1. Background:
Human papillomavirus (HPV) is responsible for most human reproductive tract viral infections. Most HPV infections are asymptomatic and self-limiting, chronic infections can progress into warts in precancerous, cervical, anogenital or oropharyngeal regions in men and women. The disease that is most frequently associated with HPV is cervical cancer. Though, majority of HPV pre-cancerous lesions have tendency to disappear on their own yet there remains a risk for every woman with HPV infection to become persistent and pre-cancerous leading to invasive cervical cancer [
1].
Cervical cancer is the leading cause of death in women. According to GLOBOCAN 2020, the incidence rate of cervical cancer was 15.6% and the mortality rate was 8.8%, worldwide. The age-specific standardized rate of cervical cancer was 13.3%. In Asia, the incidence rate of cervical cancer was 58.2 per cent. The five-year prevalence of cervical cancer was 59.5% in Asia. In India, the incidence and mortality rates were 16.2% and 9.5%, respectively. The proportion of cervical cancer in India was 7.9 per 100,000 [
2]. In India, 22% of women have undergone cervical screening examinations based on an NFHS report [
3]. According to the World Health Organization, 99% of cervical cancer cases were associated with high-risk human papillomavirus [
1]. Among the Indian population, the prevalence of cervical cancer was higher among sex workers, in an urban slum in Mumbai and HIV-positive women. HPV 16 and 18 were observed in 56% of cases in the West Indian region [
4,
5]. Human papillomavirus (HPV) is members of the papillomaviridae family are small, non-enveloped circular sds DNA virus. The DNA molecule is 8000 base pairs in size and the genome has six early (E) E1-E2, E4-E7 regions and 2 late (L) L1 and L2 regions [
6].
Papillomavirus categorised into alpha-papillomavirus, beta-papillomavirus, gamma-papillomavirus, delta-papillomavirus, and zeta- papillomavirus and theta- papillomavirus. The alpha and beta papillomaviruses were commonly detected in human samples. Alpha- papillomavirus causes mucosal and cutaneous lesions. Based on Molecular data further alpha-papillomavirus is classified as low-risk and high-risk. HPV-32, HPV-10, HPV-61, HPV-76, HPV-54, HPV-71 cause’s low-risk benign tumors (mucosal lesion and cutaneous lesion). However, HPV-2, HPV-26, HPV-34,HPV-16,HPV-18,and HPV-53 are responsible for high risk mucosal lesions. Beta-papillomavirus cause malignant cutaneous lesions. Among beta-papilloma virus, HPV 5 and 9 were most commonly associated with benign lesions in immunosuppressed patients. HPV 4, 48, 50, and 60 were responsible for cutaneous lesions. Other papilloma virus such as Delta-papilloma virus, zeta papilloma virus, eta-papillomavirus, and theta-papillomavirus were responsible for lesions in cattle, horses, and birds [
7]. Over 200 different HPV genotypes have been characterised conforming to the >10% diversity within the L1 gene sequence [
8,
9].
High-risk HPV types are 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, 53, 30, 66 and 82. Low-risk HPV types are 6, 11, 40, 42, 43, 44, 54, 61, 70, 72, 81, and CP6108. This epidemiological classification and the classification based on phylogenetic grouping were pretty close to each other. HPV16 and HPV18 are two particularly potent and obviously oncogenic [
7,
8]. Cervical cancer was attributable to one of 13 HPV types (HPV16, 18, 33, 31, 45, 56, 35, 52, 56, 58, 59, and 68) as cancer-associated agent, whereas Eight HPV types (HPV26, 53, 66, 67, 68, 70, 73, and 82) that were phylogenetically related to 12 WHO-defined high-risk (HR) HPV have consistently been identified as single HPV infections and categorized as potentially carcinogenic possible (p) HR-HPV [
10].
Andaman & Nicobar Islands are situated in the southern regions of the Bay of Bengal in the Indian Ocean, closer to Indonesia and Thailand. According to Census of India (2011), the territory’s population was 380,581, and the female population is 177,710 (46.7%). Literacy rate of Andaman and Nicobar Islands is 77.3% [
11].
