Submitted:
01 June 2023
Posted:
02 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
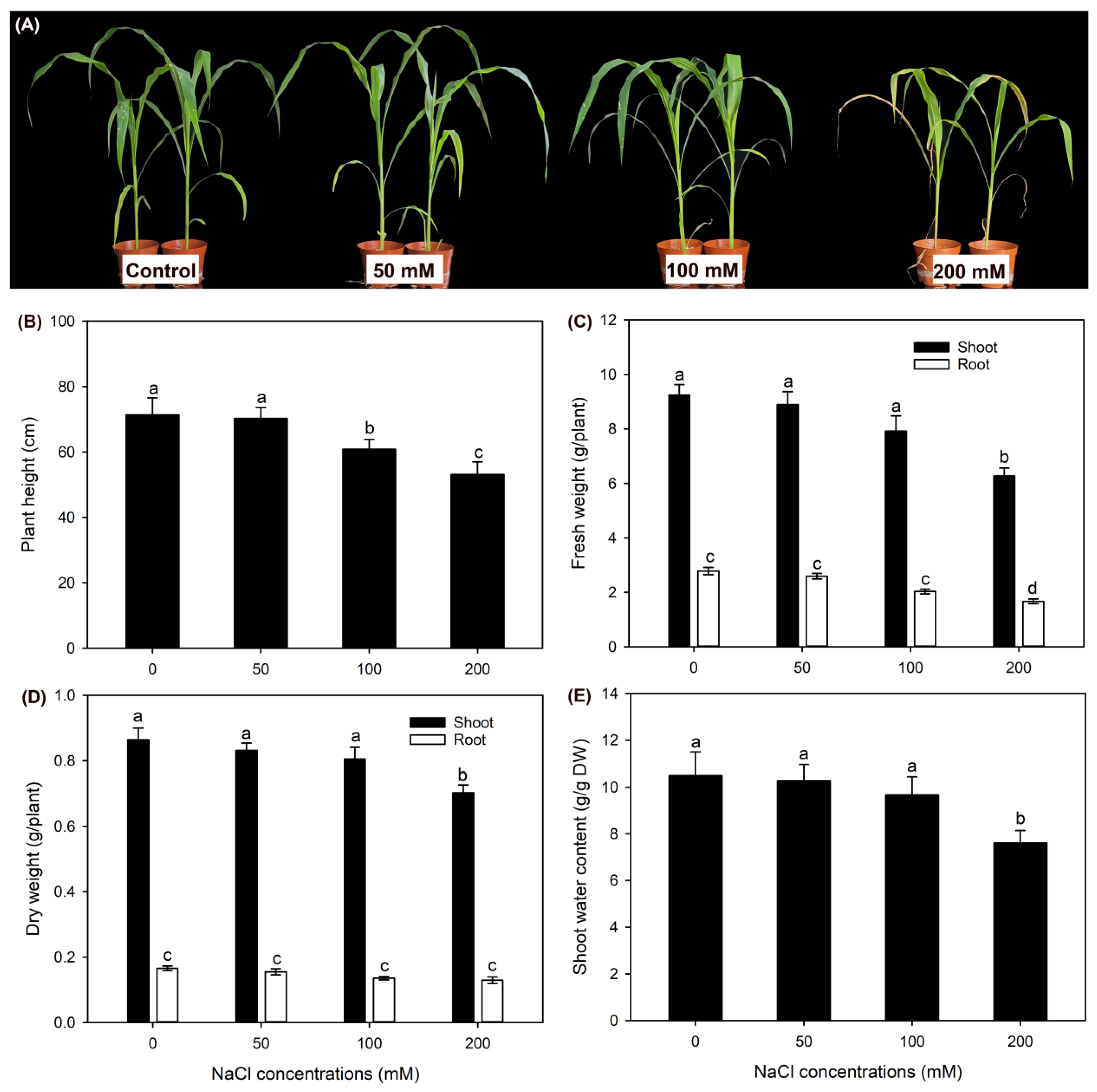
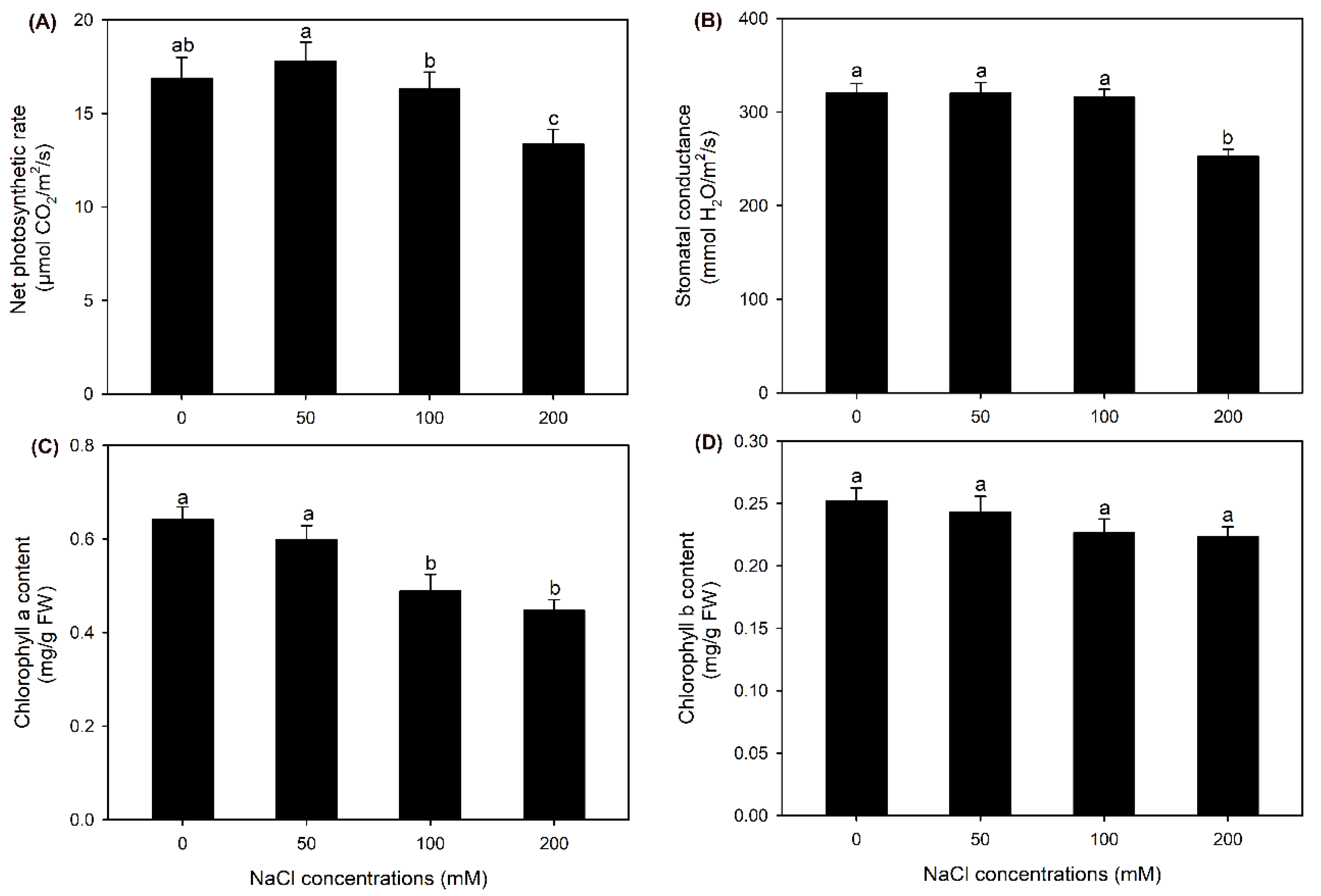
2.1. The Effect of NaCl Treatments on Growth and Photosynthesis of Sweet Sorghum
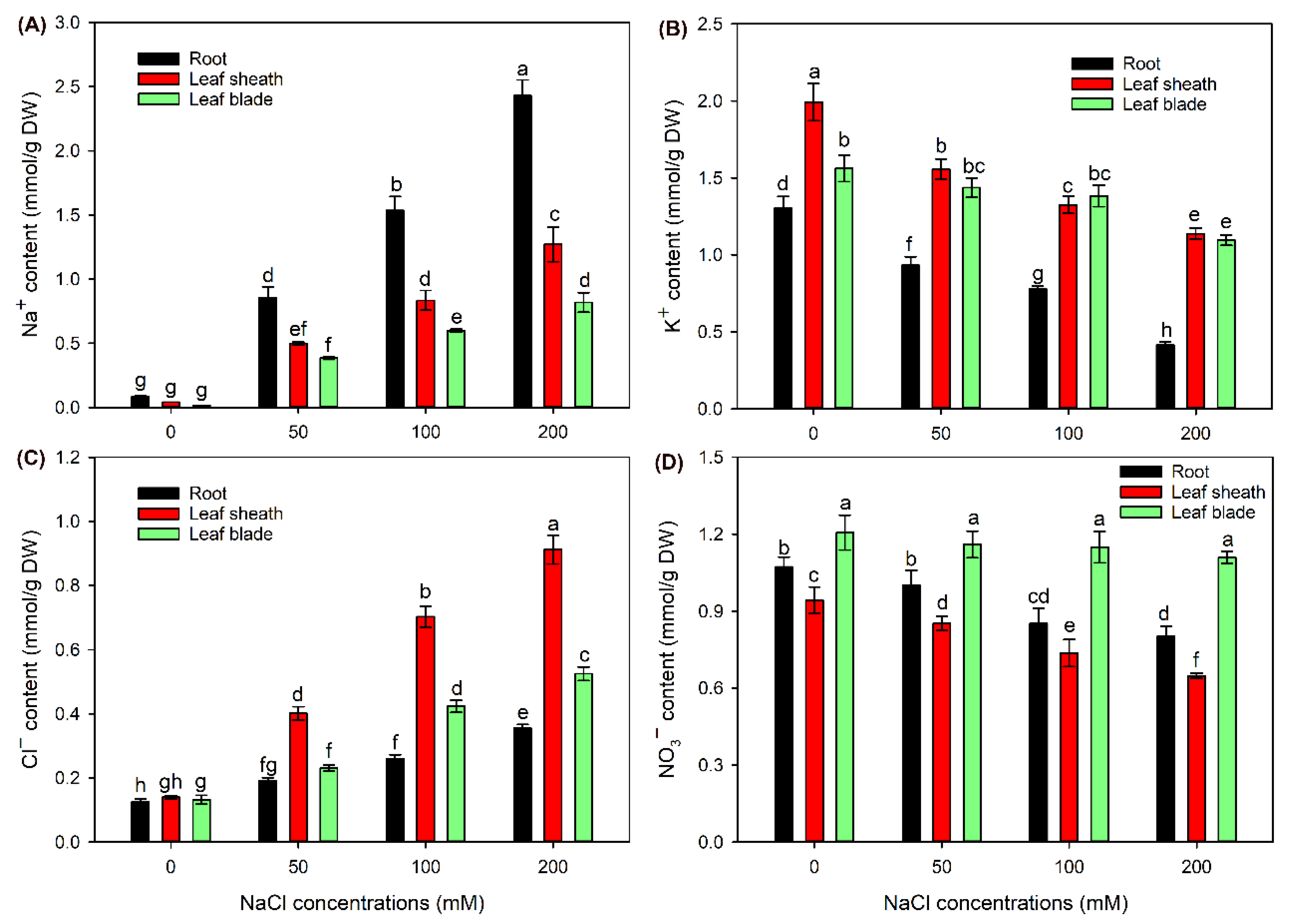
2.2. The Ion Contents in Different Tissues of Sweet Sorghum under NaCl Treatments
2.3. RNA-seq Analysis of Sweet Sorghum under NaCl Stress
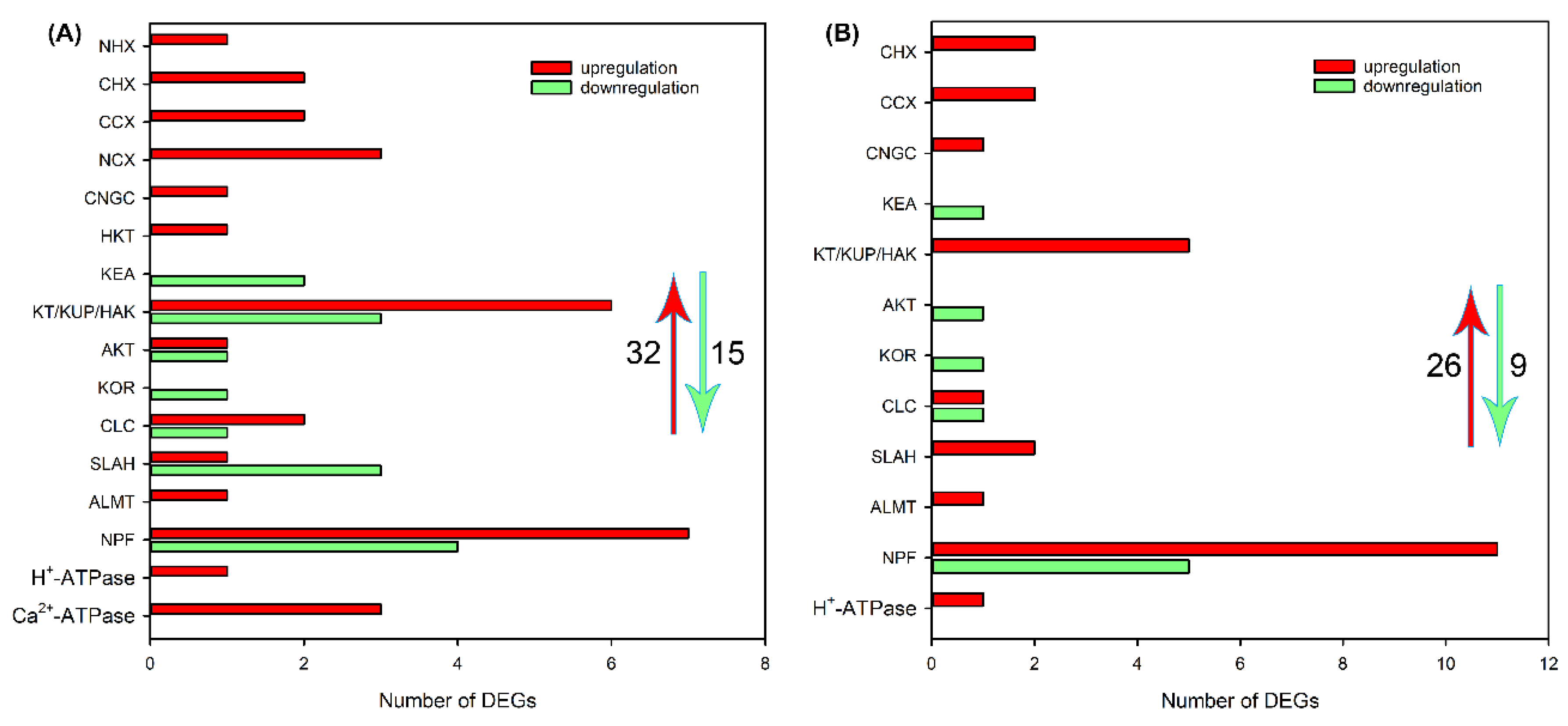
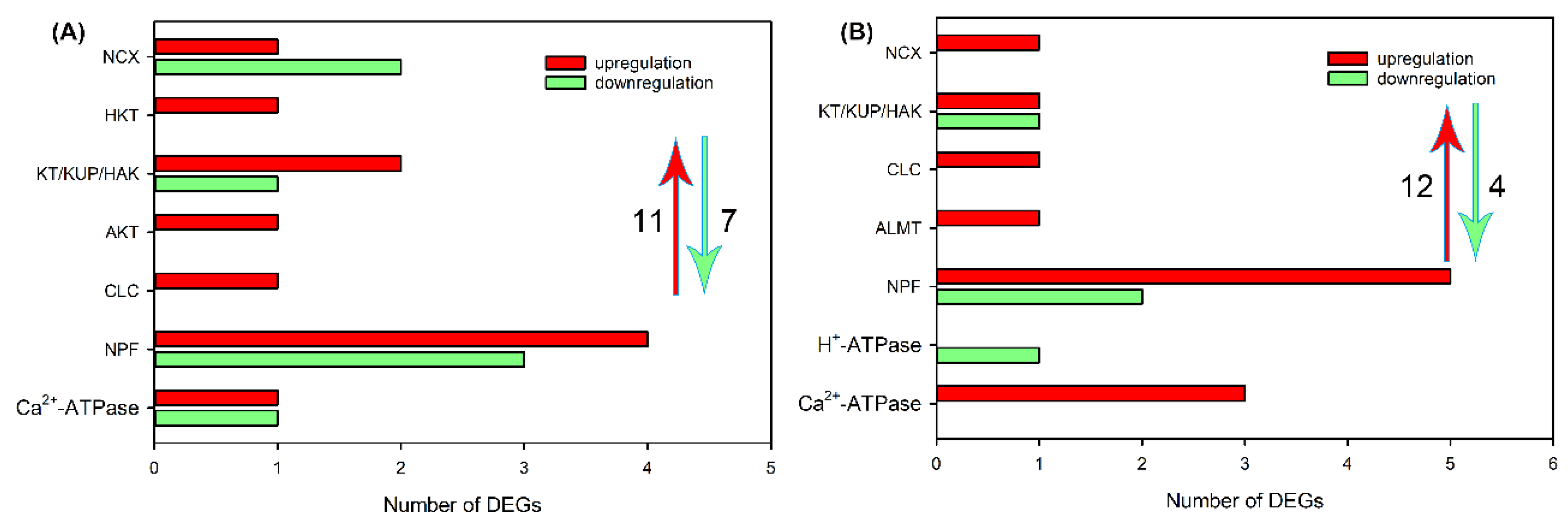
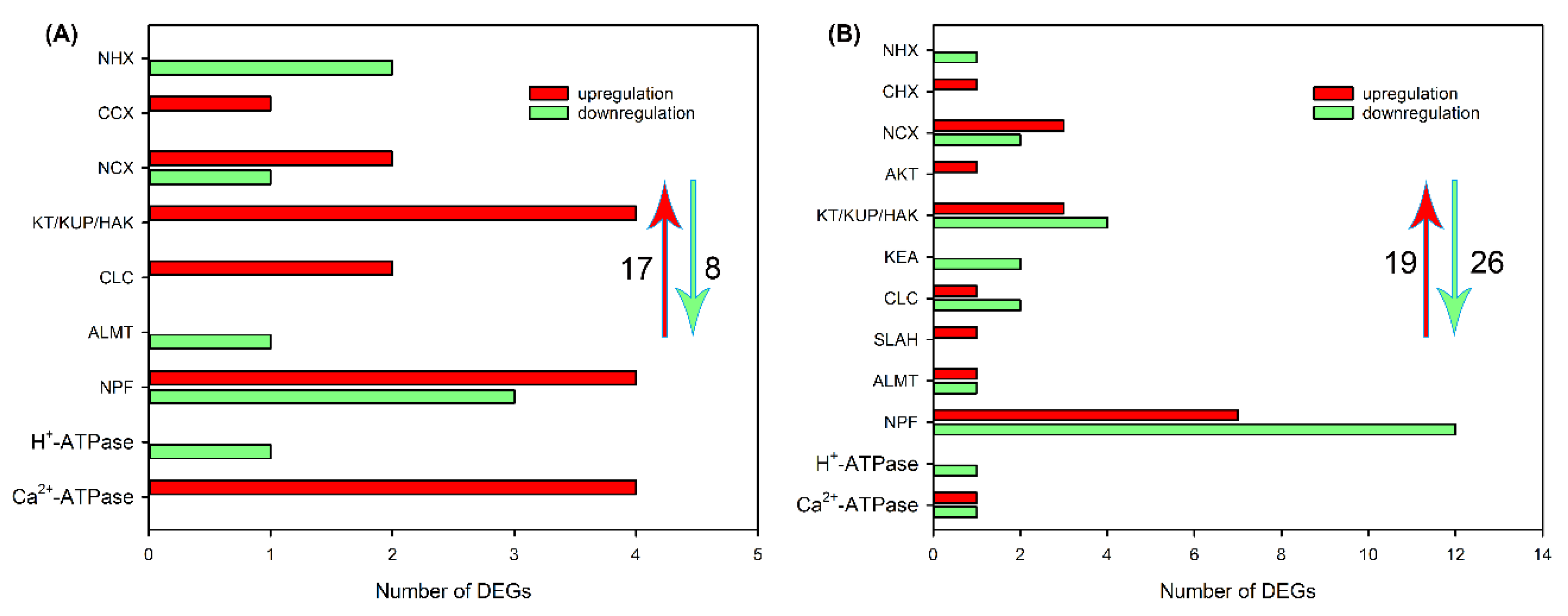
2.4. Identification of DEGs Related to Ion Transport in Roots, Leaf Sheaths and Leaf Blades after NaCl Treatment for 6 and 24 h
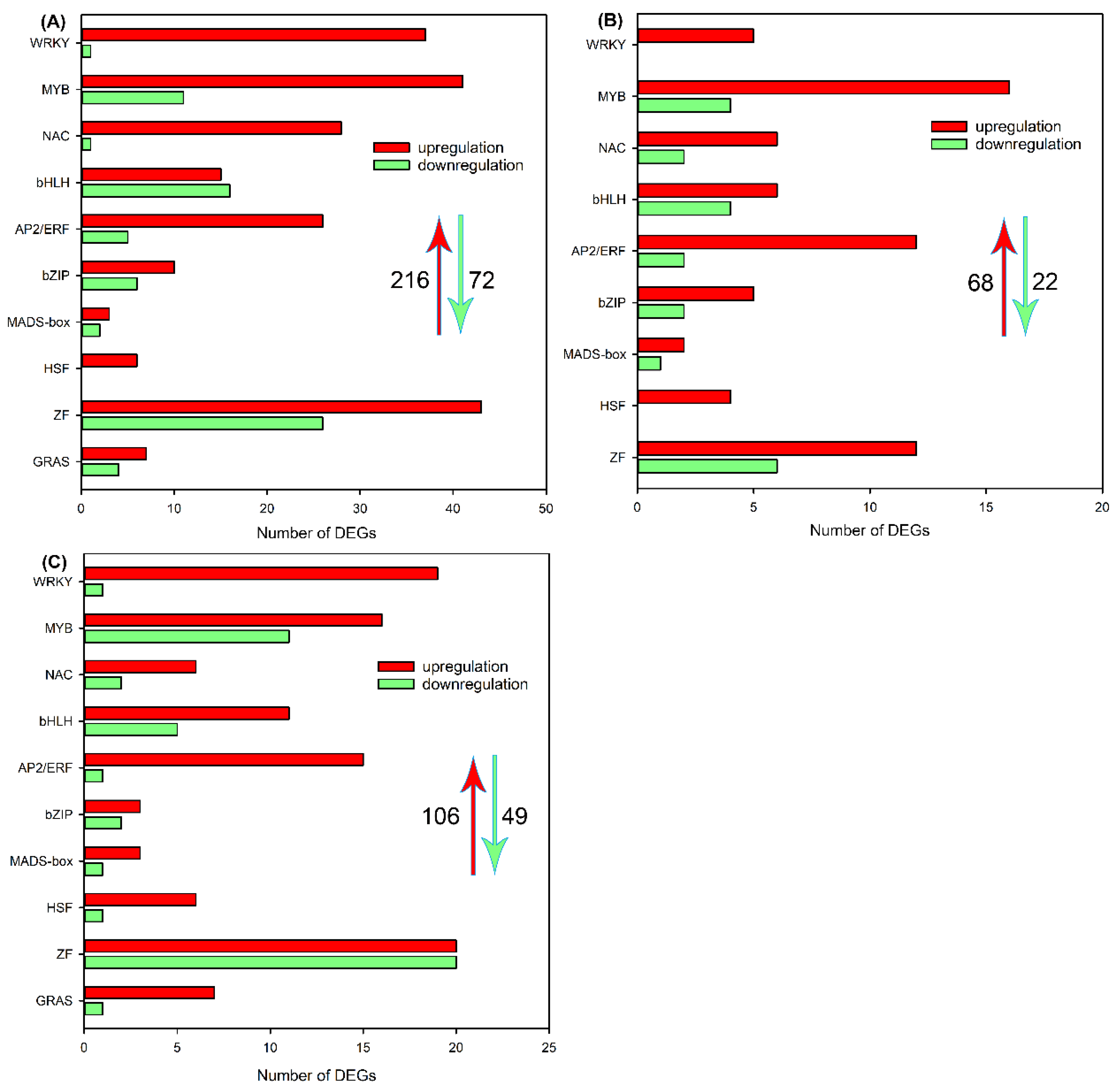
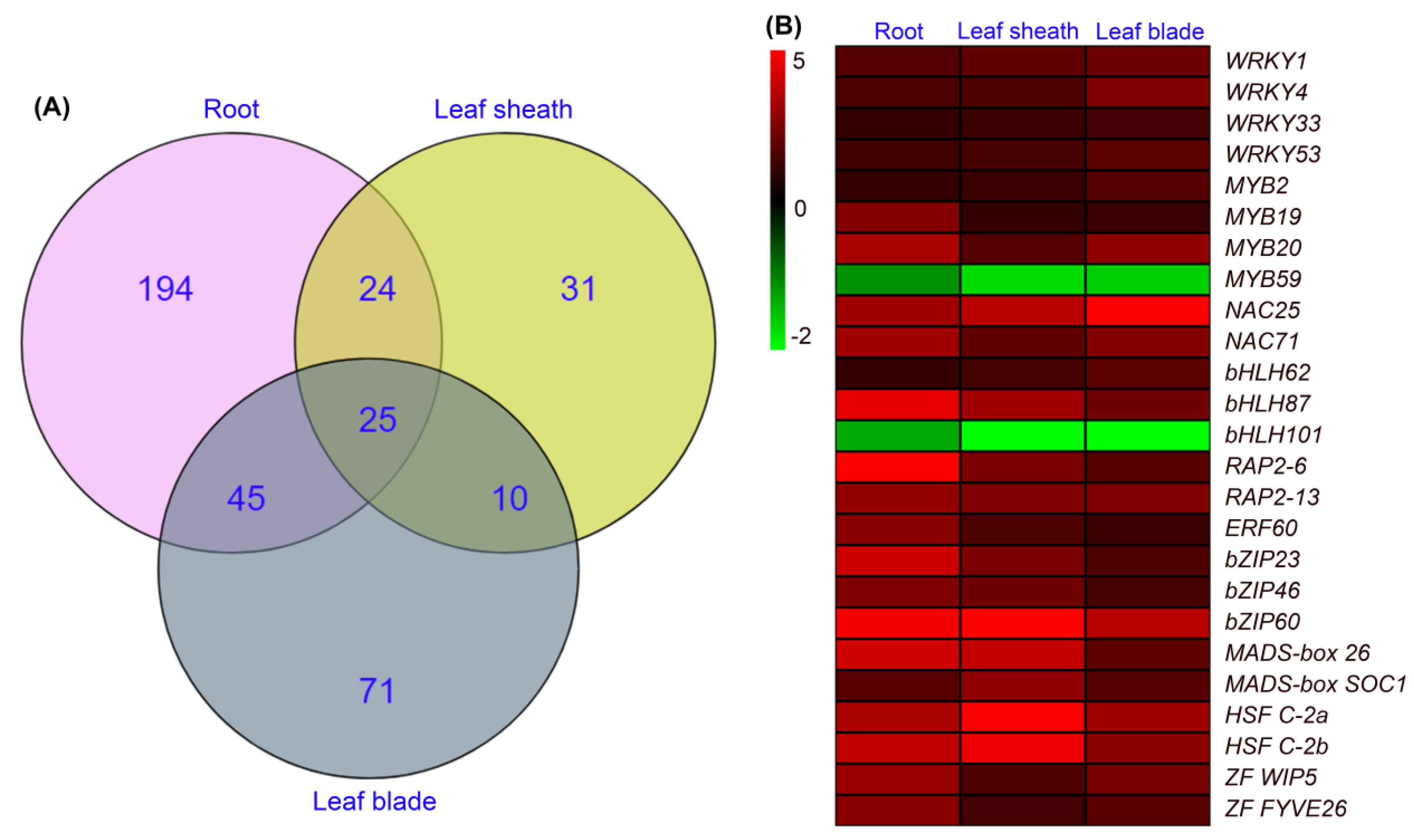
2.5. Identification of DEGs Encoding Transcription Factor in Roots, Leaf Sheaths and Leaf Blades after NaCl Treatment for 6 h
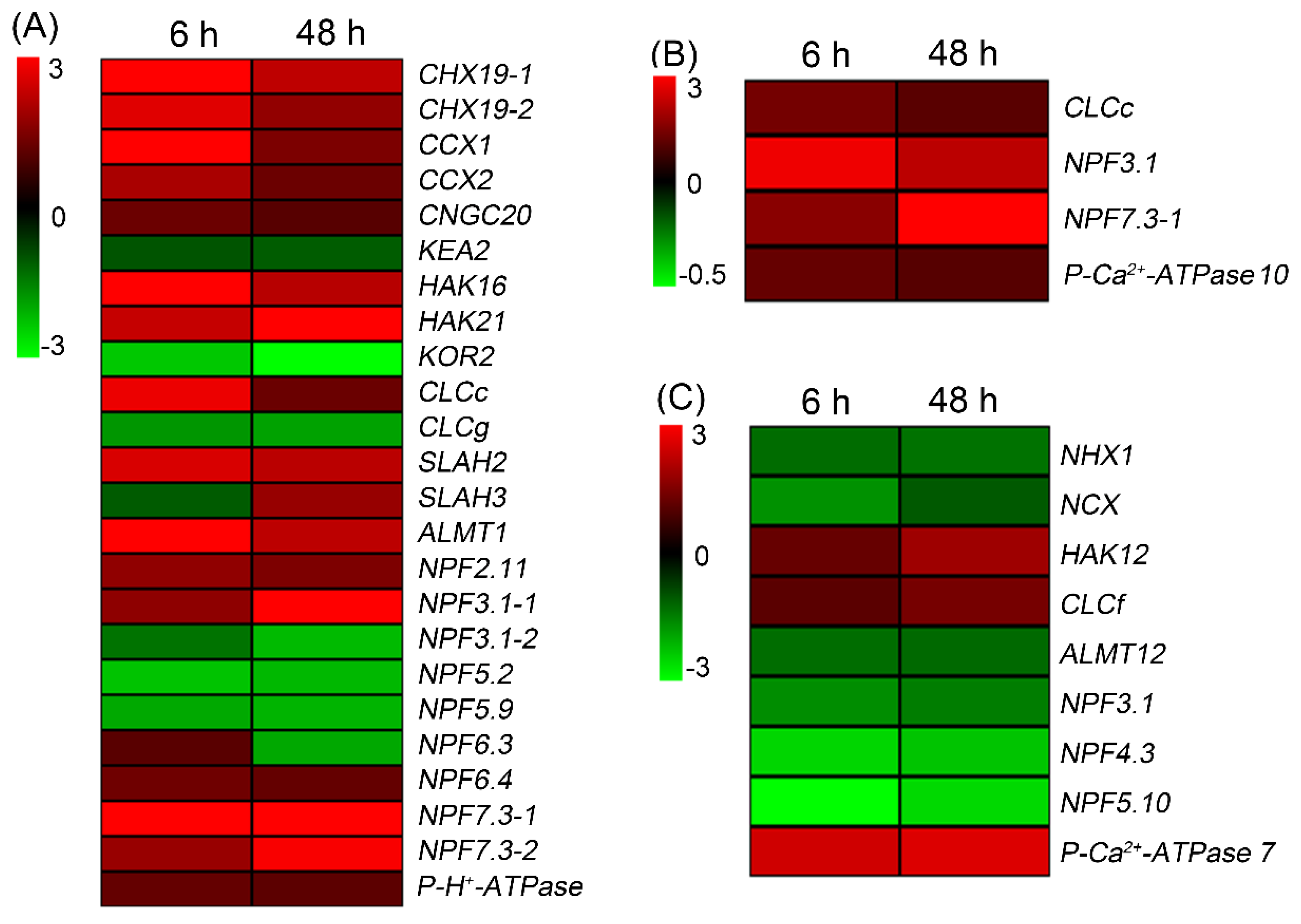
2.6. Validation of RNA-Seq Results
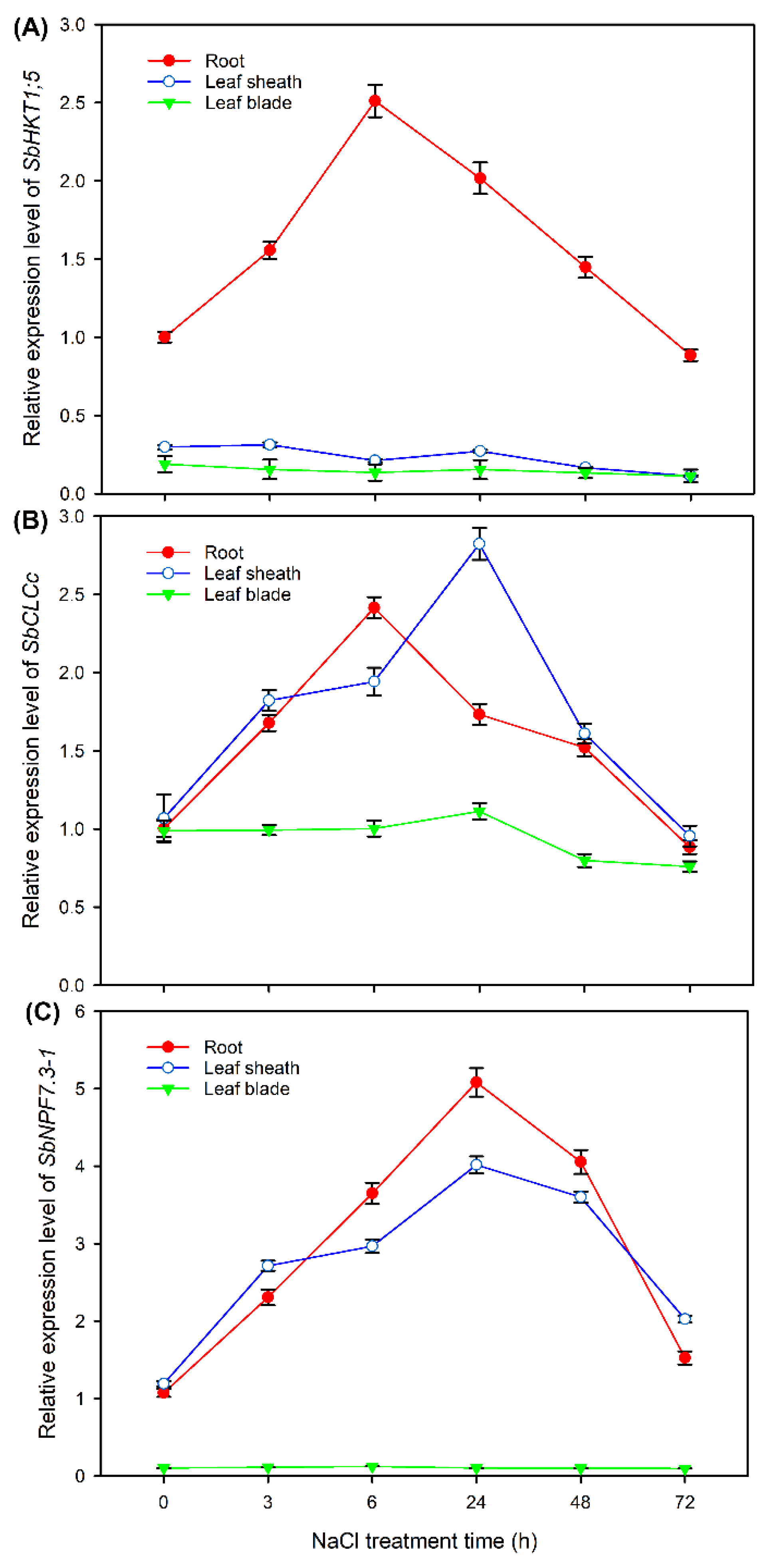
2.7. Expression Pattern of HKT1;5, CLCc and NPF7.3 in Sweet Sorghum under NaCl Treatments
3. Discussion
3.1. Sweet Sorghum Could Efficiently Exclude Na+ from Shoots and Accumulate Cl− in Leaf Sheaths under NaCl Stress
3.2. The Genes Related to Ion Transport Play Key Roles in the Salt Tolerance of Sweet Sorghum
3.3. Identification of Key Transcriptome Factors Involved in the Salt Tolerance of Sweet Sorghum
