Submitted:
05 June 2023
Posted:
06 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
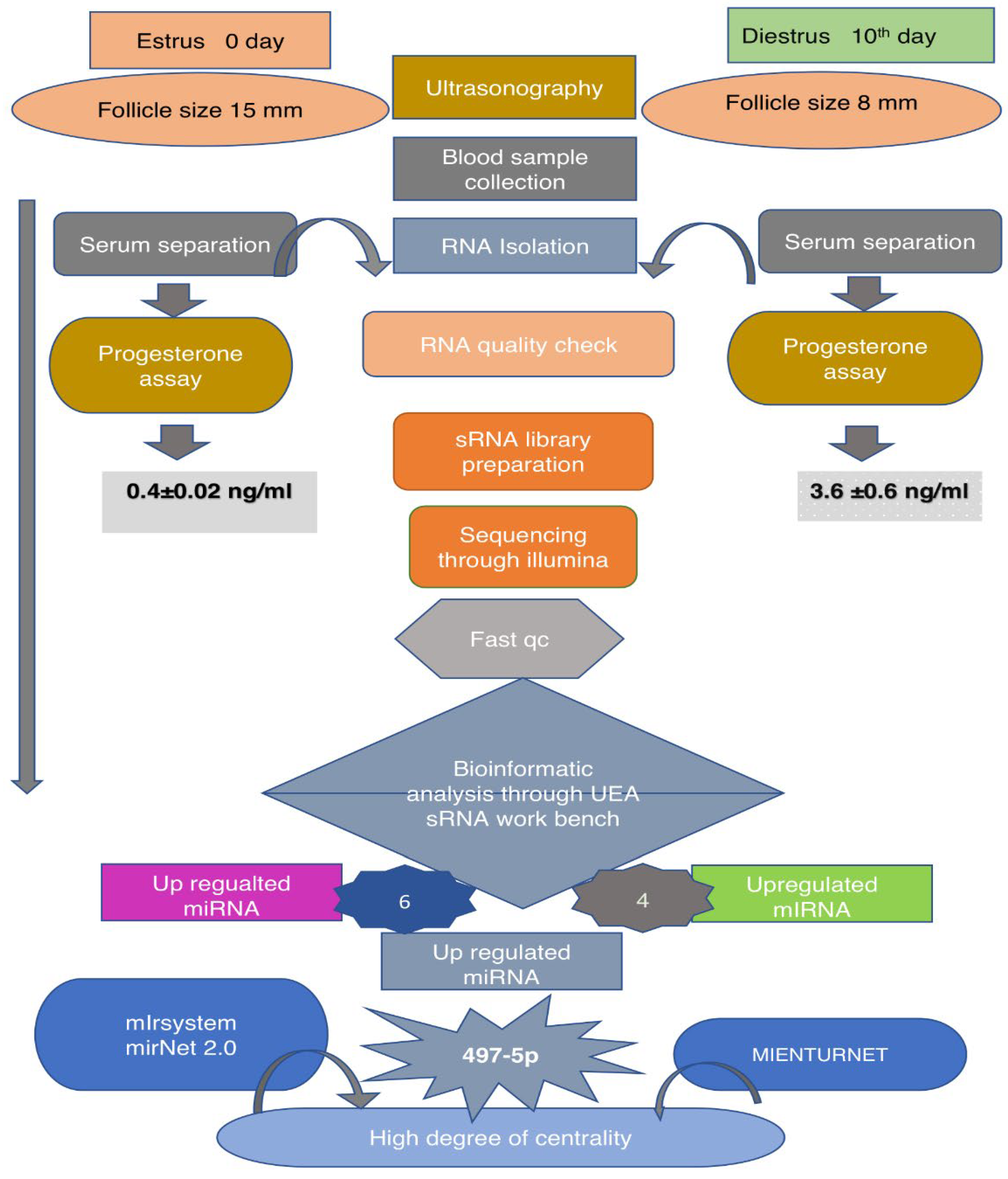
2. Results
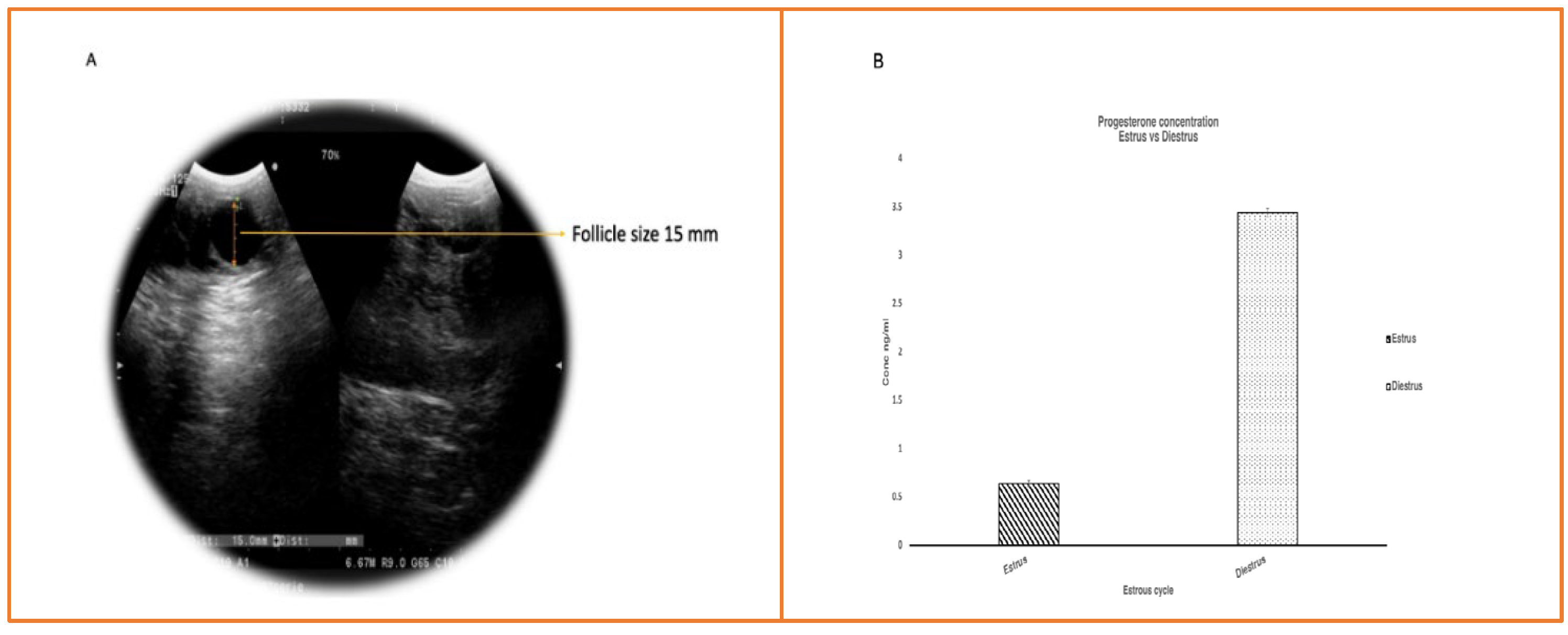
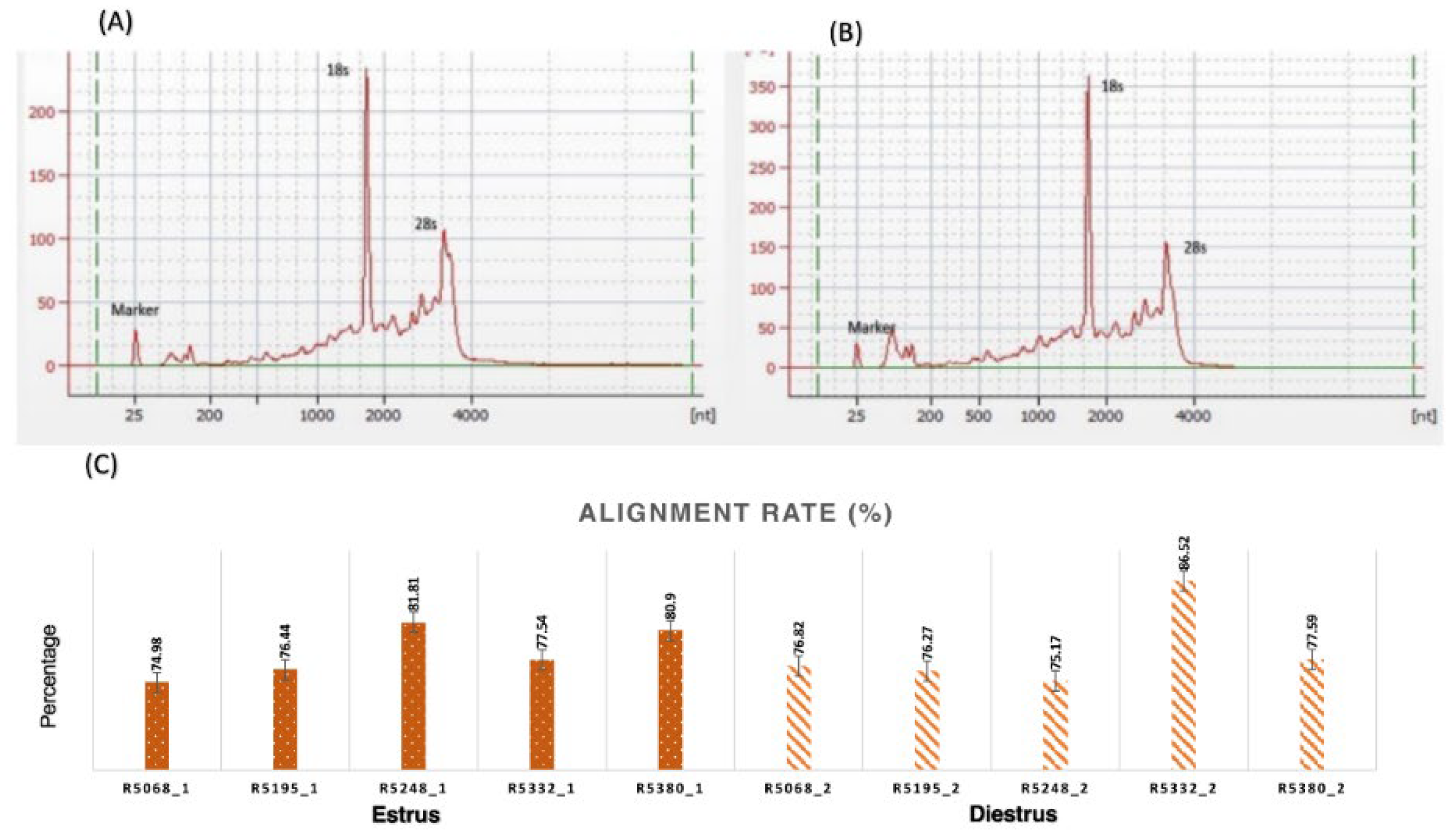
2.1. RNA quantity and quality
2.2. RNA Seq Data
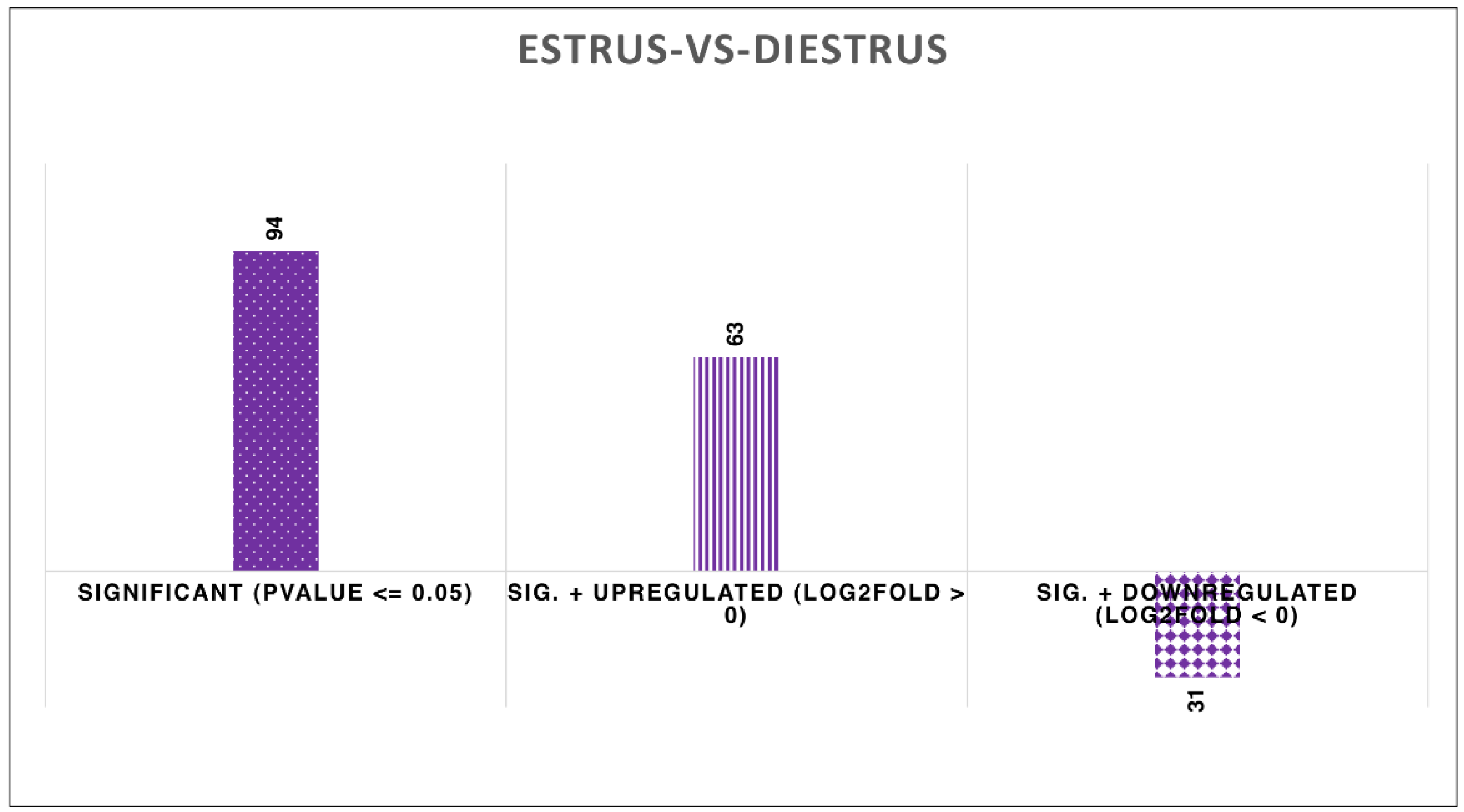
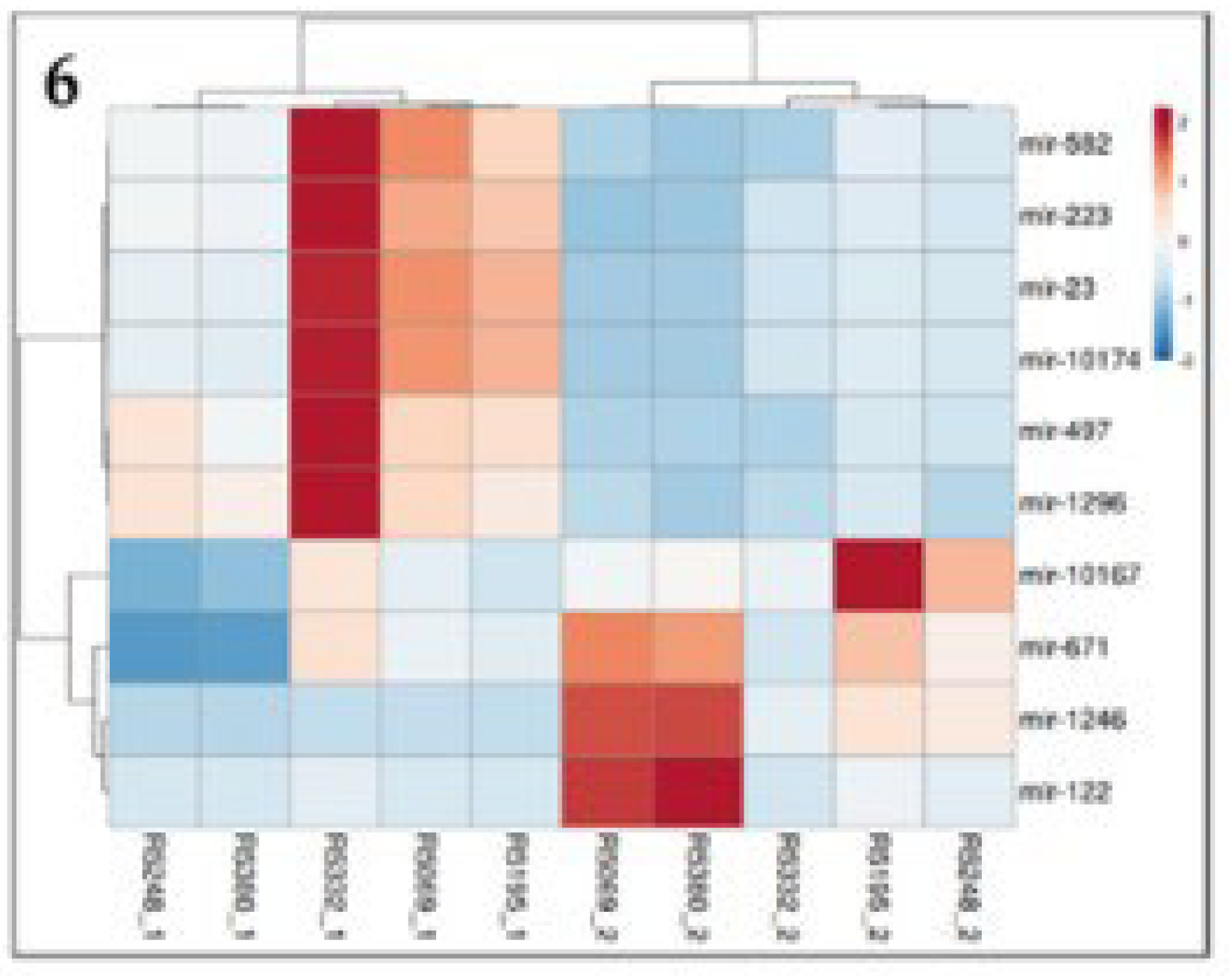
2.3. Identification of DEmiRNA
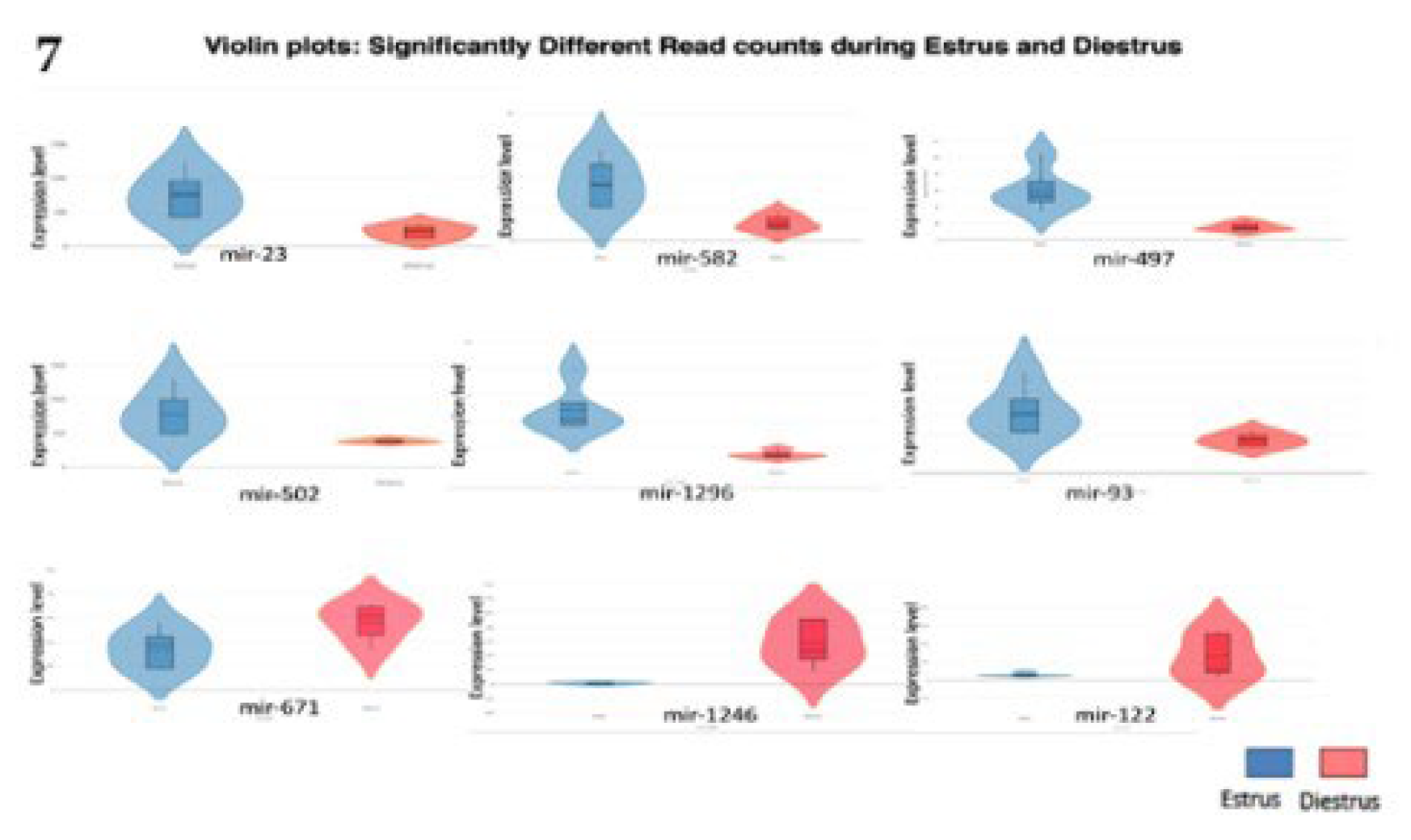
2.4. Top 10 Significantly different miRNA
2.5. Validation by qRT-PCR
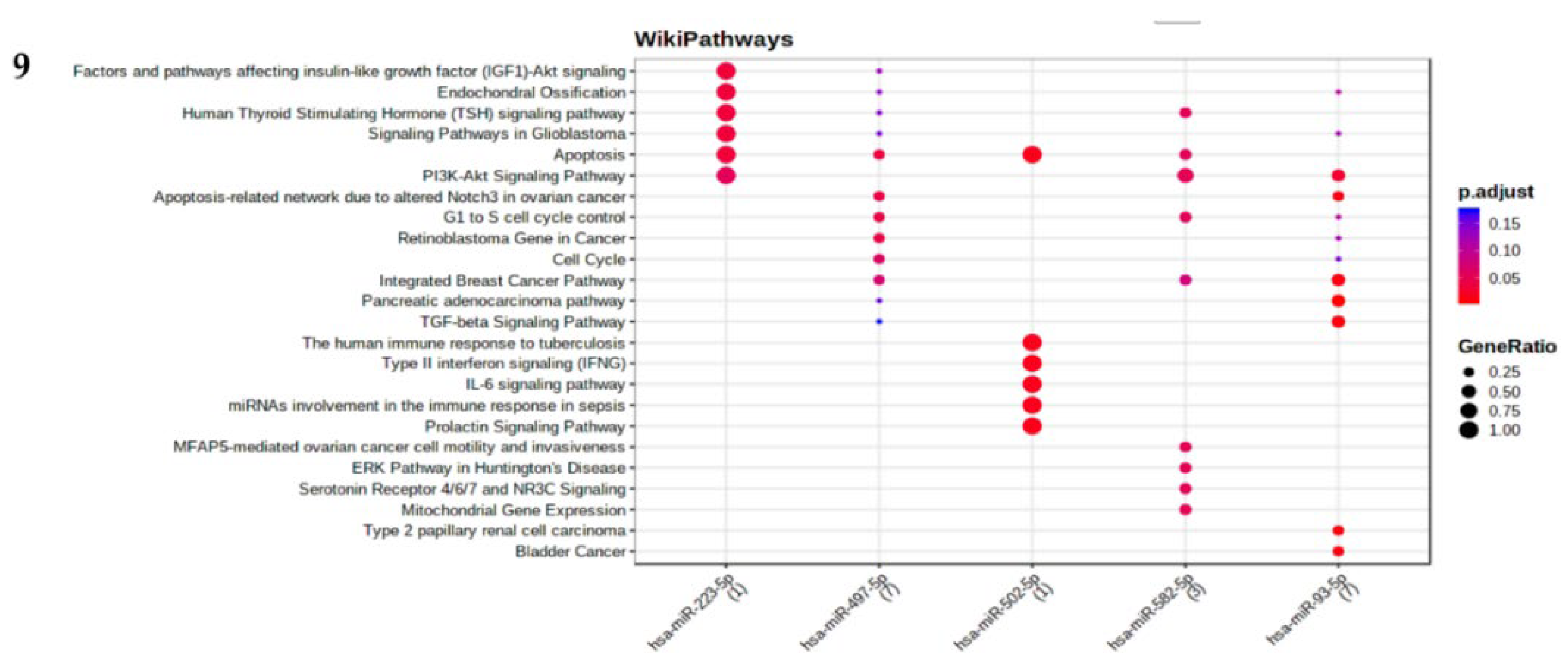
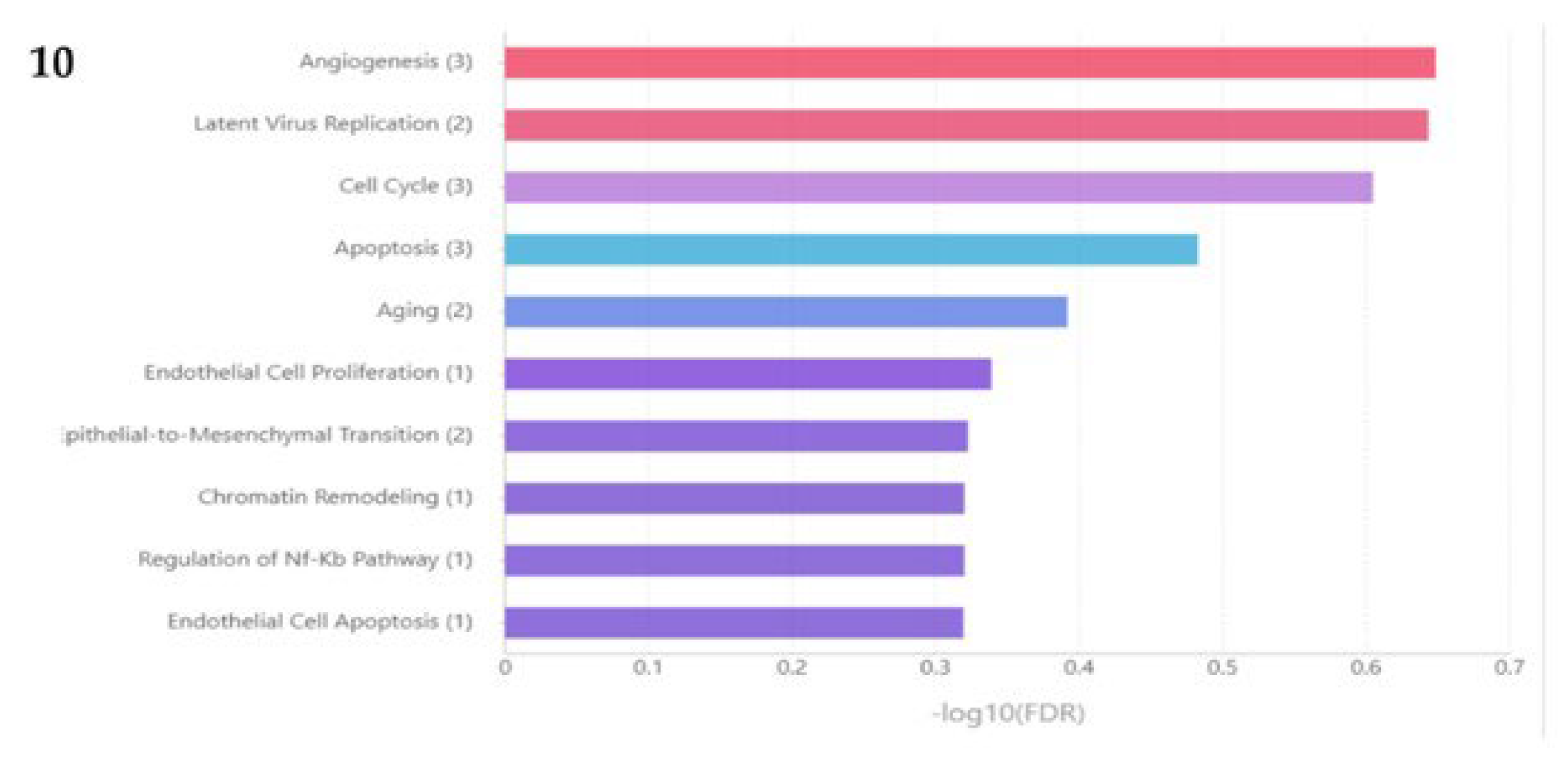