Our previous study in Andaman and Nicobar islands reported the high-risk HPV types (HPV 16 and 18). This was the first of its kind study to find HPV types in these islands [
12]. HPV variants were not studied in detail among the population in this region so far. It is necessary that the public health system should be aware of the circulating variants of local strain pattern of HPV to frame recommendation for developing appropriate broad spectrum vaccine aiming at HR-HPV variants. To our knowledge, this study was the first cross-sectional survey conducted among a large population across Andaman and Nicobar Islands. Further, the current study aims to know the variants of HPV among married women in the Andaman and Nicobar Islands.
2. Methodology
A community-based cross-sectional study was conducted among married women of reproductive age (18-59 years) residing in the South Andaman District of the Andaman and Nicobar Islands, India. Patients were excluded if there was evidence of pregnancy, severe gynaecological bleeding, hysterectomy or previous history of the disease, including cancer, warts and other cutaneous manifestations.
This study has been approved by the Institutional Human Ethics Committee of ICMR-Regional Medical Research Centre, Port Blair [IEC No: 03/RMRC/29/06/2017]. The target population was chosen via cluster sampling and the sampling units were villages or municipal wards. After stratifying the sampling units into rural/urban strata, the required sample size was determined by random selection of the required sample size’s units. Based on the Andaman population ratio the study participants were drawn from rural villages and urban wards in a ratio of 2.5:1, yielding a sample of 700 from the rural and 300 from the urban.
Initially, awareness programmes were conducted in each selected village/ward at the Anganwadi centres/ community hall. The health care team (clinician along with trained nurses) were detailed about the health issues like cervical cancer, its symptoms, and genital hygiene. In addition, the need for the study was also explained and requested for written informed consent before enrollment.
The enrolled women were called to the field clinics which were held in the sub-centres, PHCs, CHCs and District Hospitals catering the population of the particular village/wards. The cervical scrapes were collected using the standard procedure from the ectocervix or surface of the cervical portion using a cytobrush. Specimens were collected in a tube containing phosphate-buffered saline (pH 8.6) and transported to the laboratory in ICMR-Regional Medical Research Centre, Port Blair by maintaining a cold chain.
Once the samples arrived at the laboratory, the specimen tubes were vortexed, cytobrushes were discarded and tubes were centrifuged to pellet the cells, which were suspended in 1 mL of phosphate-buffered saline. Aliquots of each fresh specimen were made and stored for a short duration at −20 °C until further processing. The analysis of HPV DNA was performed in the molecular biology laboratory of ICMR-Regional Medical Research Centre, Port Blair.
The total DNA was extracted with the QIAamp DNA Minikit (Qiagen, Germany) according to the manufacturer’s instructions. The DNA was eluted in 45 μL of elution buffer. The isolated DNA was amplified with ß-globin (internal control) to ensure the purity of the DNA extractions, as described previously [
13].
To confirm the HPV infection, the DNA of all the samples were subjected to PCR amplification targeting the L1 consensus gene, by a standard procedure reported previously.8,9 The results were recorded as positive if amplicon size specific to the 450bp DNA band was observed in agarose gel electrophoresis.
PCR for detection of type specific HR-HPV 16 & 18 the predominant genotypes was done [
13,
14]. In addition, the E6 and E7 genes of HPV16, and the E6 gene of HPV18 were also amplified to identify the lineages and sub-lineages of HR-HPV 16 & 18 in the South Andaman Islands [
14].
The DNA sequence analysis was carried out to confirm the HPV types distributed in South Andaman. L1 gene PCR amplicons of all the samples negative for HPV16 and 18 as well as a few samples of HPV16 confirmed were subjected to DNA sequence analysis. In addition, the E6 gene and E7 gene PCR amplicons of HPV16, and the E6 gene of HPV18 were also subjected to DNA sequencing. DNA sequencing was carried out by the Sanger sequencing method with corresponding primer sets [
15].
