Submitted:
14 June 2023
Posted:
14 June 2023
You are already at the latest version
Abstract
Keywords:
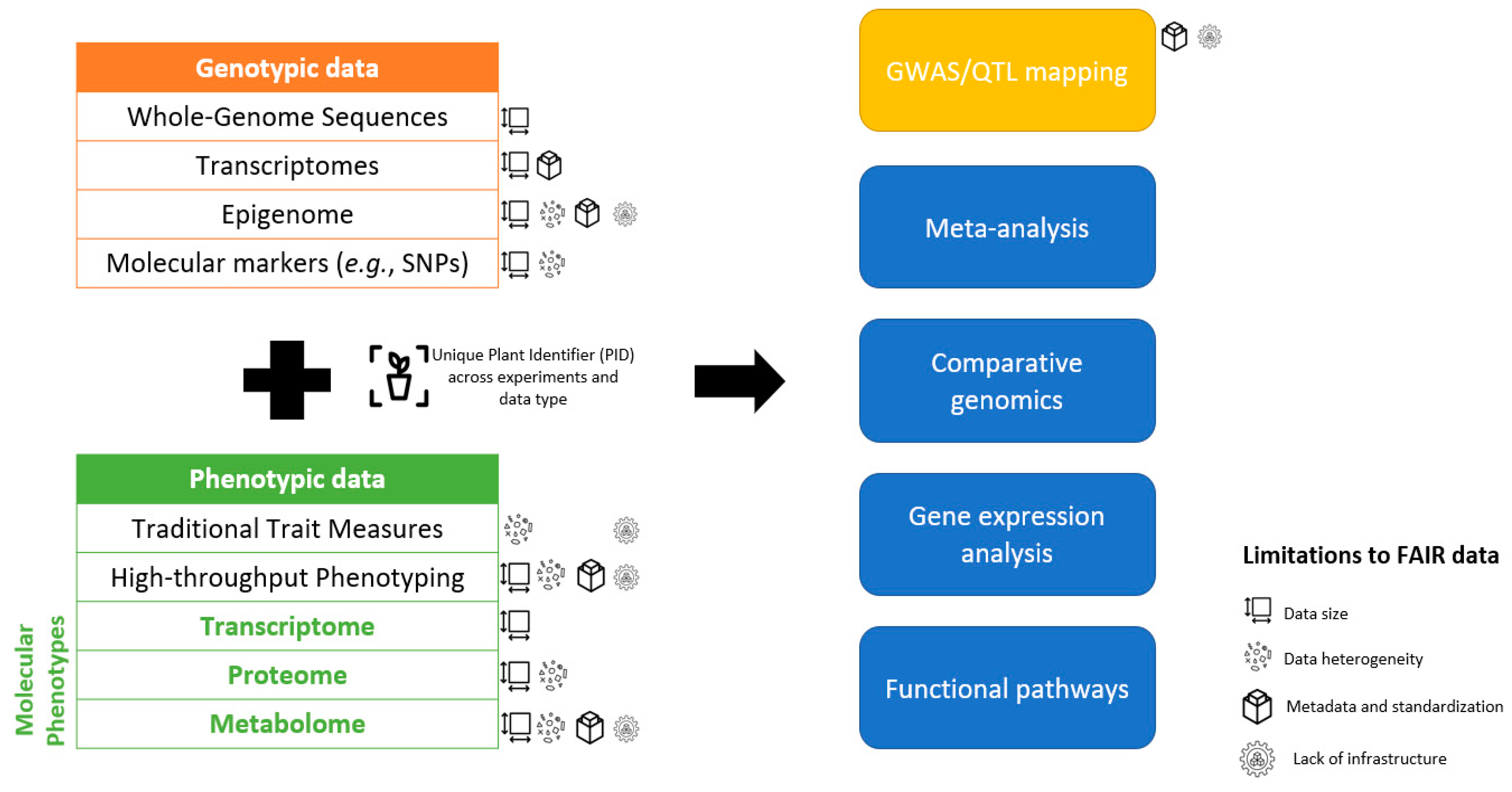
1. Introduction
2. Genomics and Transcriptomics data
2.1. Whole genome and transcriptome sequences
2.2. Genome Sequencing Strategies for Genotyping
2.3. Public repositories for genomics and transcriptomics data
2.3.1. The metadata requirements on genomics and transcriptomics data set
2.3.2. Genotyping data submission and metadata requirements
2.3.3. Crop/clad Community GGB Databases
2.3.4. Uses and Applications
3. Phenotype and Phenomics
3.1. Data types, Repositories, and Knowledge Bases
3.2. Phenotype data formats, standards and metadata
4. Association mapping (GWAS) and linkage mapping (QTL)
5. Data reusability limitations and challenges
5.1. Challenges
5.2. Resources and Funding
5.3. Implementation of FAIR data policy
6. Recommendations
- Standardization of data collection protocols: Standardizing data collection protocols and using common data formats can help to ensure that data is collected in a consistent and comparable way. Use of metadata standards and the requirement of new ontology terms will make it easier to share and compare data across different studies.
- Centralized data sharing platform: Developing and using centralized data sharing platforms, the use of standardized data models and exchange formats and the deployment of existing and emerging software components can help to facilitate the sharing of genotypic and phenotypic data among researchers. It includes the use of online databases and repositories that are specifically designed for the storage and sharing of the plant genetic and phenotypic data.
- Consistent data annotation: Consistently annotating data with relevant information such as the genotype, phenotype, and experimental treatments can help to make the data more easily searchable and usable by other researchers.
- Data quality control: More automated management of data flows and implementing data quality control such as data curation and validation can help to ensure that the data is accurate, reliable and can be used to make valid conclusions.