2.6. Functional and pathway enrichment analysis
| Category | Term | Total genes of the term |
Union_targets in the term |
miRs_in _the_term |
Score |
|---|---|---|---|---|---|
| Reactome | Axon_guidance | 266 | 78 | 5 | 4.298 |
| Reactome | Developmental_biology | 494 | 114 | 5 | 4.153 |
| KEGG | Pathways_in_cancer | 325 | 84 | 5 | 3.7 |
| Biocarta | Biocarta_mapk_pathway | 87 | 36 | 5 | 3.69 |
| Reactome | Signalling_by_ngf | 221 | 71 | 5 | 3.672 |
| KEGG | Neurotrophin_signaling_pathway | 127 | 40 | 5 | 3.334 |
| Reactome | Ngf_signalling | 136 | 46 | 5 | 3.304 |
| KEGG | MAPK_signaling_pathway | 272 | 73 | 5 | 3.254 |
| Pathway_interaction_ | Erbb1_downstream_signaling | 106 | 41 | 5 | 3.178 |
| KEGG | Insulin_signaling_pathway | 137 | 42 | 5 | 3.068 |
| KEGG | Wnt_signaling_pathway | 150 | 49 | 4 | 3.029 |
| KEGG | Axon_guidance | 129 | 43 | 5 | 2.996 |
| KEGG | Prostate_cancer | 89 | 30 | 5 | 2.798 |
| KEGG | Olfactory_transduction | 388 | 6 | 4 | 2.691 |
| Reactome | L1cam_interactions | 94 | 33 | 5 | 2.62 |
| KEGG | Erbb_signaling_pathway | 87 | 29 | 5 | 2.605 |
| KEGG | Endocytosis | 201 | 59 | 5 | 2.567 |
| PATHWAY_INTERACTION_DATABASE | Cdc42_signaling_events | 70 | 27 | 5 | 2.533 |