The DNA sequences were assembled using the MEGA11 software tool and were analysed together with worldwide diverse HPV sequences using ClustalW multiple alignments and pairwise alignment for phylogenetic analysis and were subsequently analyzed using Kimura’s two-parameter model as a method of substitution and neighbour-joining to reconstruct the phylogenetic tree. The statistical significance of the relationships obtained was estimated by bootstrap resampling analysis (1000 repetitions). A similar analysis was performed for the E6 and E7 genes of HPV16 and E6 gene HPV18.
The phylogenetic trees depicting the evolutionary relationship between taxonomic groups were generated for L1 genes of HPVs, E6 and E7 genes HPV16, and E6 gene of HPV18 sequences using molecular evolutionary genomic analyser software MEGA 11 [
16]. Genetic distances were calculated by using the Kimura 2 parameter (K2P) model at the nucleotide level and phylogenetic trees were constructed by using the neighbour-joining method. The reliability of the phylogenetic trees was tested using the bootstrap test with 1000 bootstrap replications.
3. Results:
All of the cervical samples tested positive for the β-globin gene, indicating that there were adequate cells in the samples. Out of 1000 samples screened, 50 specimens tested positive for HPV L1 gene amplification. Subsequently, type-specific PCR for HR-HPV16 and 18 identified 32 patients positive for HR-HPV16 and four (04) patients positive for HR-HPV18. DNA sequencing for the L1 region was successful for 24 samples. The molecular evolutionary genetic analysis of sequences from South Andaman and worldwide was done, and the pairwise genetic distances between the closely related HPV types from worldwide are specified in
Table 1 given below.
