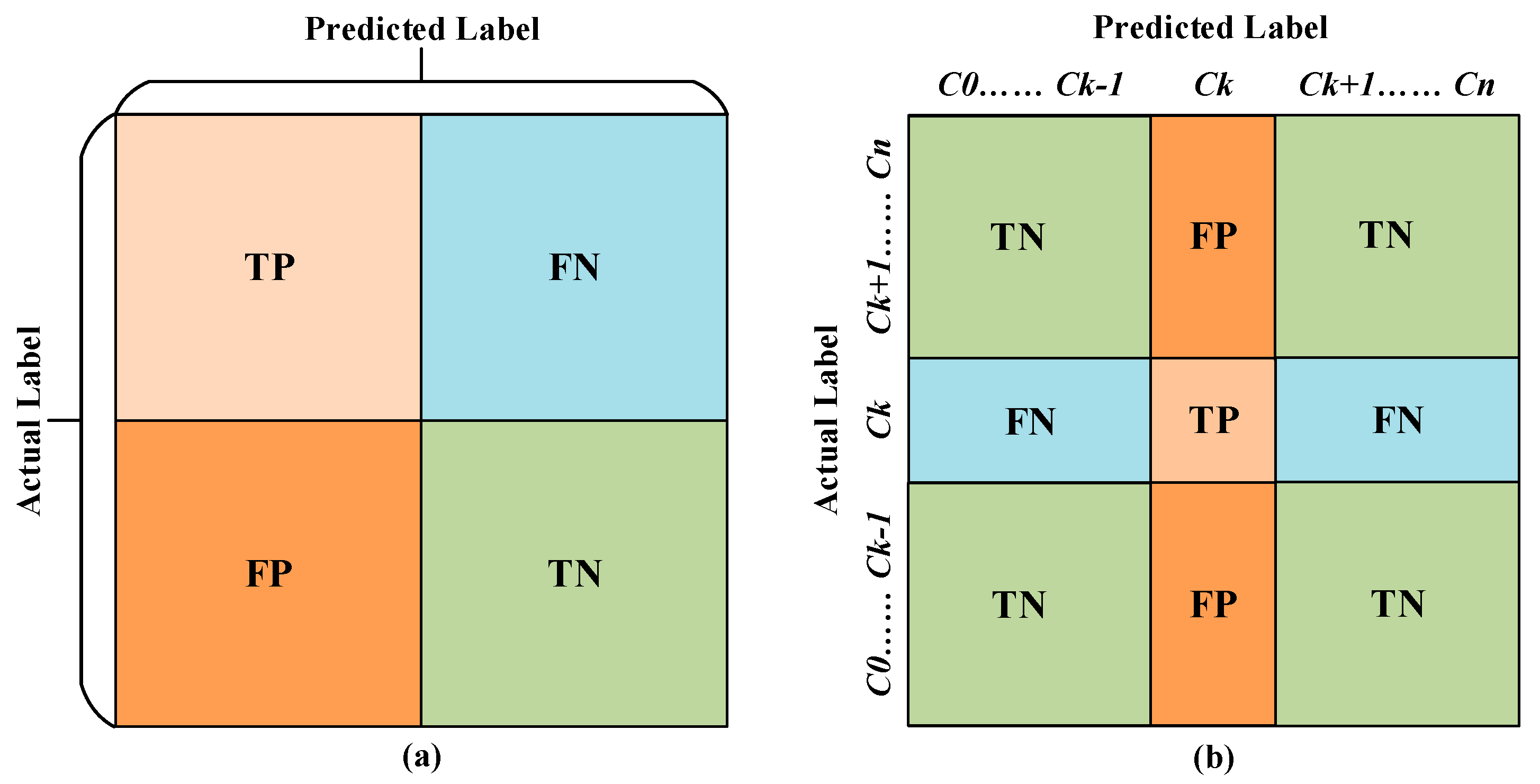
3.2. Performance Evaluation Metrics
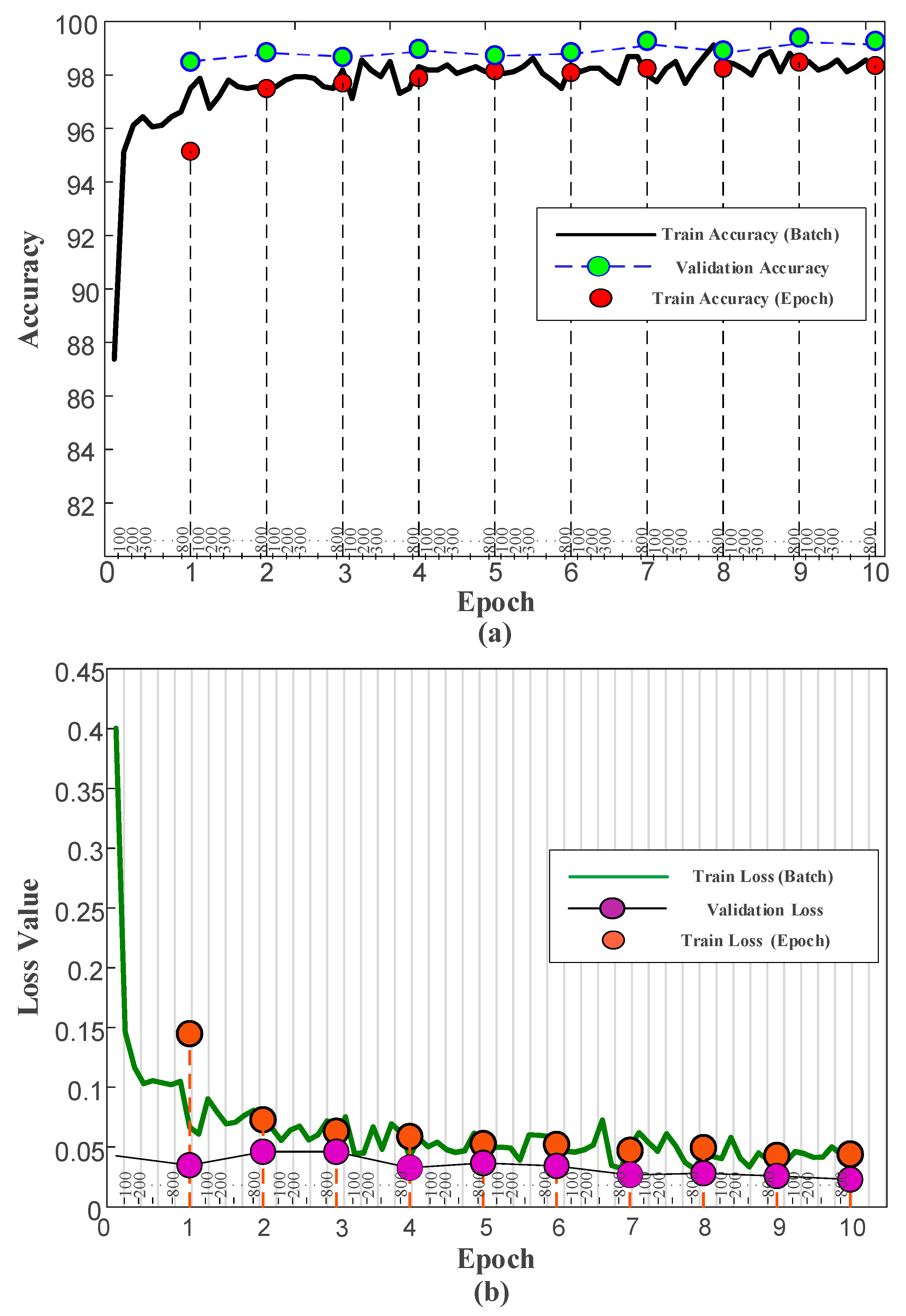
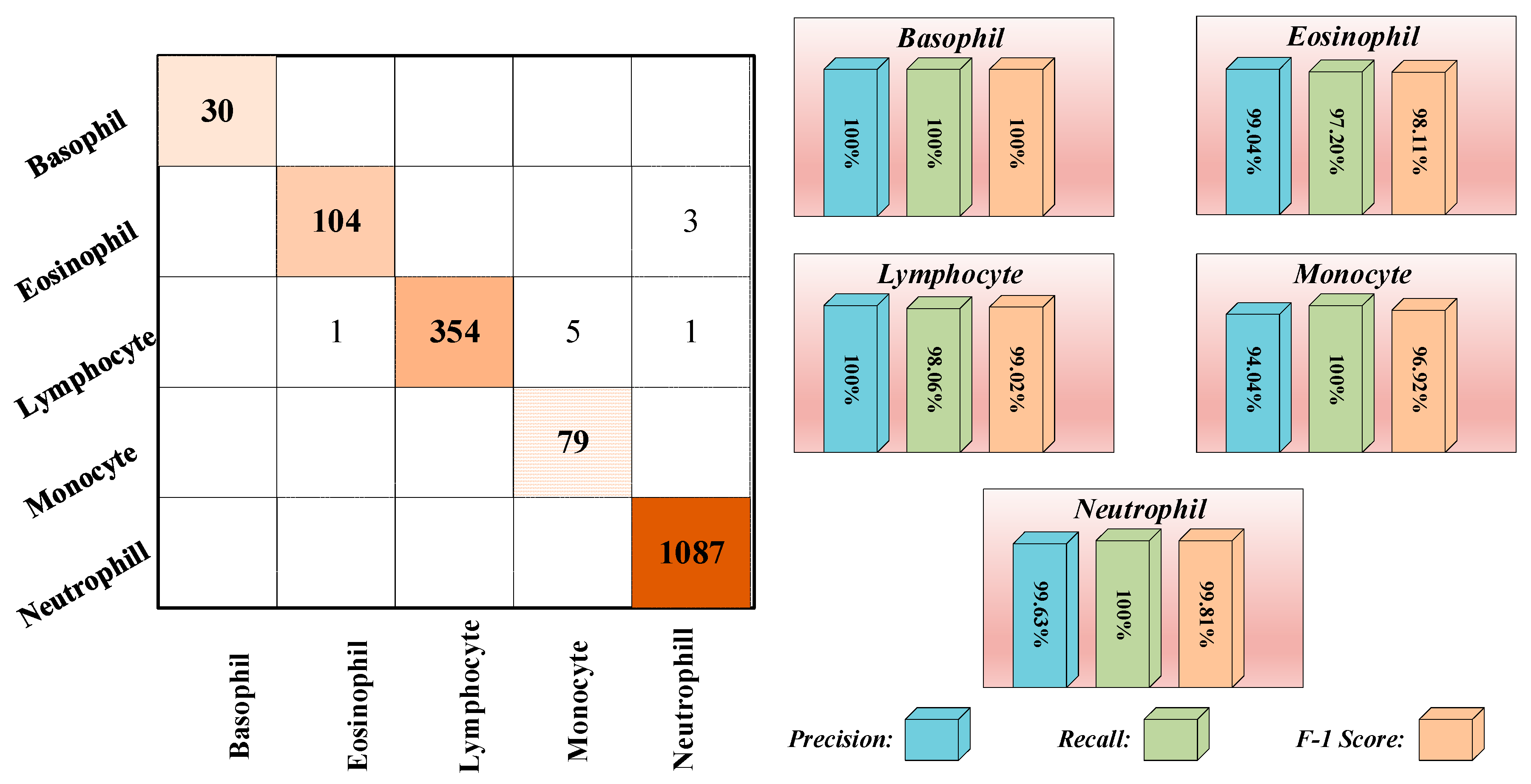
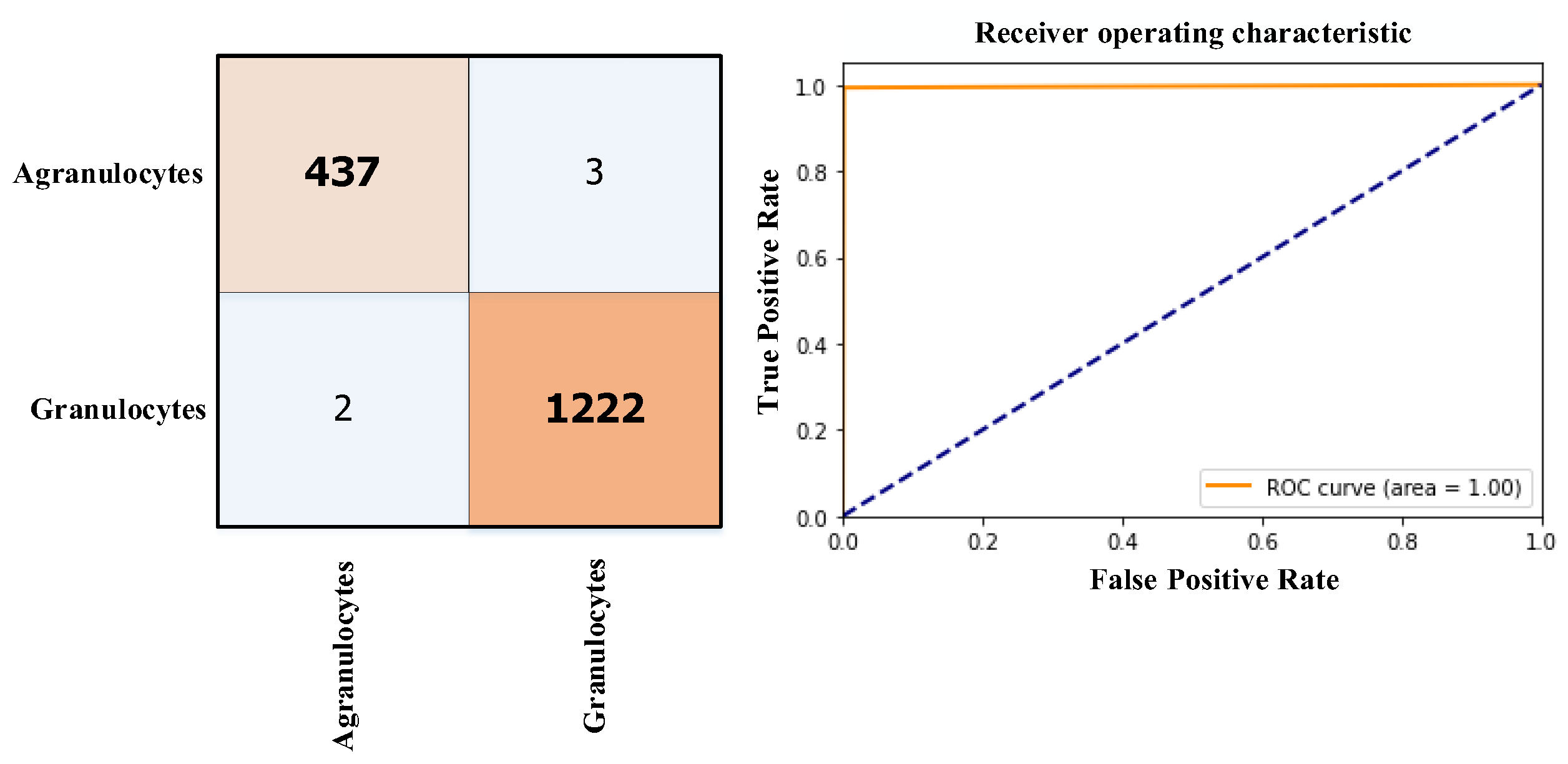
3.3. Results
4. Discussion
- The proposed model is based on vision transformers that has become popular research field. Therefore this study is an example to examine vision transformers performance in biomedical image classification.
- This model can classify WBCs images with end-to-end transformer structure. There is no need to use any feature engineering.