| KEGG | Progesterone-mediated_oocyte_maturation | 86 | 27 | 5 | 2.531 |
2.7. miRNA Target prediction and degree centrality
3. Discussion
4. Materials and Methods
4.1. Identification of Heifers
4.2. Ultrasonography confirmation
4.3. Collection of Blood Samples
4.4. Progesterone Assay Confirmation
4.5. RNA Isolation and Quality Assessment
4.6. Next generation sequencing
4.7. Differential expression analysis
4.8. Identification of known and novel microRNAs
4.9. Validation by qRT-PCR
4.10. Target genes and pathway prediction
4.11. Statistical analysis
5. Conclusions
Supplementary Materials
Author Contributions
Data Availability Statement
Acknowledgments
References
- Holman, A., J. Thompson, J. E. Routly, J. Cameron, D. N. Jones, D. Grove-White, R. F. Smith, and H. Dobson. Comparison of oestrus detection methods in dairy cattle. Veterinary Record. 2011, 169, 47-47. [CrossRef]
- Fricke PM, Carvalho PD, Giordano JO, Valenza A, Lopes G, Amundson MC. Expression and detection of estrus in dairy cows: the role of new technologies. Animal. 2014, 8, 134-43. [CrossRef]
- Ioannidis J, Donadeu FX. Circulating microRNA profiles during the bovine oestrous cycle. PloS one. 2016, 24, e0158160.