3.4. Sweet Sorghum Possesses a Strong Photosynthetic Ability under Salt Stresses
4. Materials and methods
4.1. Plant Material and Growth Conditions
4.2. Determination of Plant Height, Tissue Biomass and Shoot Water Content
4.3. Measurements of Photosynthesis-related Parameters
4.4. Measurement of Ion Contents in Tissues
4.5. Transcriptome Sequencing
4.6. Differentially Expressed Genes Analysis
4.7. Validation of RNA-sequencing Results
4.8. Analysis of Expression Pattern of HKT1;5, CLCc and NPF7.3 in Sweet Sorghum under NaCl Treatment
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, F.; Xie, P.; Sun, S, Qiao, X.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; Yang, D.; Wu, Y.; Xia, R.; Li, X.; Lu, J.; Liu, Y.; Xie, X.; Ma, D.; Xu, X.; Liang, Z.; Feng, Z.; Huang, X.; Yu, H.; Liu, G.; Wang, Y.; Li, J.; Zhang, Q.; Chen, C.; Ouyang, Y.; Xie, Q. A Gγ protein regulates alkaline sensitivity in crops. Science 2023, 379, eade8416. [CrossRef]
- Li, J.; Lei, S.; Gong, H.; Liu, Z.; Zhang, Y.; Ouyang, Z. Field performance of sweet sorghum in salt-affected soils in China: A quantitative synthesis. Environ. Res. 2023, 222, 115362. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; Plett, D.; Gilliham, M. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef]