- Data integration: Adoption of new database technologies and the development of robust data standards can facilitate the global integration of G2P data in future. Data integration from different resources such as genomics, transcriptomics, proteomics and metabolomics can help to better understand the complex relationship between genotype and phenotype.
- Community driven efforts: Community driven efforts such as open-source projects, workshops and collaborations can help to promote the sharing and use of data among researchers, which in turn will lead to better understanding of the G2P relationship. There should be encouragement on integrated science training plans that enable biologists to think quantitatively and facilitate collaboration with experts in physical, computational and engineering sciences. It can help the scientists to get familiar with the development of computational pipelines and workflows that will be essential for researchers to acquire, analyze and critically interpret G2P data.
- Data storage infrastructure, data management software and data curation tools are necessary to handle the large volumes of data in diverse formats.
- A concerted effort to make multi-omics data sets interoperable by automated biocuration with controlled ontology terms will help address this issue. Community databases address some of this issue by collecting, curating, and integrating various data of different types, from different sources, and from different but related species. However, community databases need to have sustainable funding.
- Data security, backup and recovery must be considered and implemented for sustainability.
- Data compliance with data sharing policies, privacy regulations and laws should be enforced.
Acknowledgements
References
- Scossa, F. , Alseekh, S., Fernie, A.R. Integrating multi-omics data for crop improvement. J Plant Physiol 2021, 257, 153352. [Google Scholar] [CrossRef]
- Yang, W. , Feng, H., Zhang, X.; et al. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol Plant 2020, 13, 187–214. [Google Scholar] [CrossRef]
- Wilkinson, M.D. , Dumontier, M., Aalbersberg, I.J.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016, 3, 160018. [Google Scholar] [CrossRef]
- Harper, L. , Campbell, J., Cannon, E.K.S.; et al. AgBioData consortium recommendations for sustainable genomics and genetics databases for agriculture. Database (Oxford) 2018, 2018. [Google Scholar] [CrossRef]
- Adam-Blondon, A.F. , Alaux, M., Pommier, C.; et al. Towards an open grapevine information system. Hortic Res 2016, 3, 16056. [Google Scholar] [CrossRef]
- Ekblom, R. , Wolf, J.B. A field guide to whole-genome sequencing, assembly and annotation. Evol Appl 2014, 7, 1026–1042. [Google Scholar] [CrossRef]
- Kanzi, A.M. , San, J.E., Chimukangara, B.; et al. Next Generation Sequencing and Bioinformatics Analysis of Family Genetic Inheritance. Front Genet 2020, 11, 544162. [Google Scholar] [CrossRef]
- Patterson, J. , Carpenter, E.J., Zhu, Z.; et al. Impact of sequencing depth and technology on de novo RNA-Seq assembly. BMC Genomics 2019, 20, 604. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R. , Judd, L.M., Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol 2019, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Grodzicker, T. , Williams, J., Sharp, P.; et al. Physical mapping of temperature-sensitive mutations of adenoviruses. Cold Spring Harb Symp Quant Biol. 1975, 39 Pt 1, 439-446.
- Yang, W. , Kang, X., Yang, Q.; et al. Review on the development of genotyping methods for assessing farm animal diversity. J Anim Sci Biotechnol 2013, 4, 2. [Google Scholar] [CrossRef]
- McKain, M.R. , Johnson, M.G., Uribe-Convers, S.; et al. Practical considerations for plant phylogenomics. Appl Plant Sci 2018, 6, e1038. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P. , Choudhary, M., Jat, B.S.; et al. Skim sequencing: an advanced NGS technology for crop improvement. J Genet. 2021, 100..
- Schmickl, R. , Liston, A., Zeisek, V.; et al. Phylogenetic marker development for target enrichment from transcriptome and genome skim data: the pipeline and its application in southern African Oxalis (Oxalidaceae). Mol Ecol Resour 2016, 16, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Head, S.R. , Komori, H.K., LaMere, S.A.; et al. Library construction for next-generation sequencing: overviews and challenges. Biotechniques 2014, 56, 61–64, 66, 68, passim. [Google Scholar] [CrossRef]
- Deschamps, S. , Llaca, V., May, G.D. Genotyping-by-Sequencing in Plants. Biology (Basel) 2012, 1, 460–483. [Google Scholar]
- Elshire, R.J. , Glaubitz, J.C., Sun, Q.; et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Andrews, K.R. , Good, J.M., Miller, M.R.; et al. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet 2016, 17, 81–92. [Google Scholar] [CrossRef]
- Miller, M.R. , Dunham, J.P., Amores, A.; et al. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 2007, 17, 240–248. [Google Scholar] [CrossRef]
- Danecek, P. , Auton, A., Abecasis, G.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Lyon, M.S. , Andrews, S.J., Elsworth, B.; et al. The variant call format provides efficient and robust storage of GWAS summary statistics. Genome Biol 2021, 22, 32. [Google Scholar] [CrossRef]
- Bronner, I.F. , Lorenz, S. Combined Genome and Transcriptome (G&T) Sequencing of Single Cells. Methods Mol Biol 2019, 1979, 319–362. [Google Scholar]
- Promoting best practice in nucleotide sequence data sharing. Sci Data 2020, 7, 152. [CrossRef]
- Goodstein, D.M. , Shu, S., Howson, R.; et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 2012, 40, D1178–1186. [Google Scholar] [CrossRef]