- Hussain MU. Micro-RNAs (miRNAs): genomic organisation, biogenesis and mode of action. Cell and tissue research. 2012, 349, 405-13. [CrossRef]
- Weber JA, Baxter DH, Zhang S, Huang DY, How Huang K, Jen Lee M, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clinical chemistry. 2010, 56, 1733-41. [CrossRef]
- Al-Muhtaresh HA, Al-Kafaji G. Evaluation of two-diabetes related microRNAs suitability as earlier blood biomarkers for detecting prediabetes and type 2 diabetes mellitus. Journal of clinical medicine. 2018, 26, 12. [CrossRef]
- Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs—an update. Nature reviews Clinical oncology. 2018, 15, 541-63. [CrossRef]
- Mondal S, Prakash BS, Palta P. Endocrine aspects of oestrous cycle in buffaloes (Bubalus bubalis): an overview. Asian-australasian journal of animal sciences. 2006, 27, 124-31. [CrossRef]
- Li J, Han X, Wan Y, Zhang S, Zhao Y, Fan R, Cui Q, Zhou Y. TAM 2.0: tool for MicroRNA set analysis. Nucleic acids research. 2018, 46, W180-5. [CrossRef]
- Licursi V, Conte F, Fiscon G, Paci P. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC bioinformatics. 2019, 20, 1-0.
- Singh P, Golla N, Singh P, Baddela VS, Chand S, Baithalu RK, Singh D, Onteru SK. Salivary miR-16, miR-191 and miR-223: intuitive indicators of dominant ovarian follicles in buffaloes. Molecular Genetics and Genomics. 2017, 292:935-53. [CrossRef]
- Surla GN, Kumar LK, Vedamurthy VG, Singh D, Onteru SK. Salivary TIMP1 and predicted mir-141, possible transcript biomarkers for estrus in the buffalo (Bubalus bubalis). Reproductive Biology. 2022, 22, 100641. [CrossRef]
- Hebbar A, Chandel R, Rani P, Onteru SK, Singh D. Urinary cell-free miR-99a-5p as a potential biomarker for estrus detection in buffalo. Frontiers in Veterinary Science. 2021, 17, 643910. [CrossRef]
- Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC, Chuang EY. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. 2012, e42390. [CrossRef]
- Dissen GA, Lara HE, Leyton V, Paredes A, Hill DF, Costa ME, Martinez-Serrano A, Ojeda SR. Intraovarian excess of nerve growth factor increases androgen secretion and disrupts estrous cyclicity in the rat. Endocrinology. 2000, 141, 1073-82. [CrossRef]
- Zavareh S, Gholizadeh Z, Lashkarbolouki T. Evaluation of changes in the expression of Wnt/β-catenin target genes in mouse reproductive tissues during estrous cycle: An experimental study. International Journal of Reproductive BioMedicine. 2018, 16, 69. [CrossRef]
- Kaczmarek MM, Krawczynski K, Najmula J, Reliszko ZP, Sikora M, Gajewski Z. Differential expression of genes linked to the leukemia inhibitor factor signaling pathway during the estrus cycle and early pregnancy in the porcine endometrium. Reproductive Biology. 2014, 14, 293-7. [CrossRef]
- Salilew-Wondim D, Ahmad I, Gebremedhn S, Sahadevan S, Hossain MM, Rings F, Hoelker M, Tholen E, Neuhoff C, Looft C, Schellander K. The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PloS one. 2014, 9, e106795. [CrossRef]
- Garg D, Ng SS, Baig KM, Driggers P, Segars J. Progesterone-mediated non-classical signaling. Trends in Endocrinology & Metabolism. 2017, 28, 656-68. [CrossRef]
- Migliaccio A, Piccolo D, Castoria G, Di Domenico M, Bilancio A, Lombardi M, Gong W, Beato M, Auricchio F. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. The EMBO journal. 1998, 17, 2008-18. [CrossRef]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics. 2012, 28, 2059-61. [CrossRef]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11:R106.
- Paicu C, Mohorianu I, Stocks M, Xu P, Coince A, Billmeier M, Dalmay T, Moulton V, Moxon S. miRCat2: accurate prediction of plant and animal microRNAs from next-generation sequencing datasets. Bioinformatics. 2017, 33, 2446-54. [CrossRef]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009, 4, 44-57. [CrossRef]
- Chang L, Xia J. MicroRNA Regulatory Network Analysis Using miRNet 2.0. Transcription Factor Regulatory Networks. 2022, 21, 185-204.