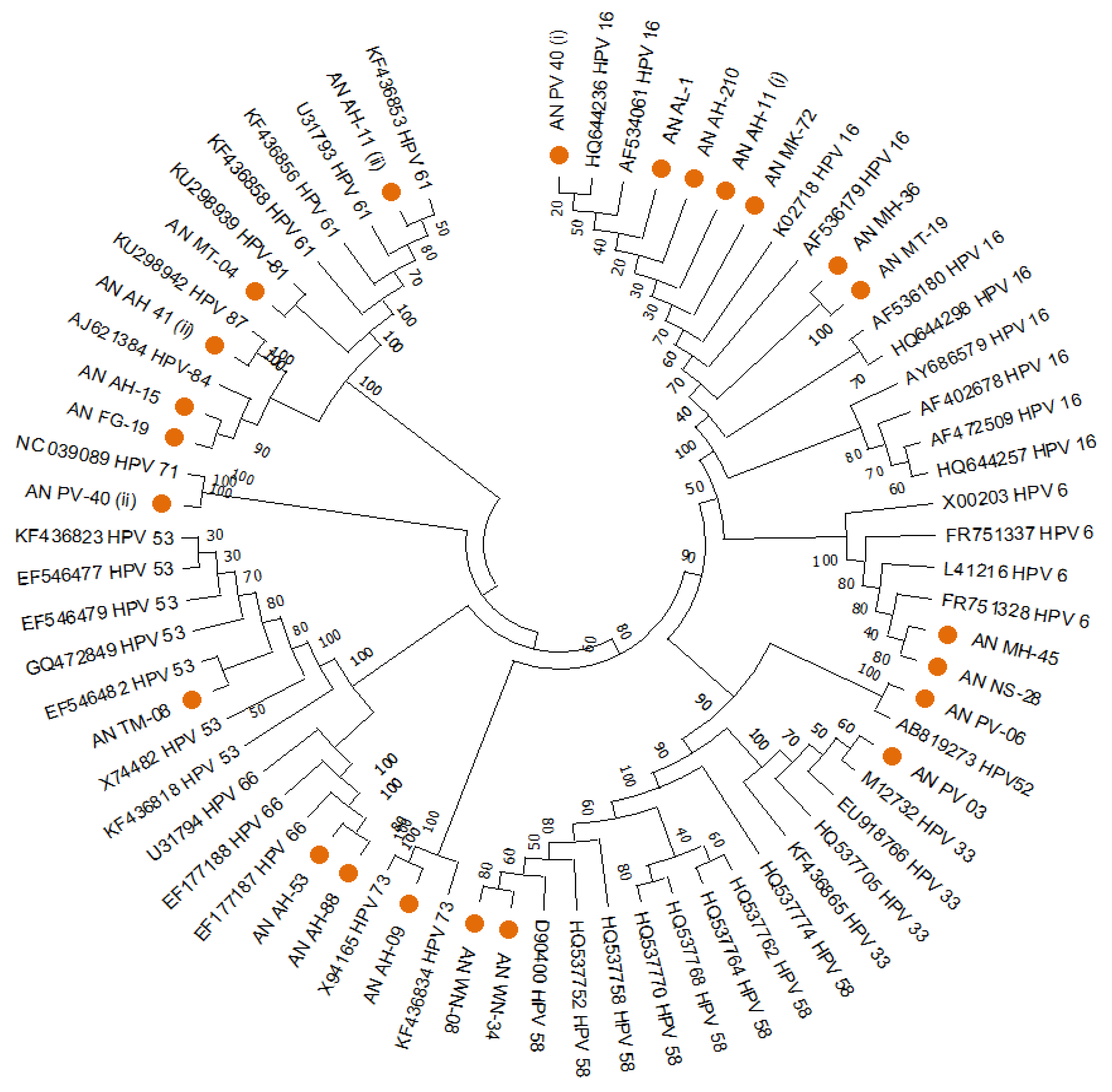
The HPVs found in South Andaman had genetic relatedness with various genotypes of worldwide, according to the phylogenetic analysis of the HPV-L1 partial gene. Out of the 24 sequences of the L1 region, 7 were clustered with HPV16, two with HPV6, one with HPV52, one with HPV33, two with HPV58, one with HPV73, two with HPV66, one with HPV53, one with HPV71, two with HPV84, one with HPV87, one with HPV81, one with HPV30, and one with HPV61. A MEGA11 software tool based Neighbour-joining phylogenetic analysis for the HPV-L1 partial gene exhibited the distribution of different HPV types in South Andaman Island (
Figure 1).
The distribution of the multiple genotypes of HPV in South Andaman could be determined by a combined analysis that uses a specific PCR and DNA sequence analysis.
Table 2 lists the genotype, risk group, frequency, and percentage of HPV detected in the South Andaman Island. The molecular evolutionary genetic analysis could identify the distribution of HPV types 16, 52, 58, 66, 33, 18, 73, 53, 30, 6, 61, 71, 81, 84 and 87. There were high-risk as well as low-risk types prevalent among the women in South Andaman Island.
Among the 50 samples identified by the HPV L1 gene, six cases had HPV co-infections. One case was found to be co-infected with 3 HPVs 16, 18 &71. Five cases were identified with co-infection with two HPVs i.e., (16 &18, 16&61, 16& 84, 18& 30 and 66 & 87). All the co-infected cases were found to have at least one High-risk type association. However, HPV type could not be identified in 2 samples due to exhaustion of specimens for repeated experiments.
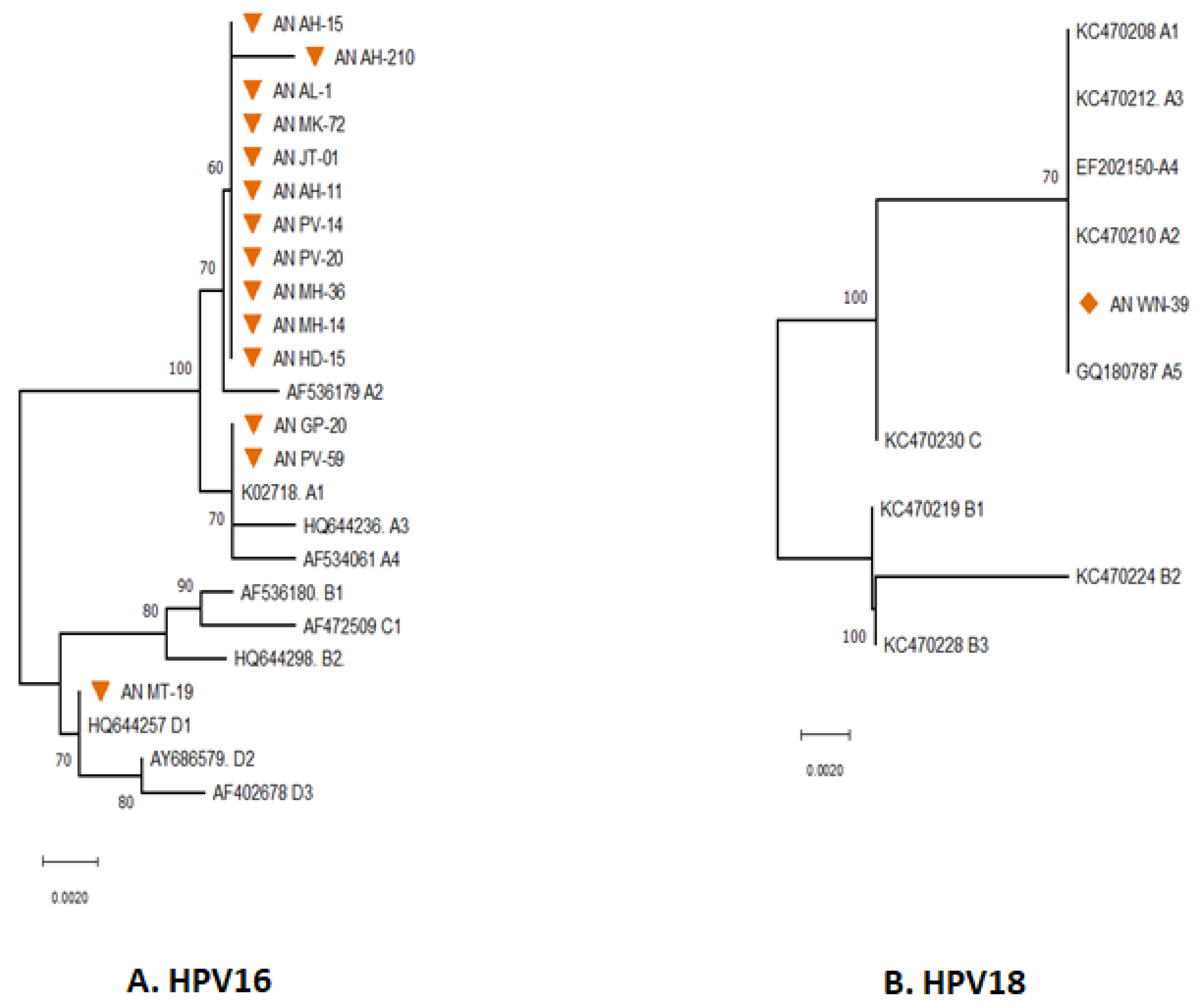