- Members, C.-N. , Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res 2021, 49, D18–D28. [Google Scholar]
- Cezard, T. , Cunningham, F., Hunt, S.E.; et al. The European Variation Archive: a FAIR resource of genomic variation for all species. Nucleic Acids Res 2022, 50, D1216–D1220. [Google Scholar] [CrossRef] [PubMed]
- Song, S. , Tian, D., Li, C.; et al. Genome Variation Map: a data repository of genome variations in BIG Data Center. Nucleic Acids Res 2018, 46, D944–D949. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y. , Song, X., Zhang, Q.; et al. Robust CRISPR/Cas9 mediated gene editing of JrWOX11 manipulated adventitious rooting and vegetative growth in a nut tree species of walnut. Scientia Horticulturae 2022, 303, 111199. [Google Scholar] [CrossRef]
- International HapMap, C. The International HapMap Project. Nature 2003, 426, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Jung, S. , Jesudurai, C., Staton, M.; et al. GDR (Genome Database for Rosaceae): integrated web resources for Rosaceae genomics and genetics research. BMC Bioinformatics 2004, 5, 130. [Google Scholar] [CrossRef] [PubMed]
- Jung, S. , Lee, T., Cheng, C.H.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed]
- Jung, S. , Staton, M., Lee, T.; et al. GDR (Genome Database for Rosaceae): integrated web-database for Rosaceae genomics and genetics data. Nucleic Acids Res 2008, 36, D1034–1040. [Google Scholar] [CrossRef]
- Yu, J. , Jung, S., Cheng, C.H.; et al. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res 2014, 42, D1229–1236. [Google Scholar] [CrossRef]
- Yu, J. , Jung, S., Cheng, C.H.; et al. CottonGen: The Community Database for Cotton Genomics, Genetics, and Breeding Research. Plants (Basel) 2021, 10. [Google Scholar]
- Grant, D. , Nelson, R.T., Cannon, S.B.; et al. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 2010, 38, D843–846. [Google Scholar] [CrossRef]
- Brown, A.V. , Conners, S.I., Huang, W.; et al. A new decade and new data at SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 2021, 49, D1496–D1501. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, M.D. , Archuleta, E., Farmer, A.; et al. The Legume Information System (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res 2005, 33, D660–665. [Google Scholar] [CrossRef]
- Dash, S. , Campbell, J.D., Cannon, E.K.; et al. Legume information system (LegumeInfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Res 2016, 44, D1181–1188. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N. , Menda, N., Edwards, J.D.; et al. The Sol Genomics Network (SGN)--from genotype to phenotype to breeding. Nucleic Acids Res 2015, 43, D1036–1041. [Google Scholar] [CrossRef]
- Foerster, H. , Bombarely, A., Battey, J.N.D.; et al. SolCyc: a database hub at the Sol Genomics Network (SGN) for the manual curation of metabolic networks in Solanum and Nicotiana specific databases. Database (Oxford) 2018, 2018. [Google Scholar] [CrossRef]
- Lawrence, C.J. MaizeGDB. Methods Mol Biol 2007, 406, 331–345. [Google Scholar]
- Portwood, J.L. , 2nd, Woodhouse, M.R., Cannon, E.K.; et al. MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Res 2019, 47, D1146–D1154. [Google Scholar] [CrossRef]
- Wegrzyn, J.L. , Lee, J.M., Tearse, B.R.; et al. TreeGenes: A forest tree genome database. Int J Plant Genomics 2008, 2008, 412875. [Google Scholar] [CrossRef] [PubMed]
- Falk, T. , Herndon, N., Grau, E.; et al. Growing and cultivating the forest genomics database, TreeGenes. Database (Oxford). 2019, 2019..
- Garcia-Hernandez, M. , Berardini, T.Z., Chen, G.; et al. TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomics 2002, 2, 239–253. [Google Scholar] [CrossRef]
- Poole, R.L. The TAIR database. Methods Mol Biol 2007, 406, 179–212. [Google Scholar] [PubMed]
- Sanderson, L.A. , Caron, C.T., Tan, R.; et al. KnowPulse: A Web-Resource Focused on Diversity Data for Pulse Crop Improvement. Front Plant Sci 2019, 10, 965. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.N. , Aleksic, J., Butano, D.; et al. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics 2012, 28, 3163–3165. [Google Scholar] [CrossRef] [PubMed]
- Kalderimis, A. , Lyne, R., Butano, D.; et al. InterMine: extensive web services for modern biology. Nucleic Acids Res 2014, 42, W468–472. [Google Scholar] [CrossRef] [PubMed]
- Tello-Ruiz, M.K. , Jaiswal, P., Ware, D. Gramene: A Resource for Comparative Analysis of Plants Genomes and Pathways. Methods Mol Biol 2022, 2443, 101–131. [Google Scholar]
- Ware, D.H. , Jaiswal, P., Ni, J.; et al. Gramene, a tool for grass genomics. Plant Physiol 2002, 130, 1606–1613. [Google Scholar] [CrossRef]
- Ware, D. Gramene. Methods Mol Biol 2007, 406, 315–329. [Google Scholar] [PubMed]
- Gladman, N. , Olson, A., Wei, S.; et al. SorghumBase: a web-based portal for sorghum genetic information and community advancement. Planta 2022, 255, 35. [Google Scholar] [CrossRef] [PubMed]
- Lyne, R. , Sullivan, J., Butano, D.; et al. Cross-organism analysis using InterMine. Genesis 2015, 53, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Fasoula, D.A. , Ioannides, I.M., Omirou, M. Phenotyping and Plant Breeding: Overcoming the Barriers. Front Plant Sci 2019, 10, 1713. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K. , Kurotani, A., Iida, K.; et al. RARGE II: an integrated phenotype database of Arabidopsis mutant traits using a controlled vocabulary. Plant Cell Physiol 2014, 55, e4. [Google Scholar] [CrossRef] [PubMed]
- Miroslaw, M. Officially Released Mutant Varieties – The FAO/IAEA Database. Plant Cell, Tissue and Organ Culture 2001, 65, 175–177. [Google Scholar]