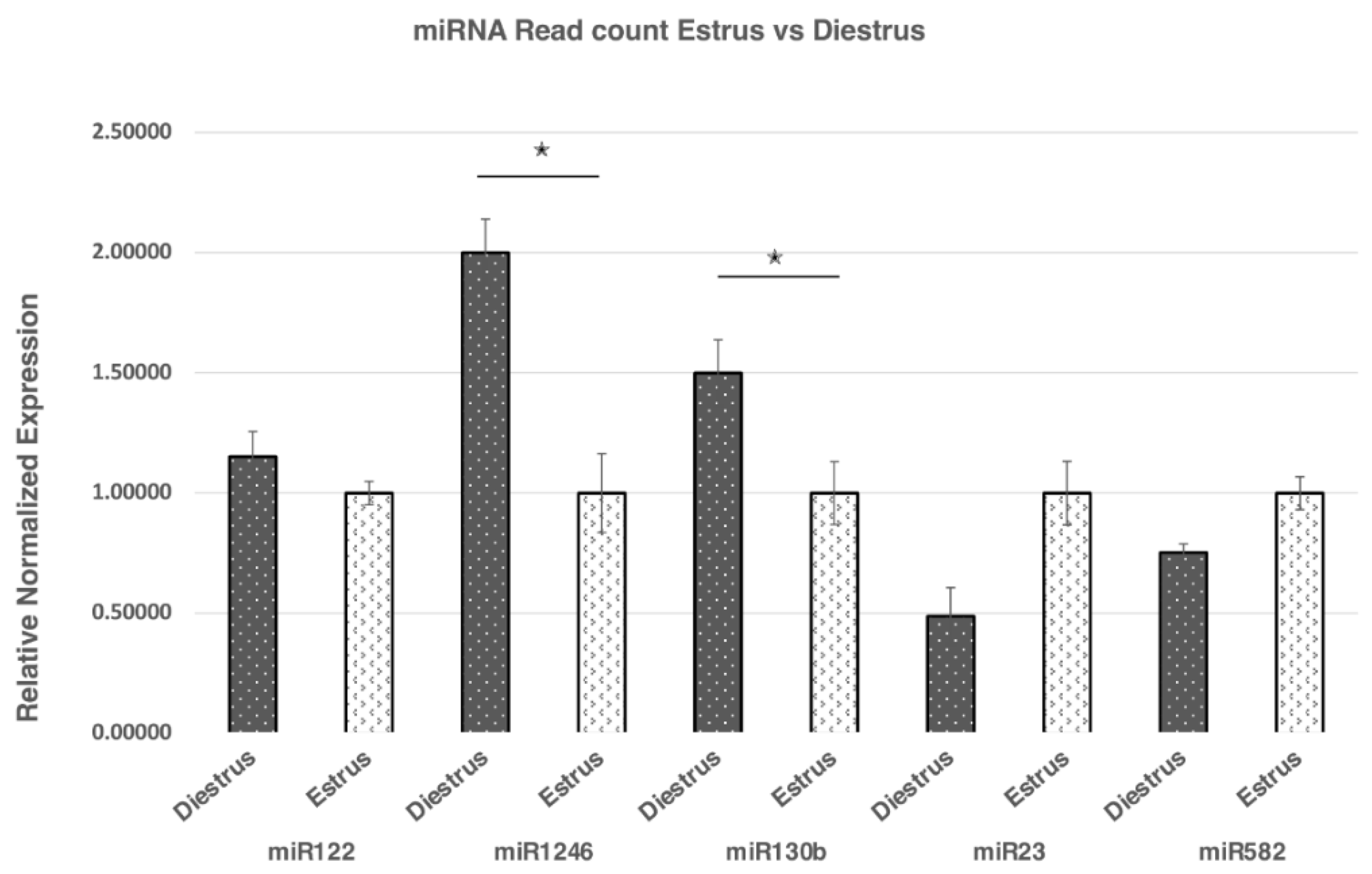
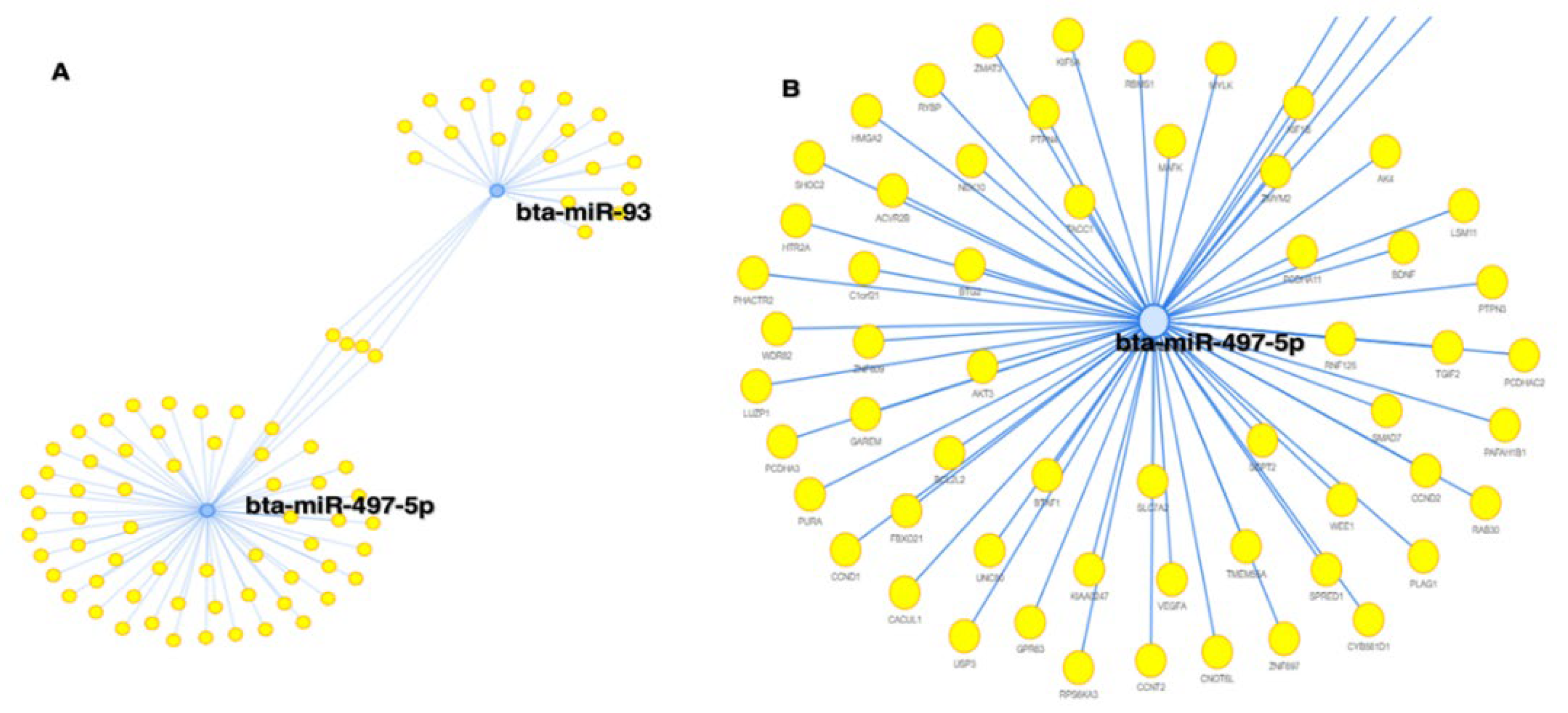
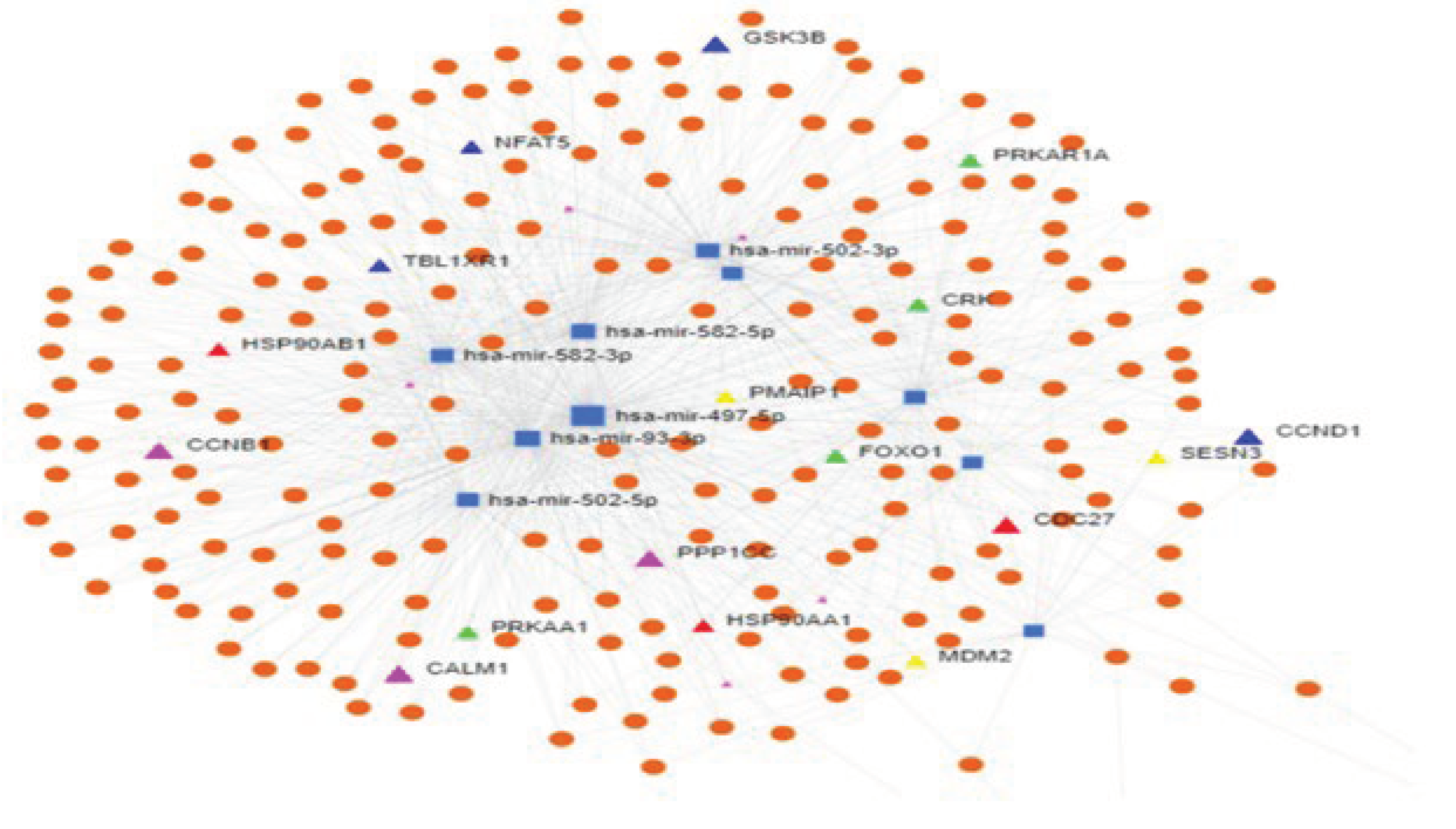
| ID | Concentration ng/ul | RIN | |
|---|---|---|---|
| 1 | 5195- Estrus | 137 | 7.4 |
| 2 | 5195-Diestrus | 128 | 7.1 |
| 3 | 5248-Estrus | 165 | 6.8 |
| 4 | 5248-Diestrus | 135 | 6.8 |
| 5 | 5069-Estrus | 257 | 6.8 |
| 6 | 5069-Diestrus | 194 | 7.0 |
| 7 | 5380-Estrus | 219 | 7.2 |
| 8 | 5380-Diestrus | 180 | 6.9 |
| 9 | 5332-Estrus | 140 | 7.1 |
| 10 | 5332-Diestrus | 115 | 7 |
| Groups | Samples | No. of Reads | Read Length | Bases | GC% | Read Alignment (%) |
|---|---|---|---|---|---|---|
| Estrus | R5068_1 | 26755063 | 50 | 1337753150 | 48 | 74.98 |
| R5195_1 | 23952660 | 50 | 1197633000 | 49 | 76.44 | |
| R5248_1 | 24501790 | 50 | 1225089500 | 48 | 81.81 | |
| R5332_1 | 48454450 | 50 | 2422722500 | 46 | 77.54 | |
| R5380_1 | 22492112 | 50 | 1124605700 | 59 | 80.9 | |
| Diestrus | R5068_2 | 22661330 | 50 | 1133066500 | 47 | 76.82 |
| R5195_2 | 24500720 | 50 | 1225036000 | 47 | 76.27 | |
| R5248_2 | 20872662 | 50 | 1043633100 | 48 | 75.17 | |
| R5332_2 | 24160605 | 50 | 1208030250 | 47 | 86.52 | |
| R5380_2 | 23551522 | 50 | 1177576100 | 47 | 77.59 |
| miRNA | FORWARD PRIMER SEQUENCE |
| 5SRNA | GCCCGATCTCGTCTGATCT |
| miR122 | CGCGTGGAGTGTGACAATGG |
| miR1246 | GAATGGATTTTTGGAGCAGGAA |
| miR130b | AGCAGGCAGTGCAATGATGA |
| miR23 | ATCACATTGCCAGGGATTTCCA |
| miR582 | TTACAGTTGTTCAACCAGTTACT |
| U6 | CTCGCTTCGGCAGCACA |
| Universal reverse primer | ATGGCGGTAAGTCCAGATACG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