- Zheng, L.Y.; Guo, X.S.; He, B.; Sun, L.J.; Peng, Y.; Dong, S.S.; Liu, T.F.; Jiang, S.; Ramachandran, S.; Liu, C.M.; Jing, H.C. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol. 2011, 12, R114. [Google Scholar] [CrossRef] [PubMed]
- Regassa, T.H.; Wortmann, C.S. Sweet sorghum as a bioenergy crop: Literature review. Biomass Bioenerg. 2014, 64, 348–355. [Google Scholar] [CrossRef]
- López-Sandin, I.; Zavala-García, F.; Levin, L.; Ruiz, H.A.; Hernández-Luna, C.E.; Gutiérrez-Soto, G. Evaluation of bioethanol production from sweet sorghum variety roger under different tillage and fertilizer treatments. BioEnerg. Res. 2021, 14, 1058–1069. [Google Scholar] [CrossRef]
- Yang, Z.; Li, J.L.; Liu, L.N.; Xie, Q.; Sui, N. Photosynthetic regulation under salt stress and salt-tolerance mechanism of sweet sorghum. Front. Plant Sci. 2020, 10, 1722. [Google Scholar] [CrossRef]
- Cui, J.; Ren, G.; Qiao, H.; Xiang, X.; Huang, H.; Chang, J. Comparative transcriptome analysis of seedling stage of two sorghum cultivars under salt stress. J. Plant Growth Regul. 2018, 37, 986–998. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Plant salt tolerance: adaptations in halophytes. Ann. Bot. 2015, 115, 327–331. [Google Scholar] [CrossRef]
- Geilfus, C.M. Review on the significance of chlorine for crop yield and quality. Plant Sci. 2018, 270, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Bazihizina, N.; Colmer, T.D.; Cuin, T.A.; Mancuso, S.; Shabala, S. Friend or foe? Chloride patterning in halophytes. Trends Plant Sci. 2019, 24, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; Nayef, M.A.; Shabala, S.; An, G.; Ma, J.F.; Horie, T. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liang, X.; Wang, L.; Cao, Y.; Song, W.; Shi, J.; Lai, J.; Jiang, C. A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 2019, 5, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Kingsbury, R. W.; Epstein, E. Salt sensitivity in wheat, a case for specific ion toxicity. Plant Physiol. 1986, 80, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.Q.; Gao, X.H.; Sun, W.; An, J.; Zhao, Y.X.; Zhang, H. Cloning and functional characterization of a cation-chloride cotransporter gene OsCCC1. Plant Mol. Biol. 2011, 75, 567–578. [Google Scholar] [CrossRef]
- Li, B.; Tester, M.; Gilliham, M. Chloride on the move. Trends Plant Sci. 2017, 22, 236–248. [Google Scholar] [CrossRef]
- Louarn, G.; Andrieu, B.; Giauffret, C. A size-mediated effect can compensate for transient chilling stress affecting maize (Zea mays) leaf extension. New Phytol. 2010, 187, 106–118. [Google Scholar] [CrossRef]
- James, R.A.; Davenport, R.J.; Munns, R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 2016, 142, 1537–1547. [Google Scholar] [CrossRef]
- Wei, X.; Liu, L.; Lu, C.; Yuan, F.; Han, G.; Wang, B. SbCASP4 improves salt exclusion by enhancing the root apoplastic barrier. Planta 2021, 254, 81. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Sui, Y.; Han, G.; Zhang, Y.; Guo, S.; Sui, N. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 2020, 102, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.N.; Wang, F.Z.; Yang, C.H.; Yuan, J.Z.; Guo, H.; Zhang, J.L.; Wang, S.M.; Ma, Q. Transcriptomic profiling identifies candidate genes involved in the salt tolerance of the xerophyte Pugionium cornutum. Genes 2019, 10, 1039. [Google Scholar] [CrossRef] [PubMed]
- Jossier, M.; Kroniewicz, L.; Dalmas, F.; Thiec, D.L.; Ephritikhine, G.; Barbier-Brygoo, H.; Vavasseur, A.; Filleur, S.; Leonhardt, N. The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 2010, 64, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Z.; Lv, X.F.; Li, J.Y.; Yi, H.Y.; Gong, J.M. Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 2012, 159, 1582–1590. [Google Scholar] [CrossRef]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.S.; Zou, Q.; Zhao, K.F. Effect of NaCl stress on ionic contents in different organs of sorghum plants. Acta Agronomica Sinica 2000, 26, 845–850. [Google Scholar]
- Teakle, N.L.; Tyerman, S.D. Mechanisms of Cl– transport contributing to salt tolerance. Plant Cell Environ. 2010, 33, 566–589. [Google Scholar] [CrossRef]