- Zheng, Y. , Zhang, N., Martin, G.B.; et al. Plant Genome Editing Database (PGED): A Call for Submission of Information about Genome-Edited Plant Mutants. Mol Plant 2019, 12, 127–129. [Google Scholar] [CrossRef]
- Shikata, M. , Hoshikawa, K., Ariizumi, T.; et al. TOMATOMA Update: Phenotypic and Metabolite Information in the Micro-Tom Mutant Resource. Plant Cell Physiol 2016, 57, e11. [Google Scholar] [CrossRef]
- Li, M. , Xia, L., Zhang, Y.; et al. Plant editosome database: a curated database of RNA editosome in plants. Nucleic Acids Res 2019, 47, D170–D174. [Google Scholar] [CrossRef]
- McGill, B.J. , Enquist, B.J., Weiher, E.; et al. Rebuilding community ecology from functional traits. Trends Ecol Evol 2006, 21, 178–185. [Google Scholar] [CrossRef]
- Violle, V. , Navas, M., Vile, D.; et al. Let the concept of trait be functional! Oikos 2007, 116, 882–892. [Google Scholar] [CrossRef]
- Schneider, F.D. , Fichtmueller, D., Gossner, M.M.; et al. Towards an ecological trait-data standard. Methods in Ecology and Evolution 2019, 10, 2006–2019. [Google Scholar] [CrossRef]
- Allan, E. , Manning, P., Alt, F.; et al. Land use intensification alters ecosystem multifunctionality via loss of biodiversity and changes to functional composition. Ecol Lett 2015, 18, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Diaz, S. , Quetier, F., Caceres, D.M.; et al. Linking functional diversity and social actor strategies in a framework for interdisciplinary analysis of nature's benefits to society. Proc Natl Acad Sci U S A 2011, 108, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Lavorel, S. , Grigulis, K. How fundamental plant functional trait relationships scale-up to trade-offs and synergies in ecosystem services. Journal of ecology. 2012, 100, 128-140..
- Ni, J. , Pujar, A., Youens-Clark, K.; et al. Gramene QTL database: development, content and applications. Database (Oxford) 2009, 2009; bap005. [Google Scholar]
- Singh, K. , Batra, R., Sharma, S.; et al. WheatQTLdb: a QTL database for wheat. Mol Genet Genomics 2021, 296, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.B. , Wright, I.J., Lusk, C.H. Predicting leaf physiology from simple plant and climate attributes: a global GLOPNET analysis. Ecol Appl 2007, 17, 1982–1988. [Google Scholar] [CrossRef]
- Kissling, W.D. , Walls, R., Bowser, A.; et al. Towards global data products of Essential Biodiversity Variables on species traits. Nat Ecol Evol 2018, 2, 1531–1540. [Google Scholar] [CrossRef]
- Peat, H.J. , Fitter, A.H. A comparative study of the distribution and density of stomata in the British flora. Biol J Linn Soc Lond 1994, 52, 377–393. [Google Scholar] [CrossRef]
- Poschlod, P. , Kleyer, M., Jackel, A.-K.; et al. BIOPOP — A database of plant traits and internet application for nature conservation. Folia Geobotanica 2003, 38, 263–271. [Google Scholar] [CrossRef]
- Garcia-Recio, A. , Santos-Gomez, A., Soto, D.; et al. GRIN database: A unified and manually curated repertoire of GRIN variants. Hum Mutat 2021, 42, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Kühn, I. , Walter Durka, Klotz, S. BiolFlor: a new plant-trait database as a tool for plant invasion ecology. Diversity and Distributions 2004, 10, 363–365. [Google Scholar]
- Kleyer, M. , Bekker, R.M., Knevel, I.C.; et al. The LEDA Traitbase: a database of life-history traits of the Northwest European flora. Journal of ecology 2008, 96, 1266–1274. [Google Scholar] [CrossRef]
- Tavsanoglu, C. , Pausas, J.G. A functional trait database for Mediterranean Basin plants. Sci Data 2018, 5, 180135. [Google Scholar] [CrossRef] [PubMed]
- Falster, D. , Gallagher, R., Wenk, E.H.; et al. AusTraits, a curated plant trait database for the Australian flora. Sci Data 2021, 8, 254. [Google Scholar] [CrossRef] [PubMed]
- Houle, D. , Govindaraju, D.R., Omholt, S. Phenomics: the next challenge. Nat Rev Genet 2010, 11, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Hati, A.J. , Singh, R.R. Artificial Intelligence in Smart Farms: Plant Phenotyping for Species Recognition and Health Condition Identification Using Deep Learning. AI 2021, 2, 274–289. [Google Scholar] [CrossRef]
- Saleem, M.H. , Potgieter, J., Mahmood Arif, K. Plant Disease Detection and Classification by Deep Learning. Plants (Basel) 2019, 8. [Google Scholar]
- Zhang, C. , Zhou, L., Xiao, Q.; et al. End-to-End Fusion of Hyperspectral and Chlorophyll Fluorescence Imaging to Identify Rice Stresses. Plant Phenomics 2022, 2022, 9851096. [Google Scholar] [CrossRef]
- Sandhu, K.S. , Mihalyov, P.D., Lewien, M.J.; et al. Combining Genomic and Phenomic Information for Predicting Grain Protein Content and Grain Yield in Spring Wheat. Front Plant Sci 2021, 12, 613300. [Google Scholar] [CrossRef]
- Araus, J.L. , Kefauver, S.C., Zaman-Allah, M.; et al. Translating High-Throughput Phenotyping into Genetic Gain. Trends Plant Sci 2018, 23, 451–466. [Google Scholar] [CrossRef]
- Steinbach, D. , Alaux, M., Amselem, J.; et al. GnpIS: an information system to integrate genetic and genomic data from plants and fungi. Database (Oxford) 2013, 2013, bat058. [Google Scholar]
- Pommier, C. , Michotey, C., Cornut, G.; et al. Applying FAIR Principles to Plant Phenotypic Data Management in GnpIS. Plant Phenomics 2019, 2019, 1671403. [Google Scholar] [CrossRef] [PubMed]
- Brookes, A.J. , Robinson, P.N. Human genotype-phenotype databases: aims, challenges and opportunities. Nat Rev Genet 2015, 16, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Cobo-Simón, I. (2022) Cartograplant: Cyberinfrastructure to Improve Forest Health and Productivity in the Context of a Changing Climate. Plant and Animal Genome XXIX Conference. San Diego (CA).
