Submitted:
05 June 2023
Posted:
06 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
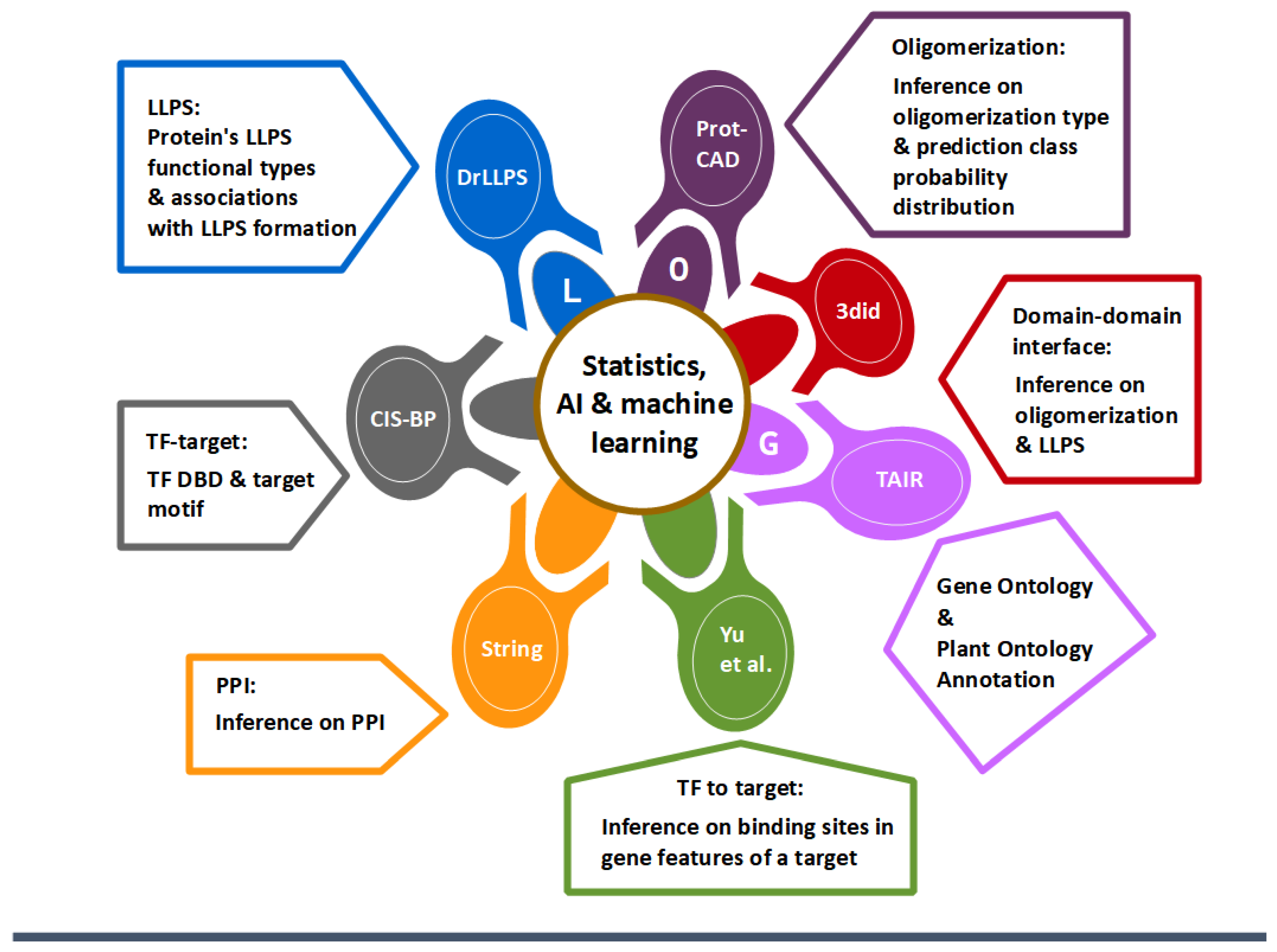
2.1. Database Modules
2.2. DDI Module
2.3. LLPS Module
2.4. GO Analysis Module
2.5. TF-Target Module
2.6. PPI Module
2.7. TF to Target Module
2.8. Proof of Concept of Search Algorithms
2.9. Demonstration of the Program Usage
2.9.1. Predictions of Important Features- LLPS Subcellular Type, LLPS Factor Type, GO Type, Gene Association Type, Signaling Pathway Type, Subcellular Location, Oligomerization Feature, and Oligomerization Type
2.9.2. Predictions of Oligomerization Type and Correlations Analysis
2.9.3. Prediction of DNA Binding Motif Types and Association Study
3. Results
3.1. Prediction of Important Features
3.2. Prediction and Extraction of Important Features from TF-LLPS Factor Data
3.3. Prediction and Association Study on LLPS Factor Interacting with TFs
4. Discussions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verde, I., Abbott, A. et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 2013. 45, 487–494. [CrossRef]
- Cao, K., Zhou, Z., Wang, Q. et al. Genome-wide association study of 12 agronomic traits in peach. Nat Commun. 2016. 7, 13246. [CrossRef]
- Mark W E J Fiers, Liesbeth Minnoye, Sara Aibar, Carmen Bravo González-Blas, Zeynep Kalender Atak, Stein Aerts, Mapping gene regulatory networks from single-cell omics data, Briefings in Functional Genomics. 2018. Volume 17, Issue 4, Pages 246–254. [CrossRef]
- Li, Y., Li, Q., Beuchat, G., Zeng, H., Zhang, C., and Chen, L.Q. Combined analyses of translatome and transcriptome in Arabidopsis reveal new players responding to magnesium deficiency. J. Integr. Plant Biol. 2021. 63: 2075– 2092.
- Qi Su, Sohum Mehta, Jin Zhang. Liquid-liquid phase separation: Orchestrating cell signaling through time and space. Molecular Cell. 2021. Volume 81, Issue 20. Pages 4137-4146. [CrossRef]
- Peng PH, Hsu KW, Wu KJ. Liquid-liquid phase separation (LLPS) in cellular physiology and tumor biology. Am J Cancer Res. 2021. 15;11(8):3766-3776. 7.