The majority of the HPV types distributed in South Andaman were found to be HPV16 followed by HPV18. The distribution of various sub-lineages in South Andaman Island was revealed by phylogenetic analysis of the HPV16 partial E6 gene (
Figure 2 A). Thirteen of the fourteen were found to be associated with lineage A, and the remaining one was associated with lineage D. Further analysis to identify the HPV16 sub-lineages revealed that 11 sequences had close genetic relatedness with reference sequence AF536179, which belongs to the sub-lineage type A2. Two sequences had close genetic relatedness with K02718 of the sub-lineage type A1. One sequence had close genetic relatedness with HQ644257, which is of the sub-lineage type D1.
The partial E6gene sequence HPV16 were analysed to understand the lineages circulating in South Andaman. The phylogenetic analysis revealed that all the HPV16 identified in South Andaman Island were found to be Lineage-A. The majority of HPV16 sequences were grouped with AF536179 which belongs to the sub-lineage A2 (K2P = 0.002%). One isolate (MT-19) identified from South Andaman was grouped with the HQ644257 which belongs to the D1 lineage (K2P = 0.00%). The pairwise genetic distance between the two isolates from South Andaman and the European isolate (K02718) was found that the South Andaman isolates belong to the A1 lineage (0.000%).
Analysis to identify the HPV18 sub-lineages revealed that the partial E6 gene of HPV18 had close genetic relatedness with reference sequences of HPV18 Lineages A. The E6 gene of HPV18 identified from South Andaman was associated with the lineage A1 to A5 (K2P = 0.00%). However, there were no genetic differences within the lineages of the E6 partial gene region sequenced to identify the sub-lineage. E7 gene of both HPV16 & 18 did not show major lineage discrimination, hence E7 was not considered significant.