- Sansone, S.A. , McQuilton, P., Rocca-Serra, P.; et al. FAIRsharing as a community approach to standards, repositories and policies. Nat Biotechnol 2019, 37, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Bulow, L. , Schindler, M., Choi, C.; et al. PathoPlant: a database on plant-pathogen interactions. In Silico Biol 2004, 4, 529–536. [Google Scholar] [PubMed]
- Bulow, L. , Schindler, M., Hehl, R. PathoPlant: a platform for microarray expression data to analyze co-regulated genes involved in plant defense responses. Nucleic Acids Res 2007, 35, D841–845. [Google Scholar] [CrossRef] [PubMed]
- Wu, W. , Wu, Y., Hu, D.; et al. PncStress: a manually curated database of experimentally validated stress-responsive non-coding RNAs in plants. Database (Oxford) 2020, 2020. [Google Scholar] [CrossRef]
- Global Burden of Disease Cancer, C. , Fitzmaurice, C., Abate, D.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2019, 5, 1749–1768. [Google Scholar]
- Dhondt, S. , Wuyts, N., Inze, D. Cell to whole-plant phenotyping: the best is yet to come. Trends Plant Sci 2013, 18, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Diaz, B.P. , Knowles, B., Johns, C.T.; et al. Seasonal mixed layer depth shapes phytoplankton physiology, viral production, and accumulation in the North Atlantic. Nat Commun 2021, 12, 6634. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.P. , D'Eustachio, P., Berardini, T.Z.; et al. Modeling biochemical pathways in the gene ontology. Database (Oxford) 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Poux, S. , Gaudet, P. Best Practices in Manual Annotation with the Gene Ontology. Methods Mol Biol 2017, 1446, 41–54. [Google Scholar] [PubMed]
- Chibucos, M.C. , Tyler, B.M. Common themes in nutrient acquisition by plant symbiotic microbes, described by the Gene Ontology. BMC Microbiol 2009, 9 (Suppl. 1). S6. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.E. , Geniza, M., Hanumappa, M.; et al. De novo transcriptome assembly and analyses of gene expression during photomorphogenesis in diploid wheat Triticum monococcum. PLoS One 2014, 9, e96855. [Google Scholar] [CrossRef] [PubMed]
- Vining, K.J. , Romanel, E., Jones, R.C.; et al. The floral transcriptome of Eucalyptus grandis. New Phytol 2015, 206, 1406–1422. [Google Scholar] [CrossRef] [PubMed]
- Fennell, A.Y. , Schlauch, K.A., Gouthu, S.; et al. Short day transcriptomic programming during induction of dormancy in grapevine. Front Plant Sci 2015, 6, 834. [Google Scholar] [CrossRef]
- Gupta, P. , Geniza, M., Naithani, S.; et al. Chia (Salvia hispanica) Gene Expression Atlas Elucidates Dynamic Spatio-Temporal Changes Associated With Plant Growth and Development. Front Plant Sci 2021, 12, 667678. [Google Scholar] [CrossRef]
- Godoy, F. , Kuhn, N., Munoz, M.; et al. The role of auxin during early berry development in grapevine as revealed by transcript profiling from pollination to fruit set. Hortic Res 2021, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y. , Xu, Q.W., Wang, R.; et al. PRIDE Inspector Toolsuite: Moving Toward a Universal Visualization Tool for Proteomics Data Standard Formats and Quality Assessment of ProteomeXchange Datasets. Mol Cell Proteomics 2016, 15, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Kosova, K. , Vitamvas, P., Urban, M.O.; et al. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front Plant Sci 2018, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Jarnuczak, A.F. , Vizcaino, J.A. Using the PRIDE Database and ProteomeXchange for Submitting and Accessing Public Proteomics Datasets. Curr Protoc Bioinformatics 2017, 59, 13–31, 11-13 31 12. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S. , Watanabe, Y., Moriya, Y.; et al. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res 2017, 45, D1107–D1111. [Google Scholar] [CrossRef]
- Moriya, Y. , Kawano, S., Okuda, S.; et al. The jPOST environment: an integrated proteomics data repository and database. Nucleic Acids Res 2019, 47, D1218–D1224. [Google Scholar] [CrossRef]
- Chen, T. , Ma, J., Liu, Y.; et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res 2022, 50, D1522–D1527. [Google Scholar] [CrossRef]
- Ma, J. , Chen, T., Wu, S.; et al. iProX: an integrated proteome resource. Nucleic Acids Res 2019, 47, D1211–D1217. [Google Scholar] [CrossRef]
- Sharma, V. , Eckels, J., Taylor, G.K.; et al. Panorama: a targeted proteomics knowledge base. J Proteome Res 2014, 13, 4205–4210. [Google Scholar] [CrossRef]