- Yan G. Zhao, Hong Zhang. Phase Separation in Membrane Biology: The Interplay between Membrane-Bound Organelles and Membraneless Condensates. Developmental Cell. Volume 55. Issue 1. 200. Pages 30-44.
- Nesterov SV, Ilyinsky NS, Uversky VN. Liquid-liquid phase separation as a common organizing principle of intracellular space and biomembranes providing dynamic adaptive responses. Biochim Biophys Acta Mol Cell Res. 2021. 1868(11):119102.
- Li J, Zhang M, Ma W, Yang B, Lu H, Zhou F, Zhang L. Post-translational modifications in liquid-liquid phase separation: a comprehensive review. Mol Biomed. 2022. 11;3(1):13. [CrossRef]
- Stoyle CL, Stephens PE, Humphreys DP, Heywood S, Cain K, Bulleid NJ. IgG light chain-independent secretion of heavy chain dimers: consequence for therapeutic antibody production and design. Biochem J. 2017. 7;474(18):3179-3188. [CrossRef]
- Tan, W., Cheng, S., Li, Y. et al. Phase separation modulates the assembly and dynamics of a polarity-related scaffold-signaling hub. Nat Commun. 2022. 13, 7181. [CrossRef]
- Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009. 10(9):990-6. [CrossRef]
- Koehler Lydia C., Grese Zachary R., Bastos Alliny C. S., Mamede Lohany D., Heyduk Tomasz, Ayala Yuna M., TDP-43 Oligomerization and Phase Separation Properties Are Necessary for Autoregulation, Frontiers in Neuroscience. 2022. (16).
- Stein A, Aloy P. Novel peptide-mediated interactions derived from high-resolution 3-dimensional structures. PLoS Comput Biol. 2010. 20;6(5):e1000789. [CrossRef]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006. 6;45(22):6873-88.
- Puranik S, Acajjaoui S, Conn S, Costa L, Conn V, Vial A, Marcellin R, Melzer R, Brown E, Hart D, Theißen G, Silva CS, Parcy F, Dumas R, Nanao M, Zubieta C. Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell. 2014. 26(9):3603-15. [CrossRef]
- Sayou, C., Nanao, M., Jamin, M. et al. A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat Commun. 2016. 7, 11222.