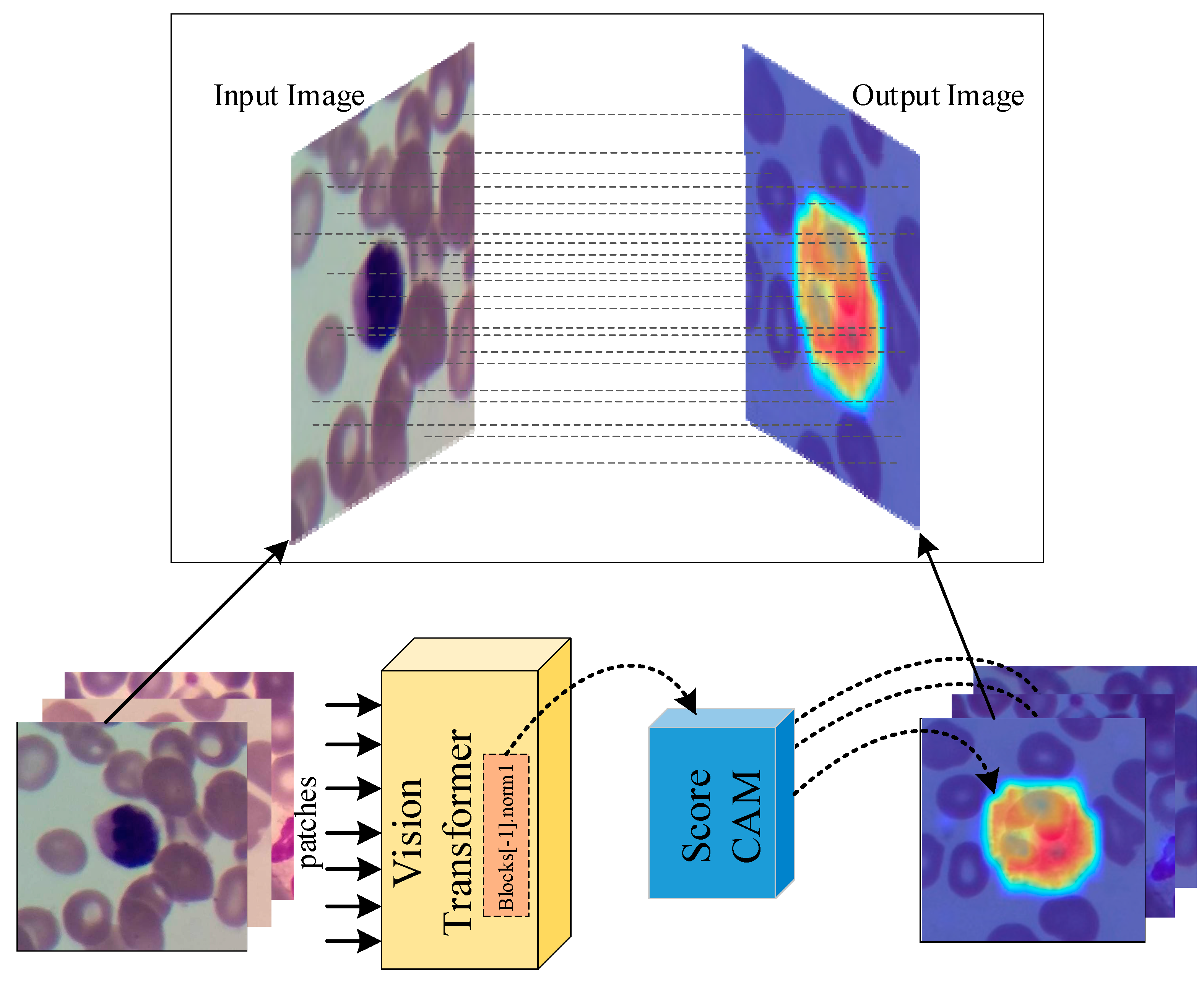
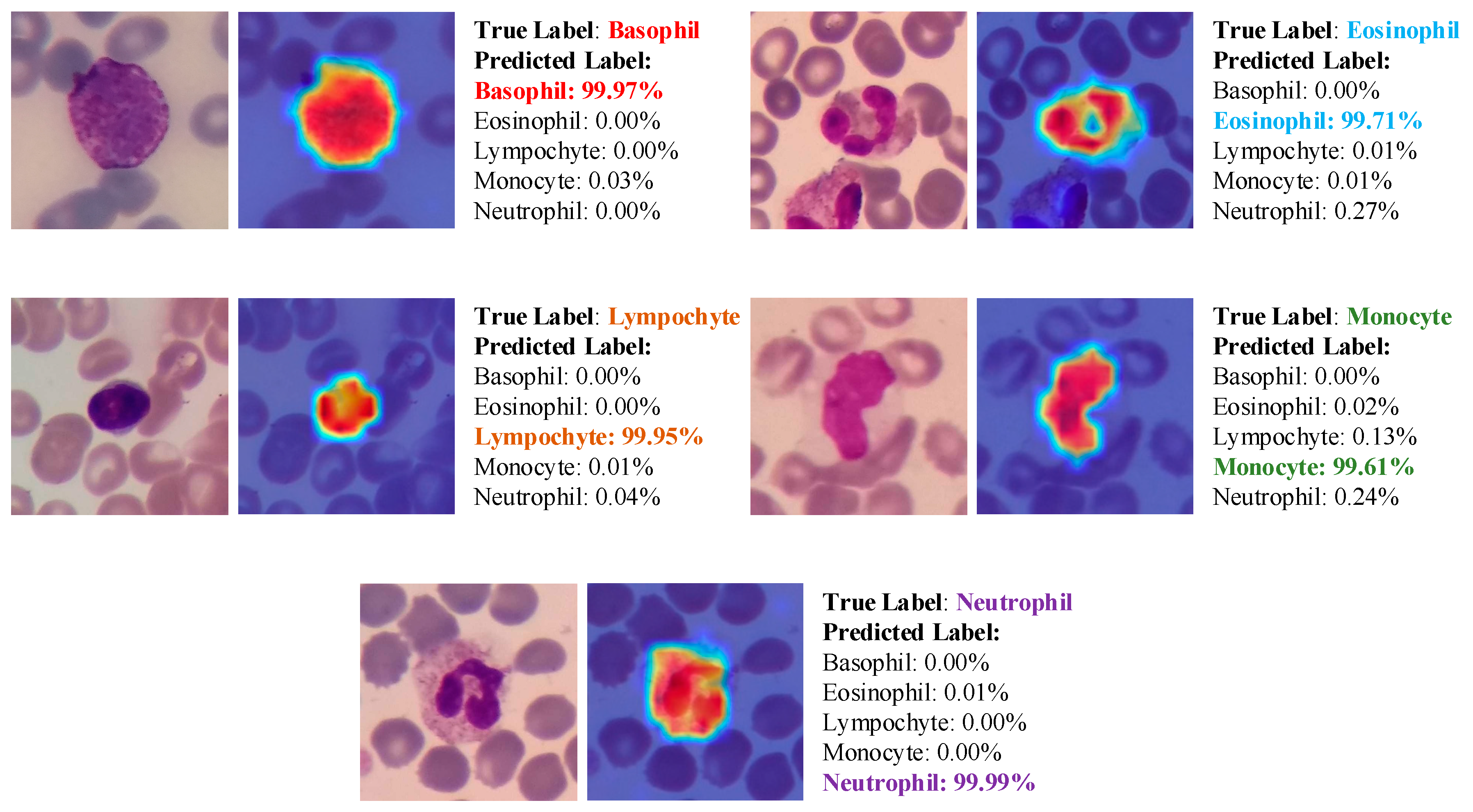
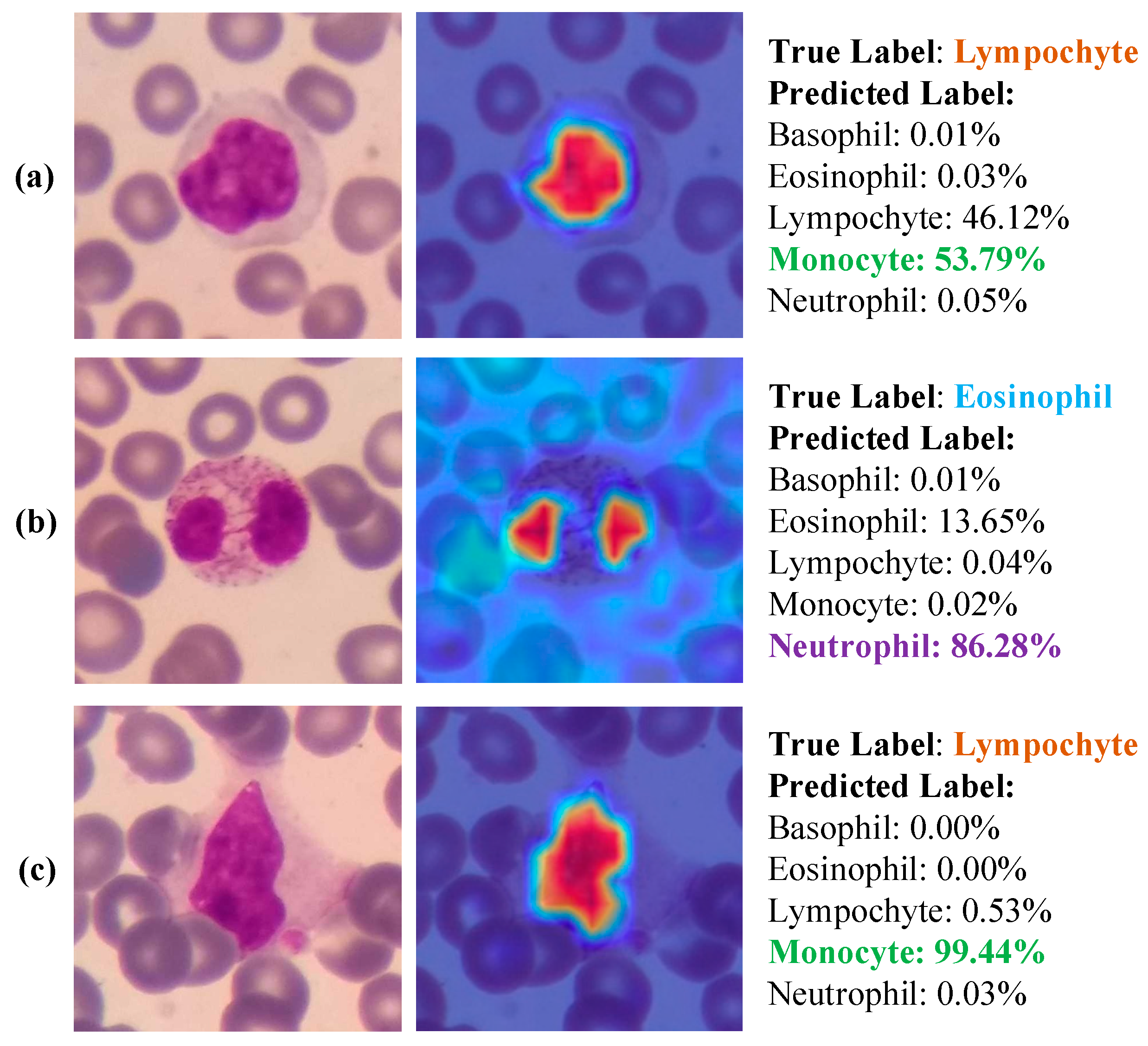
- Due to the explainable structure, the proposed model presents focused regions during the classification process. According to these results, experts can validate model performance.