- Cui, Y.N.; Li, X.T.; Yuan, J.Z.; Wang, F.Z.; Guo, H.; Xia, Z.R.; Wang, S.M.; Ma, Q. Chloride is beneficial for growth of the xerophyte Pugionium cornutum by enhancing osmotic adjustment capacity under salt and drought stresses. J. Exp. Bot. 2020, 71, 4215–4231. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Skerrett, I.M. Root ion channels and salinity. Sci. Hortic-Amsterdam 1999, 78, 175–235. [Google Scholar] [CrossRef]
- Reich, M.; Aghajanzadeh, T.; Helm, J.; Parmar, S.; Hawkesford, M.J.; De Kok, L.J. Chloride and sulfate salinity differently affect biomass, mineral nutrient composition and expression of sulfate transport and assimilation genes in Brassica rapa. Plant Soil 2017, 411, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Spielmeyer, W.; Lagudah, E.S.; James, R.A.; Platten, J.D.; Dennis, E.S.; Munns, R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006, 142, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.N.; Lin, Z.R.; Cai, M.M.; Liu, R.W.; Wang, S.M.; Ma, Q. PcCLCg is involved in the accumulation of Cl− in shoots for osmotic adjustment and salinity resistance in the Cl−-tolerant xerophyte Pugionium cornutum. Plant Soil. 2023. [Google Scholar] [CrossRef]
- Apse, M.P.; Blumwald, E. Na+ transport in plants. FEBS Lett. 2007, 581, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.T.; Agorio, A.; Jossier, M.; Depre, S.; Thomine, S.; Filleur, S. Characterization of the chloride channel-like, AtCLCg, involved in chloride tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2016, 57, 764–775. [Google Scholar] [CrossRef]