- Kato M, Hata N, Banerjee N, Futcher B, Zhang MQ. Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol. 2004. 5(8):R56. [CrossRef]
- Sanchez-Burgos I, Espinosa JR, Joseph JA, Collepardo-Guevara R. RNA length has a non-trivial effect in the stability of biomolecular condensates formed by RNA-binding proteins. PLoS Comput Biol. 2022. 2;18(2):e1009810.
- Qifang Xu, Roland?L Dunbrack, Jr., The protein common assembly database (ProtCAD) a comprehensive structural resource of protein complexes, Nucleic Acids Res. 2022. gkac937. [CrossRef]
- Roberto Mosca, Arnaud Céol, Amelie Stein, Roger Olivella, Patrick Aloy, 3did: a catalog of domain-based interactions of known three-dimensional structure, Nucleic Acids Res. 2014. Volume 42, Issue D1, 1.Pages D374–D379. [CrossRef]
- Kurotani A, Yamada Y, Shinozaki K, Kuroda Y, Sakurai T. Plant-PrAS: a database of physicochemical and structural properties and novel functional regions in plant proteomes. Plant Cell Physiol. 2015 Jan;56(1):e11. [CrossRef]
- Xue H, Zhang Q, Wang P, Cao B, Jia C, Cheng B, Shi Y, Guo WF, Wang Z, Liu ZX, Cheng H. qPTMplants: an integrative database of quantitative post-translational modifications in plants. Nucleic Acids Res. 2022 Jan 7;50(D1):D1491-D1499. [CrossRef]
- Ning W, Guo Y, Lin S, Mei B, Wu Y, Jiang P, Tan X, Zhang W, Chen G, Peng D, Chu L, Xue Y. DrLLPS: a data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res. 2020. 8;48(D1):D288-D295. [CrossRef]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, Miller N, Mueller LA, Mundodi S, Reiser L, Tacklind J, Weems DC, Wu Y, Xu I, Yoo D, Yoon J, Zhang P. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003. 1;31(1):224-8. [CrossRef]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000. 25(1):25-9.
- R. L. Walls*, Cooper*, L. D., Elser, J. L., Gandolfo, M. A., Mungall, C. J., Smith, B., Stevenson, D. W., and Jaiswal, P., “The Plant Ontology Facilitates Comparisons of Plant Development Stages Across Species”, Frontiers in Plant Science. 2019. vol. 10.
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey MG, Huang E, Mukherjee T, Chen X, Reece-Hoyes JS, Govindarajan S, Shaulsky G, Walhout AJ, Bouget FY, Ratsch G, Larrondo LF, Ecker JR, Hughes TR. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014. 11;158(6):1431-43. [CrossRef]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019. 8;47(D1):D607-D613. [CrossRef]
- Pujato M, Kieken F, Skiles AA, Tapinos N, Fiser A. Prediction of DNA binding motifs from 3D models of transcription factors; identifying TLX3 regulated genes. Nucleic Acids Res. 2014. 42(22) : 13500-12.
- Yu, CP., Lin, JJ. & Li, WH. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci Rep. 2016. 6, 25164. [CrossRef]
- Wang Y, Wang Q, Huang H, Huang W, Chen Y, McGarvey PB, Wu CH, Arighi CN, UniProt Consortium. A crowdsourcing open platform for literature curation in UniProt. Plos Biology. 2021. 19(12):e3001464.
- Jaina Mistry, Sara Chuguransky, Lowri Williams, Matloob Qureshi, Gustavo A Salazar, Erik L L Sonnhammer, Silvio C E Tosatto, Lisanna Paladin, Shriya Raj, Lorna J Richardson, Robert D Finn, Alex Bateman, Pfam: The protein families database in 2021, Nucleic Acids Res. 2021. Volume 49, Issue D1, 8, Pages D412–D419. [CrossRef]
- Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, Gough J, Haft DH, Letunić I, Marchler-Bauer A, Mi H, Natale DA, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A. InterPro in 2022. Nucleic Acids Res. 2022. [CrossRef]
- H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne, The Protein Data Bank. Nucleic Acids Res. 2000. 28: 235-242 . [CrossRef]
- Berardini, T.Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E. and Huala, E. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. genesis. 2015. 53: 474-485. [CrossRef]
- Eibe Frank, Mark A. Hall, and Ian H. Witten. The WEKA Workbench. Online Appendix for "Data Mining: Practical Machine Learning Tools and Techniques", Morgan Kaufmann, Fourth Edition, 2016.