- Due to its high level of classification accuracy, it has the potential to be utilized in clinical applications.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Dulaimi, K.A.K.; Banks, J.; Chandran, V.; Tomeo-Reyes, I.; Nguyen Thanh, K. Classification of White Blood Cell Types from Microscope Images: Techniques and Challenges. In Microscopy science: Last approaches on educational programs and applied research (Microscopy Book Series, 8); 2018; pp. 17–25. [Google Scholar]
- Ahmad, Z.; Shah, T.A.; Reddy, K.P.; Ghosh, S.; Panpatil, V.; Kottoru, S.K.; Rayees, S.; Rao, D.R. Immunology in Medical Biotechnology. In Fundamentals and Advances in Medical Biotechnology; Springer, 2022; pp. 179–207. [Google Scholar]
- Kiboneka, A.N. Basic Concepts in Clinical Immunology: A Review. 2021. [Google Scholar] [CrossRef]
- Tripathi, C. Tolerance and Autoimmunity. In An Interplay of Cellular and Molecular Components of Immunology; CRC Press, 2022; pp. 207–216. ISBN 1003286429. [Google Scholar]
- Otieno, F.; Kyalo, C. Perspective Chapter: Macrophages Plasticity and Immune Metabolism. In Basic and Clinical Aspects of Interferon Gamma; IntechOpen, 2022; ISBN 1803558865. [Google Scholar] [CrossRef]
- Wechsler, M.E.; Munitz, A.; Ackerman, S.J.; Drake, M.G.; Jackson, D.J.; Wardlaw, A.J.; Dougan, S.K.; Berdnikovs, S.; Schleich, F.; Matucci, A. Eosinophils in Health and Disease: A State-of-the-Art Review. In Proceedings of the Mayo Clinic Proceedings; Elsevier, 2021; Vol. 96; pp. 2694–2707. [Google Scholar] [CrossRef]
- Santos, A.F.; Alpan, O.; Hoffmann, H. Basophil Activation Test: Mechanisms and Considerations for Use in Clinical Trials and Clinical Practice. Allergy 2021, 76, 2420–2432. [Google Scholar] [CrossRef] [PubMed]
- Parente, J. Diagnostics for White Blood Cell Abnormalities: Leukocytosis and Leukopenia. Physician Assist Clin 2019, 4, 625–635. [Google Scholar] [CrossRef]
- Agnello, L.; Giglio, R.V.; Bivona, G.; Scazzone, C.; Gambino, C.M.; Iacona, A.; Ciaccio, A.M.; lo Sasso, B.; Ciaccio, M. The Value of a Complete Blood Count (CBC) for Sepsis Diagnosis and Prognosis. Diagnostics 2021, 11, 1881. [Google Scholar] [CrossRef]
- Wang, Q.; Bi, S.; Sun, M.; Wang, Y.; Wang, D.; Yang, S. Deep Learning Approach to Peripheral Leukocyte Recognition. PLoS One 2019, 14, e0218808. [Google Scholar] [CrossRef]
- Khamael, A.-D.; Banks, J.; Nugyen, K.; Al-Sabaawi, A.; Tomeo-Reyes, I.; Chandran, V. Segmentation of White Blood Cell, Nucleus and Cytoplasm in Digital Haematology Microscope Images: A Review–Challenges, Current and Future Potential Techniques. IEEE Rev Biomed Eng 2020, 14, 290–306. [Google Scholar]
- H Mohamed, E.; H El-Behaidy, W.; Khoriba, G.; Li, J. Improved White Blood Cells Classification Based on Pre-Trained Deep Learning Models. J. Commun. Softw. Syst. 2020, 16, 37–45. [Google Scholar] [CrossRef]
- Patil, A.M.; Patil, M.D.; Birajdar, G.K. White Blood Cells Image Classification Using Deep Learning with Canonical Correlation Analysis. Irbm 2021, 42, 378–389. [Google Scholar] [CrossRef]
- Basnet, J.; Alsadoon, A.; Prasad, P.W.C.; Aloussi, S. al; Alsadoon, O.H. A Novel Solution of Using Deep Learning for White Blood Cells Classification: Enhanced Loss Function with Regularization and Weighted Loss (ELFRWL). Neural Process Lett 2020, 52, 1517–1553. [Google Scholar] [CrossRef]