- Desiere, F. , Deutsch, E.W., King, N.L.; et al. The PeptideAtlas project. Nucleic Acids Res 2006, 34, D655–658. [Google Scholar] [CrossRef]
- Deutsch, E.W. The PeptideAtlas Project. Methods Mol Biol 2010, 604, 285–296. [Google Scholar]
- Tsugawa, H. , Rai, A., Saito, K.; et al. Metabolomics and complementary techniques to investigate the plant phytochemical cosmos. Nat Prod Rep 2021, 38, 1729–1759. [Google Scholar] [CrossRef]
- Members, M.S.I.B. , Sansone, S.A., Fan, T.; et al. The metabolomics standards initiative. Nat Biotechnol 2007, 25, 846–848. [Google Scholar] [CrossRef]
- Sumner, L.W. , Amberg, A., Barrett, D.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Vinaixa, M. , Schymanski, E.L., Neumann, S.; et al. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends in Analytical Chemistry 2016, 78, 23–35. [Google Scholar] [CrossRef]
- Salek, R.M. , Neumann, S., Schober, D.; et al. COordination of Standards in MetabOlomicS (COSMOS): facilitating integrated metabolomics data access. Metabolomics 2015, 11, 1587–1597. [Google Scholar] [CrossRef]
- Steinbeck, C. , Conesa, P., Haug, K.; et al. MetaboLights: towards a new COSMOS of metabolomics data management. Metabolomics 2012, 8, 757–760. [Google Scholar] [CrossRef]
- Considine, E.C. , Salek, R.M. A Tool to Encourage Minimum Reporting Guideline Uptake for Data Analysis in Metabolomics. M. A Tool to Encourage Minimum Reporting Guideline Uptake for Data Analysis in Metabolomics. Metabolites 2019, 9. [Google Scholar]
- Schorn, M.A. , Verhoeven, S., Ridder, L.; et al. A community resource for paired genomic and metabolomic data mining. Nat Chem Biol 2021, 17, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L. , Jaiswal, P. The Plant Ontology: A Tool for Plant Genomics. Methods Mol Biol 2016, 1374, 89–114. [Google Scholar] [PubMed]
- Cooper, L. , Walls, R.L., Elser, J.; et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol 2013, 54, e1. [Google Scholar] [CrossRef] [PubMed]
- Avraham, S. , Tung, C.W., Ilic, K.; et al. The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res 2008, 36, D449–454. [Google Scholar] [CrossRef] [PubMed]
- Warman, C. , Sullivan, C.M., Preece, J.; et al. A cost-effective maize ear phenotyping platform enables rapid categorization and quantification of kernels. Plant J 2021, 106, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Oellrich, A. , Walls, R.L., Cannon, E.K.; et al. An ontology approach to comparative phenomics in plants. Plant Methods 2015, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L. , Meier, A., Laporte, M.A.; et al. The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics. Nucleic Acids Res 2018, 46, D1168–D1180. [Google Scholar] [CrossRef]
- Tello-Ruiz, M.K. , Naithani, S., Gupta, P.; et al. Gramene 2021: harnessing the power of comparative genomics and pathways for plant research. Nucleic Acids Res 2021, 49, D1452–D1463. [Google Scholar] [CrossRef]
- Naithani, S. , Partipilo, C.M., Raja, R.; et al. FragariaCyc: A Metabolic Pathway Database for Woodland Strawberry Fragaria vesca. Front Plant Sci 2016, 7, 242. [Google Scholar] [CrossRef]
- Naithani, S. , Raja, R., Waddell, E.N.; et al. VitisCyc: a metabolic pathway knowledgebase for grapevine (Vitis vinifera). Front Plant Sci 2014, 5, 644. [Google Scholar] [CrossRef]
- Gupta, P. , Naithani, S., Preece, J.; et al. Plant Reactome and PubChem: The Plant Pathway and (Bio)Chemical Entity Knowledgebases. Methods Mol Biol 2022, 2443, 511–525. [Google Scholar]
- Naithani, S. , Gupta, P., Preece, J.; et al. Plant Reactome: a knowledgebase and resource for comparative pathway analysis. Nucleic Acids Res 2020, 48, D1093–D1103. [Google Scholar]
- Jaiswal, P. , Usadel, B. Plant Pathway Databases. Methods Mol Biol 2016, 1374, 71–87. [Google Scholar]
- Naithani, S. , Jaiswal, P. Pathway Analysis and Omics Data Visualization Using Pathway Genome Databases: FragariaCyc, a Case Study. Methods Mol Biol 2017, 1533, 241–256. [Google Scholar]
- Kattge, J. , Ogle, K., Bönisch, G.; et al. A generic structure for plant trait databases. Methods in Ecology and Evolution 2011, 2, 202–213. [Google Scholar] [CrossRef]
- Kattge, J. , Bonisch, G., Diaz, S.; et al. TRY plant trait database - enhanced coverage and open access. Glob Chang Biol 2020, 26, 119–188. [Google Scholar] [CrossRef] [PubMed]
- van Kleunen, M. , Pysek, P., Dawson, W.; et al. The Global Naturalized Alien Flora (GloNAF) database. Ecology 2019, 100, e02542. [Google Scholar] [CrossRef]
- Lee, Y.H. Meta-analysis of genetic association studies. Ann Lab Med 2015, 35, 283–287. [Google Scholar] [CrossRef]