- Degtyareva AO, Antontseva EV, Merkulova TI. Regulatory SNPs: Altered Transcription Factor Binding Sites Implicated in Complex Traits and Diseases. Int J Mol Sci. 2021. 16;22(12):6454. [CrossRef]
- Zheng Z, Goncearenco A, Berezovsky IN. Nucleotide binding database NBDB--a collection of sequence motifs with specific protein-ligand interactions. Nucleic Acids Res. 2016. 4;44(D1):D301-7. [CrossRef]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, Dunker AK, Gough J. D²P²: database of disordered protein predictions. Nucleic Acids Res. 2013. 41(Database issue):D508-16. [CrossRef]
- Schaefer U, Schmeier S, Bajic VB. TcoF-DB: dragon database for human transcription co-factors and transcription factor interacting proteins. Nucleic Acids Res. 2011. 39(Database issue):D106-10. [CrossRef]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006. 140(3):818-29. [CrossRef]
- Li Y, Wei L, Wang C, Zhao J, Han S, Zhang Y, Du W. LPInsider: a webserver for lncRNA-protein interaction extraction from the literature. BMC Bioinformatics. 2022. 23(1):135. [CrossRef]
- Yuan J, Wu W, Xie C, Zhao G, Zhao Y, Chen R. NPInter v2.0: an updated database of ncRNA interactions. Nucleic Acids Res. 2014. 42(Database issue):D104-8. [CrossRef]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013. 41(Database issue):D36-42. [CrossRef]
- Adam Frankish, Mark Diekhans, Anne-Maud Ferreira, Rory Johnson, Irwin Jungreis, Jane Loveland, Jonathan M Mudge, Cristina Sisu, James Wright, Joel Armstrong, If Barnes, Andrew Berry, Alexandra Bignell, Silvia Carbonell Sala, Jacqueline Chrast, Fiona Cunningham, Tomás Di Domenico, Sarah Donaldson, Ian T Fiddes, Carlos García Girón, Jose Manuel Gonzalez, Tiago Grego, Matthew Hardy, Thibaut Hourlier, Toby Hunt, Osagie G Izuogu, Julien Lagarde, Fergal J Martin, Laura Martínez, Shamika Mohanan, Paul Muir, Fabio C P Navarro, Anne Parker, Baikang Pei, Fernando Pozo, Magali Ruffier, Bianca M Schmitt, Eloise Stapleton, Marie-Marthe Suner, Irina Sycheva, Barbara Uszczynska-Ratajczak, Jinuri Xu, Andrew Yates, Daniel Zerbino, Yan Zhang, Bronwen Aken, Jyoti S Choudhary, Mark Gerstein, Roderic Guigó, Tim J P Hubbard, Manolis Kellis, Benedict Paten, Alexandre Reymond, Michael L Tress, Paul Flicek, GENCODE reference annotation for the human and mouse genomes, Nucleic Acids Res. 2019. Volume 47, Issue D1, Pages D766–D773.
- Shane Neph, M. Scott Kuehn, Alex P. Reynolds, Eric Haugen, Robert E. Thurman, Audra K. Johnson, Eric Rynes, Matthew T. Maurano, Jeff Vierstra, Sean Thomas, Richard Sandstrom, Richard Humbert, John A. Stamatoyannopoulos, BEDOPS: high-performance genomic feature operations, Bioinformatics. 2012. Volume 28, Issue 14. Pages 1919–1920. [CrossRef]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Larsson P, Longden I, McLaren W, Overduin B, Pritchard B, Riat HS, Rios D, Ritchie GR, Ruffier M, Schuster M, Sobral D, Spudich G, Tang YA, Trevanion S, Vandrovcova J, Vilella AJ, White S, Wilder SP, Zadissa A, Zamora J, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernández-Suarez XM, Herrero J, Hubbard TJ, Parker A, Proctor G, Vogel J, Searle SM. Ensembl 2011. Nucleic Acids Res. 2011. 39(Database issue):D800-6. [CrossRef]
- Lê S, Josse J, Husson F. “FactoMineR: A Package for Multivariate Analysis.” Journal of Statistical Software. 2008. 25(1), 1–18. [CrossRef]
- Van Rossum, G., & Drake, F. L. Python 3 Reference Manual. Scotts Valley, CA: CreateSpace. 2009.