- Teng, X.X.; Cao, W.L.; Lan, H.X.; Tang, H.J.; Bao, Y.M.; Zhang, H.S. OsNHX1, an Na+/H+ antiporter gene, can enhance salt tolerance in rice plant through more effective accumulation of toxic Na+ in leaf mesophyll and bundle sheath cells. Acta Physiol. Plant. 2007, 39, 113–125. [Google Scholar] [CrossRef]
- Bao, A.K.; Du, B.Q.; Touil, L.; Kang, P.; Wang, Q.L.; Wang, S.M. Co-expression of tonoplast cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol. J. 2016, 14, 964–975. [Google Scholar] [CrossRef]
- Lin, S.H.; Kuo, H.F.; Canivenc, G.; Lin, C.S.; Lepetit, M.; Hsu, P.K.; Tillard, P.; Lin, H.L.; Wang, Y.Y.; Tsai, C.B.; Gojon, A.; Tsay, Y.F. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef]
- Song, Y.; Li, S.; Zheng, H.; Han, G.; Sun, X.; Yang, W.; Wang, H.; Zhuang, K.; Kong, F.; Meng, Q.; Sui, N. SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sweet sorghum. Theor. Appl. Genet. 2022, 135, 201–206. [Google Scholar] [CrossRef]
- Nouri, M.Z.; Moumeni, A.; Komatsu, S. Abiotic stresses: Insight into gene regulation and protein expression in photosynthetic pathways of plants. Int. J. Mol. Sci. 2015, 16, 20392–20416. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Costa, J.M.; Zarrouka, O.; Pinheiro, C.; Lopes, C.M.; Pereira, J.S. Controlling stomatal aperture in semi-arid regions: The dilemma of saving water or being cool? Plant Sci. 2016, 251, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Yang, Z.; Liu, M.; Wang, B. Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 2015, 16, 534. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Yue, L.J.; Zhang, J.L.; Wu, G.Q.; Bao, A.K.; Wang, S.M. Sodium chloride improves photosynthesis and water status in the succulent xerophyte Zygophyllum xanthoxylum. Tree Physiol. 2012, 32, 4–13. [Google Scholar] [CrossRef]
- Inskeep, W.P.; Bloom, P.R. Extinction coefficients of chlorophyll-a and chlorophyll-b in N,N-dimethylformamide and 80-percent acetone. Plant Physiol. 1985, 77, 483–485. [Google Scholar] [CrossRef]
- Wang, S.M.; Zhang, J.L.; Flowers, T.J. Low-affinity Na+ uptake in the halophyte Suaeda maritima. Plant Physiol. 2007, 145, 559–571. [Google Scholar] [CrossRef]
- Drechsler, N.; Zheng, Y.; Bohner, A.; Nobmann, B.; von Wiren, N.; Kunze, R.; Rausch, C. Nitrate-dependent control of shoot K homeostasis by the nitrate transporter1/peptide transporter family member NPF7.3/NRT1.5 and the stelar K+ outward rectifier SKOR in Arabidopsis. Plant Physiol. 2015, 169, 2832–2847. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- De Vega, J.J.; Teshome, A.; Klaas, M.; Grant, J.; Finnan, J.; Barth, S. Physiological and transcriptional response to drought stress among bioenergy grass Miscanthus species. Biotechnol. Biofuels 2021, 14, 60. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