- Dehghan, A. Genome-Wide Association Studies. Methods Mol Biol 2018, 1793, 37–49. [Google Scholar] [PubMed]
- Khan, S.U. , Saeed, S., Khan, M.H.U.; et al. Advances and Challenges for QTL Analysis and GWAS in the Plant-Breeding of High-Yielding: A Focus on Rapeseed. Biomolecules 2021, 11. [Google Scholar] [CrossRef]
- Buniello, A. , MacArthur, J.A.L., Cerezo, M.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Togninalli, M. , Seren, U., Freudenthal, J.A.; et al. AraPheno and the AraGWAS Catalog 2020: a major database update including RNA-Seq and knockout mutation data for Arabidopsis thaliana. Nucleic Acids Res 2020, 48, D1063–D1068. [Google Scholar] [PubMed]
- Kraft, P. , Zeggini, E., Ioannidis, J.P. Replication in genome-wide association studies. Stat Sci 2009, 24, 561–573. [Google Scholar] [CrossRef]
- Pinu, F.R. , Beale, D.J., Paten, A.M.; et al. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9. [Google Scholar] [CrossRef]
- Sumner, L.W. , Styczynski, M., McLean, J.; et al. Introducing the USA Plant, Algae and Microbial Metabolomics Research Coordination Network (PAMM-NET). Metabolomics 2015, 11, 3–5. [Google Scholar] [CrossRef]
- Kodra, D. , Pousinis, P., Vorkas, P.A.; et al. Is Current Practice Adhering to Guidelines Proposed for Metabolite Identification in LC-MS Untargeted Metabolomics? A Meta-Analysis of the Literature. J Proteome Res 2022, 21, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M. , Meyer, S.W., Heyman, H.M.; et al. Generation of a Collision Cross Section Library for Multi-Dimensional Plant Metabolomics Using UHPLC-Trapped Ion Mobility-MS/MS. Metabolites 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Jeliazkova, N. , Apostolova, M.D., Andreoli, C.; et al. Towards FAIR nanosafety data. Nat Nanotechnol 2021, 16, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.R. , Pauvert, C., Kishore, D.; et al. Toward FAIR Representations of Microbial Interactions. mSystems 2022, 7, e0065922. [Google Scholar] [CrossRef] [PubMed]
- Iturbide, M. , Fernandez, J., Gutierrez, J.M.; et al. Implementation of FAIR principles in the IPCC: the WGI AR6 Atlas repository. Sci Data 2022, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Mons, B. , Neylon, C., Velterop, J.; et al. Cloudy, increasingly FAIR; revisiting the FAIR Data guiding principles for the European Open Science Cloud. Information Services & Use 2017, 37, 49–56. [Google Scholar]
| Database name | NCBI | DRA | ENA | GSA | IBDC | AGDR † | DRYAD ¥ | Zenodo ‡, ¥ | FigShare |
| Genome sequence data | + | + | + | + | + | + | + | + | + |
| WGS annotations | + | ? | ? | ? | ? | ? | ? | ? | + |
| Genotyping data | + | ? | ? | ? | ? | ? | ? | ? | + |
| Transcriptome sequence data | + | + | + | ? | ? | ? | + | + | + |
| fq.gz | + | + | + | + | + | + | + | + | + |
| BAM | + | + | + | + | + | + | + | + | + |
| SFF | + | + | + | + | + | - | + | + | + |
| HDF | + | + | + | + | + | - | + | + | + |
| VCF | + | + | + | ? | ? | ? | + | + | + |
| INSDC-Source | + | + | + | a | b | c | d | e | f |
| Species/Crop | Database | Database URL |
|---|---|---|
| Arabidopsis | TAIR | https://www.arabidopsis.org/ |
| Cassava | CassavaBase | https://www.cassavabase.org/ |
| Citrus | Citrus Genome Database | https://www.citrusgenomedb.org/ |
| Citrus/Diaphorina citri/Ca. Liberibacter asiaticus | Citrus Greening | https://www.citrusgreening.org/ |
| Cotton | CottonGen | https://www.cottongen.org/ |
| Cucurbit | Cucurbit Genomics | http://cucurbitgenomics.org/ |
| Forest trees | TreeGenes | https://treegenesdb.org |
| Hardwood Genomics | http://www.hardwoodgenomics.org/ | |
| Grains | GrainGenes | https://wheat.pw.usda.gov |
| Gramene | https://www.gramene.org/ | |
| SorghumBase | https://www.sorghumbase.org/ | |
| Triticeae toolbox, T3 | https://wheat.triticeaetoolbox.org/ | |
| WheatIS | https://wheatis.org | |
| KitBase | http://kitbase.ucdavis.edu/ | |
| Legumes | KnowPulse | https://knowpulse.usask.ca/ |
| Legume Information System | https://www.legumeinfo.org/ | |
| PeanutBase | https://peanutbase.org | |
| Pulses | Pulse Crop Database | https://www.pulsedb.org/ |
| Soybase | https://www.soybase.org/ | |
| Maize | MaizeGDB | https://maizegdb.org/ |
| Musa | MusaBase | https://www.musabase.org/ |
| Rosaceae | Genome Database for Rosaceae | https://www.rosaceae.org/ |
| Solanaceae | Sol Genomics | https://solgenomics.net/ |
| Sweet Potato | SweetPotatoBase | https://www.sweetpotatobase.org/ |