- Cheuque, C.; Querales, M.; León, R.; Salas, R.; Torres, R. An Efficient Multi-Level Convolutional Neural Network Approach for White Blood Cells Classification. Diagnostics 2022, 12, 248. [Google Scholar] [CrossRef]
- Sharma, S.; Gupta, S.; Gupta, D.; Juneja, S.; Gupta, P.; Dhiman, G.; Kautish, S. Deep Learning Model for the Automatic Classification of White Blood Cells. Comput Intell Neurosci 2022, 2022. [Google Scholar] [CrossRef]
- Jung, C.; Abuhamad, M.; Alikhanov, J.; Mohaisen, A.; Han, K.; Nyang, D. W-Net: A CNN-Based Architecture for White Blood Cells Image Classification. arXiv 2019, arXiv:1910.01091 2019. [Google Scholar]
- Rustam, F.; Aslam, N.; de La Torre Díez, I.; Khan, Y.D.; Mazón, J.L.V.; Rodríguez, C.L.; Ashraf, I. White Blood Cell Classification Using Texture and RGB Features of Oversampled Microscopic Images. In Proceedings of the Healthcare; MDPI, 2022. Vol. 10, p. 2230. [CrossRef]
- Chola, C.; Muaad, A.Y.; bin Heyat, M.B.; Benifa, J.V.B.; Naji, W.R.; Hemachandran, K.; Mahmoud, N.F.; Samee, N.A.; Al-Antari, M.A.; Kadah, Y.M. BCNet: A Deep Learning Computer-Aided Diagnosis Framework for Human Peripheral Blood Cell Identification. Diagnostics 2022, 12, 2815. [Google Scholar] [CrossRef]
- Bhojanapalli, S.; Chakrabarti, A.; Glasner, D.; Li, D.; Unterthiner, T.; Veit, A. Understanding Robustness of Transformers for Image Classification. In Proceedings of the Proceedings of the IEEE/CVF international conference on computer vision; 2021; pp. 10231–10241. [Google Scholar]
- Bazi, Y.; Bashmal, L.; Rahhal, M.M. al; Dayil, R. al; Ajlan, N. al Vision Transformers for Remote Sensing Image Classification. Remote Sens (Basel) 2021, 13, 516. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Y.; Guo, T.; Xu, C.; Deng, Y.; Liu, Z.; Ma, S.; Xu, C.; Xu, C.; Gao, W. Pre-Trained Image Processing Transformer. In Proceedings of the Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2021; pp. 12299–12310. [Google Scholar]
- Kouzehkanan, Z.M.; Saghari, S.; Tavakoli, S.; Rostami, P.; Abaszadeh, M.; Mirzadeh, F.; Satlsar, E.S.; Gheidishahran, M.; Gorgi, F.; Mohammadi, S. A Large Dataset of White Blood Cells Containing Cell Locations and Types, along with Segmented Nuclei and Cytoplasm. Sci Rep 2022, 12, 1123. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M. Transformers: State-of-the-Art Natural Language Processing. In Proceedings of the Proceedings of the 2020 conference on empirical methods in natural language processing: system demonstrations; 2020; pp. 38–45. [Google Scholar]
- Guo, M.-H.; Liu, Z.-N.; Mu, T.-J.; Hu, S.-M. Beyond Self-Attention: External Attention Using Two Linear Layers for Visual Tasks. IEEE Trans Pattern Anal Mach Intell 2022. [Google Scholar] [CrossRef]
- Wang, L.; He, Y.; Li, L.; Liu, X.; Zhao, Y. A Novel Approach to Ultra-Short-Term Multi-Step Wind Power Predictions Based on Encoder–Decoder Architecture in Natural Language Processing. J Clean Prod 2022, 354, 131723. [Google Scholar] [CrossRef]