- Chen L, Cui Y, Yao Y, An L, Bai Y, Li X, Yao X, Wu K. Genome-wide identification of WD40 transcription factors and their regulation of the MYB-bHLH-WD40 (MBW) complex related to anthocyanin synthesis in Qingke (Hordeum vulgare L. var. nudum Hook. f.). BMC Genomics. 2023. 4;24(1):166. [CrossRef]
- Buhrman K, Aravena-Calvo J, Ross Zaulich C, Hinz K, Laursen T. Anthocyanic Vacuolar Inclusions: From Biosynthesis to Storage and Possible Applications. Front Chem. 2022. 28;10:913324. [CrossRef]
- Ma M, Ru Y, Chuang LS, Hsu NY, Shi LS, Hakenberg J, Cheng WY, Uzilov A, Ding W, Glicksberg BS, Chen R. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics. 2015.16 Suppl 8(Suppl 8):S3. [CrossRef]
- Terrile MC, Tebez NM, Colman SL, Mateos JL, Morato-López E, Sánchez-López N, Izquierdo-Álvarez A, Marina A, Calderón Villalobos LIA, Estelle M, Martínez-Ruiz A, Fiol DF, Casalongué CA, Iglesias MJ. S-Nitrosation of E3 Ubiquitin Ligase Complex Components Regulates Hormonal Signalings in Arabidopsis. Front Plant Sci. 2022. 4;12:794582. [CrossRef]
- Zhu S, Gu J, Yao J, Li Y, Zhang Z, Xia W, Wang Z, Gui X, Li L, Li D, Zhang H, Liu C. Liquid-liquid phase separation of RBGD2/4 is required for heat stress resistance in Arabidopsis. Dev Cell. 2022. 14;57(5):583-597.e6. [CrossRef]
- Feng C, Cai XW, Su YN, Li L, Chen S, He XJ. Arabidopsis RPD3-like histone deacetylases form multiple complexes involved in stress response. J Genet Genomics. 2021. 20;48(5):369-383. [CrossRef]
- Truebestein L, Leonard TA. Coiled-coils: The long and short of it. Bioessays. 2016. 38(9):903-16. [CrossRef]
- Dang M, Li T, Song J. ATP and nucleic acids competitively modulate LLPS of the SARS-CoV2 nucleocapsid protein. Commun Biol. 2023. 21;6(1):80. [CrossRef]
- Dang M, Li T, Zhou S, Song J. Arg/Lys-containing IDRs are cryptic binding domains for ATP and nucleic acids that interplay to modulate LLPS. Commun Biol. 2022. 1;5(1):1315. [CrossRef]
- Yueying Zhang, Minglei Yang, Susan Duncan, Xiaofei Yang, Mahmoud A S Abdelhamid, Lin Huang, Huakun Zhang, Philip N Benfey, Zoë A E Waller, Yiliang Ding, G-quadruplex structures trigger RNA phase separation, Nucleic Acids Res. 2019. Volume 47, Issue 22, Pages 11746–11754. [CrossRef]
- Erin M. Langdon, Amy S. Gladfelter, Chapter Four - Probing RNA Structure in Liquid–Liquid Phase Separation Using SHAPE-MaP, Editor(s): Elizabeth Rhoades, Methods in Enzymology, Academic Press, Volume 611, Pages 67-79, 2018. [CrossRef]
- Haibo Zhu, Hao Fu, Tianyu Cui, Lin Ning, Huaguo Shao, Yehan Guo, Yanting Ke, Jiayi Zheng, Hongyan Lin, Xin Wu, Guanghao Liu, Jun He, Xin Han, Wenlin Li, Xiaoyang Zhao, Huasong Lu, Dong Wang, Kongfa Hu, Xiaopei Shen, RNAPhaSep: a resource of RNAs undergoing phase separation, Nucleic Acids Res. 2022. Volume 50, Issue D1. Pages D340–D346. [CrossRef]

| Amino acid group letter | Amino acid | Amino acid features |
| P | R,K,S,T | Positive or polar-uncharged |
| N | D,E,N,Q | Negative or polar-uncharged |
| H | A,V,I,L,M | Hydrophobic |
| R | F,W,Y | Ring structures |
| S | C,G,P,H | Special properties |
| DNA group letter | DNA/ambiguous DNA |
| G | G |
| Z | R,S,K,B,D,V |
| X | A,C,T,Y,W,M,H |
| N | N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