| Vaccinium | Genome Database for Vaccinium | https://www.vaccinium.org/ |
| Yam | YamBase | https://www.yambase.org/ |
| Comparative genomic database used by multiple communities | ||
| A comparative genomic database for ~300 plant species | Phytozome | https://phytozome-next.jgi.doe.gov/ |
| A comparative genomic databasehosting 118 genomesfrom models, crops,fruits, vegetables, etc. | Gramene | https://www.gramene.org/ |
| Others | AgBase | https://agbase.arizona.edu/ |
| Bio-Analytic Resource | https://bar.utoronto.ca/ | |
| Category | Databases | URLs |
|---|---|---|
|
Species-specific mutant collections |
Database of image and genome (MaizeDIG) |
https://maizedig.maizegdb.org/ |
| Mutant Variety Database | https://nucleus.iaea.org/sites/mvd/SitePages/Home.aspx | |
| Plant Genome Editing Database | http://plantcrispr.org/cgi-bin/crispr/index.cgi | |
| RIKEN Arabidopsis Genome Encyclopedia (RARGE) |
http://rarge-v2.psc.riken.jp/line | |
| TOMATOMA | https://tomatoma.nbrp.jp/index.jsp | |
| Plant Editosome | https://ngdc.cncb.ac.cn/ped/ | |
| Traits and QTL | Gramene QTL | https://archive.gramene.org/qtl/ |
| Wheatqtl | http://www.wheatqtldb.net/ | |
| GLOPNET | http://bio.mq.edu.au/~iwright/glopian.htm | |
| TRY database | https://www.try-db.org/TryWeb/Home.php | |
| Ecological Flora of the Britain and Ireland |
http://ecoflora.org.uk/ | |
| BIOPOP | http://www.landeco.uni-oldenburg.de/Projects/biopop/main.htm | |
| FloraWeb | https://www.floraweb.de/ | |
| USDA GRIN | https://www.ars-grin.gov/ | |
| BiolFlor | https://wiki.ufz.de/biolflor/index.jsp | |
| LEDA | https://uol.de/en/landeco/research/leda | |
| USDA PLANTS | https://plants.usda.gov/home | |
| BROT | https://www.uv.es/jgpausas/brot.htm | |
| AusTraits | https://austraits.org/ | |
| Community Databases in Table 2 and Supplementary Table S3 |
||
| Phenomics | GnpIS | https://urgi.versailles.inra.fr/gnpis |
| PGP Repository | https://edal-pgp.ipk-gatersleben.de/ | |
| Cartograplant | https://cartograplant.org/ | |
| AgData commons Plants & Crops: |
https://data.nal.usda.gov/ag-data-commons-hierarchy/plants-crops | |
| PathoPlant | http://www.pathoplant.de/ | |
| PncStress | http://bis.zju.edu.cn/pncstress/ | |
| Indian Crop Phenome DB (ICPD) | https://ibdc.rcb.res.in/icpd/ | |
| Gene Expression | Ozone Stress Responsive Gene Database |
https://www.osrgd.com |
| EBI-Plant Expression Atlas | https://www.ebi.ac.uk/gxa/plant/experiments | |
| CoNeKT | https://conekt.sbs.ntu.edu.sg/ | |
|
Protein, peptides and proteomes |
Expath | http://expath.itps.ncku.edu.tw/ |
| Proteome Xchange | https://www.proteomexchange.org | |
| Plant Proteome Database | http://ppdb.tc.cornell.edu/ | |
| PlantMWpIDB | https://plantmwpidb.com/ | |
| Heat Shock Proteins database | http://hsfdb.bio2db.com/ | |
| WallProtDB | https://www.polebio.lrsv.ups-tlse.fr/WallProtDB/ | |
| Aramemnon | http://aramemnon.botanik.uni-koeln.de/ | |
| PhosPhAt | https://phosphat.uni-hohenheim.de/db.html | |
| Database of Phospho-sites in Plants |
http://dbppt.biocuckoo.org/browse.php | |
| Plant Protein Phosphorylation Database |
https://www.p3db.org/home | |
| qPTMplants | http://qptmplants.omicsbio.info/ | |
| Plant PTM viewer | https://www.psb.ugent.be/webtools/ptm-viewer/ | |
| PlaPPISite | http://zzdlab.com/plappisite/index.ph | |
| M. truncatula Small Secreted Peptide Database |
https://mtsspdb.zhaolab.org/database | |
| PlantPepDB | http://14.139.61.8/PlantPepDB/index.php | |
| Arabidopsis PeptideAtlas | http://www.peptideatlas.org/builds/arabidopsis/ | |
| Indian Structural Data Archive | https://isda.rcb.ac.in/ | |
|
Metabolites, biochemical, and small chemical entities |
Antimicrobial plant peptides (PhytAMP) |
http://phytamp.pfba-lab-tun.org/main.php |
| PubChem | https://pubchem.ncbi.nlm.nih.gov | |
| ChEBI | https://www.ebi.ac.uk/chebi | |
| Metabolomics Workbench | https://www.metabolomicsworkbench.org | |
|
Secondary Knowledgebase |
MetaboLights | https://www.ebi.ac.uk/metabolights/index |
| PoDP | https://pairedomicsdata.bioinformatics.nl/ | |
| Plant Reactome pathway knowledgebase |
https://plantreactome.gramene.org | |
| MetaCyc | https://metacyc.org | |
| PMN | https://plantcyc.org/data | |
| KEGG pathways | https://www.genome.jp/kegg/pathway.html | |
| PlantPathMarks (PPMdb) | http://ppmdb.easyomics.org/ | |
| The Bio-Analytic Resource (BAR) | https://bar.utoronto.ca | |
| The protein-protein interaction database for Maize (PPIM) |
https://mai.fudan.edu.cn/ppim/ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