- Thakur, P.S.; Khanna, P.; Sheorey, T.; Ojha, A. Explainable Vision Transformer Enabled Convolutional Neural Network for Plant Disease Identification: PlantXViT. arXiv 2022, arXiv:2207.07919. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An Image Is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv arXiv:2010.11929, 2020.
- Lee, S.H.; Lee, S.; Song, B.C. Vision Transformer for Small-Size Datasets. arXiv 2021, arXiv:2112.13492 2021. [Google Scholar]
- Ekanayake, I.U.; Meddage, D.P.P.; Rathnayake, U. A Novel Approach to Explain the Black-Box Nature of Machine Learning in Compressive Strength Predictions of Concrete Using Shapley Additive Explanations (SHAP). Case Stud. Constr. Mater. 2022, 16, e01059. [Google Scholar] [CrossRef]
- Liang, Y.; Li, S.; Yan, C.; Li, M.; Jiang, C. Explaining the Black-Box Model: A Survey of Local Interpretation Methods for Deep Neural Networks. Neurocomputing 2021, 419, 168–182. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-Weighted Visual Explanations for Convolutional Neural Networks. In Proceedings of the Proceedings of the IEEE/CVF conference on computer vision and pattern recognition workshops; 2020; pp. 24–25. [Google Scholar]
- Tavakoli, S.; Ghaffari, A.; Kouzehkanan, Z.M.; Hosseini, R. New Segmentation and Feature Extraction Algorithm for Classification of White Blood Cells in Peripheral Smear Images. Sci Rep 2021, 11, 19428. [Google Scholar] [CrossRef] [PubMed]
- KATAR, O.; KILINÇER, İ.F. Automatic Classification of White Blood Cells Using Pre-Trained Deep Models. Sak. Univ. J. Comput. Inf. Sci. 2022, 5, 462–476. [Google Scholar] [CrossRef]
- AKALIN, F.; YUMUŞAK, N. Detection and Classification of White Blood Cells with an Improved Deep Learning-Based Approach. Turk. J. Electr. Eng. Comput. Sci. 2022, 30, 2725–2739. [Google Scholar] [CrossRef]
- Leng, B.; Wang, C.; Leng, M.; Ge, M.; Dong, W. Deep Learning Detection Network for Peripheral Blood Leukocytes Based on Improved Detection Transformer. Biomed Signal Process Control 2023, 82, 104518. [Google Scholar] [CrossRef]
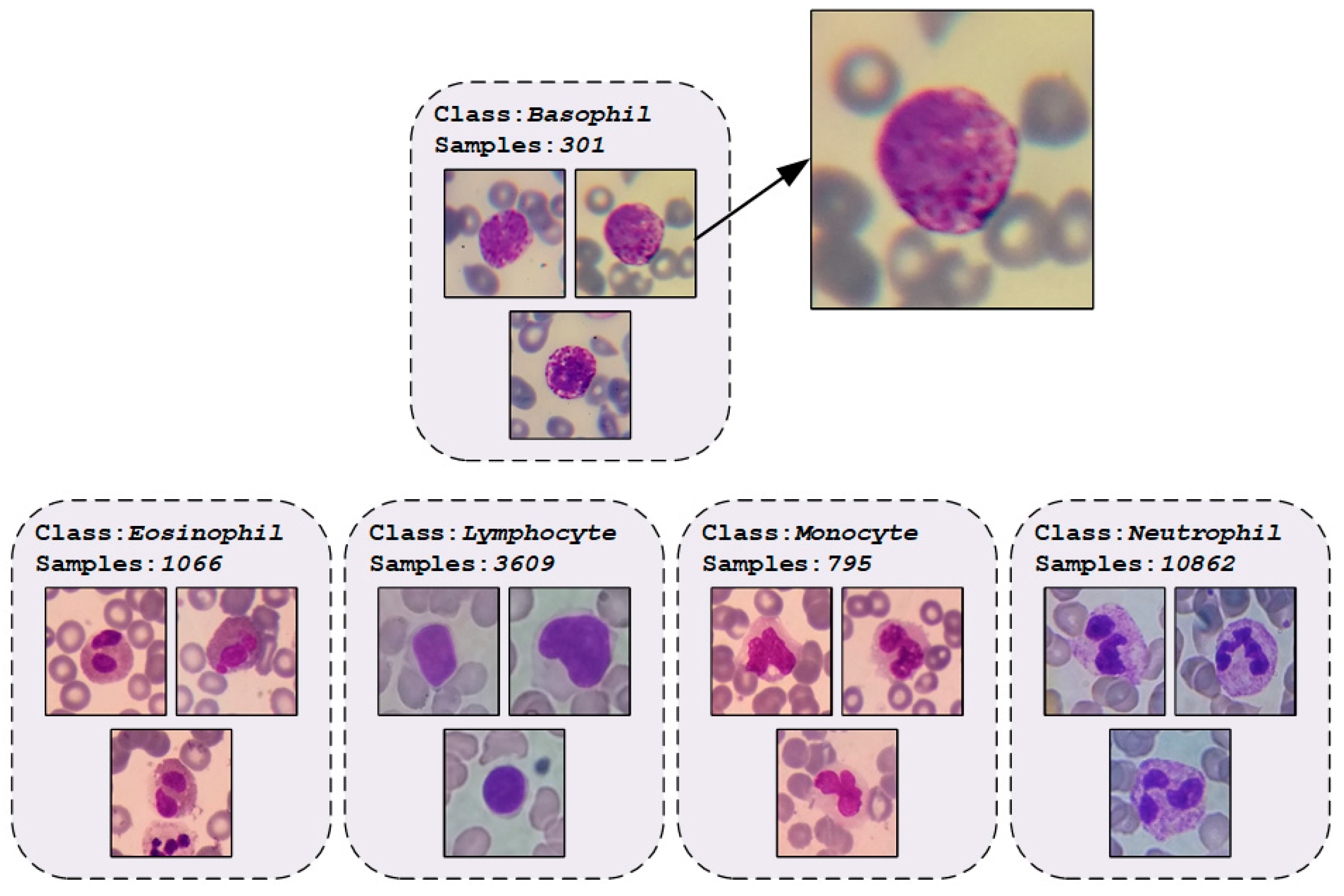

| Phase | Basophil | Eosinophil | Lymphocyte | Monocyte | Neutrophil | Total |
|---|---|---|---|---|---|---|
| Training | 241 | 852 | 2,887 | 637 | 8,688 | 13,305 |
| Validation | 30 | 107 | 361 | 79 | 1,087 | 1,664 |
| Testing | 30 | 107 | 361 | 79 | 1,087 | 1,664 |
| Total | 301 | 1,066 | 3,609 | 795 | 10,862 | 16,633 |
| Study | Year | Number of Class | Method | Explainability | Performance |
|---|---|---|---|---|---|
| Tavakoli et al. [33] | 2021 | 5 (Basophil, Eosinophil, Lymphocyte, Monocyte, Neutrophil) | SVM | Black-box | Acc=94.65% |
| Katar and Kilincer [34] | 2022 | 5 (Basophil, Eosinophil, Lymphocyte, Monocyte, Neutrophil) | CNN | Grad-CAM | Acc=98.86% |
| Akalin and Yumusak [35] | 2022 | 5 (Basophil, Eosinophil, Lymphocyte, Monocyte, Neutrophil) | Hybrid | Black-box | Acc=98.00% |
| Leng et al. [36] | 2023 | 3 (Eosinophil, Monocyte, Neutrophil) | DETR | Black-box | mAP=96.10% |
| The proposed study | 2023 | 2 (Granulocytes and Agranulocytes) | ViT | Score-CAM | Acc=99.75% |
| 5 (Basophil, Eosinophil, Lymphocyte, Monocyte, Neutrophil) | Acc=99.40% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