4. Discussion:
The current study provided the diversity of HPV among women in Andaman and Nicobar Island. It is essential to comprehend the spectrum of HPV genotypes because data on the distribution of HPV genotypes is relevant to vaccine development. The key etiological cause of cervical cancer is prolonged HR-HPV infection. HPV16 and HPV18 cause more than 70 per cent of cervical cancer cases, with the remaining cervical cancers caused by other HR-HPV genotypes [
17].
In India, a study found that HPV was 94.6% in women with ICC (Invasive Cervical Cancer), 86.5% in those with HSIL (High-grade Squamous Intraepithelial Lesions), and 65.4% in those with LSIL (Low-grade Squamous Intraepithelial Lesions). The majority of invasive cervical cancers were caused by HPV16 (64.8 %), followed by HPV18, 45, 33, 35, 58, 59, and 31. HPV 35 looked to be more common in the South, whereas HPV16 and HPV45 appeared to be more common in the North [
18,
19]. Also HPV16, HPV18 and HPV 58 was common among marginalised urban women in India [
19].
The phylogenetic analysis revealed the presence of 15 HPV genotypes in the South Andaman District, including High-Risk types 16, 18, 33, 52, 53, 58, 66 and 73 and Low-Risk types 6, 30, 61, 71, 81, 84 and 87. Globally, the most prevalent genotypes were HPV-16 and 18. Other HR-HPV genotypes were HPV-59, 73, 45, 31, 33, 52, 35, 39, and 68 [
20]. Prevalence of HPV type 16 and 18, in Asia is 55.1% and 13.8%. Other types such as 31, 33, 35, 39, 52, 58, and 59 were also reported in Asia. HPV prevalence was associated with invasive cervical cancer [
21]. In India, the prevalence of cervical cancer associated with HPV 16 and HPV18 are 69.7% and 13.5%. Prevalence of other HR-HPV types such as HPV45, 33, 35, 58, 31, 59, and 52 are 5.1%, 4%, 2.5%, 2.4%, 2.3%, 1.4%, and 1.4%. [
22].
HR-HPV infection was associated with age, the number of sexual partners, the use of hormonal contraception, and smoking. The strongest correlation between HPV 16 and aberrant cytology was found across all age groups [
23]. Many studies showed the predominance of HR-HPV types i.e., HPV16, followed by HPV31, HPV58, HPV68, HPV18, and HPV56 with cytological abnormalities [
24].
HPV16 was the most prevalent strain in Central India. The other prevalent high-risk HPV types were types 18, 31, 35, 45, 56, and 59. Oral and vulva/vagina malignancies were exclusively related to HPV16 [
25]. Another study from Madhya Pradesh detected HPV16 as a highly prevalent type found among ICC cases followed by HPV18, 45, 66, 35, and 56. Less prevalent types were HPV31, 51, 58, 59, 67, 82, and JEB2 [
26].
HPV16 genetic variation may have a significant impact on cervical cancer risk. However, the global burden of cervical cancer associated with various sub-lineages is predominantly driven by past HPV16 sub-lineage distribution. HPV-16 A1 sub-lineage was globally widespread. However, sub-lineages A3 and A4 were common in Asia. Sub-lineages A3, A4 and lineage D were common in regions like East Asia and North America. Sub lineage A4 were associated with more severity disease status than A1-3 sub lineage in Chinese females and higher risk of cancer. These lineages were highly cancer-risk associated [
27]. The current study found sub-lineages A1, A2 and D1 to be prevailing in South Andaman. The majority of the isolates from the current study belonged to A2 sub-lineage. HPV18 sub-lineage distribution in China belonged to A1 to A7 [
28,
29]. Another study from Iran found that the prevalence of sub-lineage A4 was high compared to other sub-lineages [
30]. In this study, the South Andaman isolates belonged to lineage A.
Diversity in the distribution of HPV types gives rise to a challenge to vaccine strategies. Molecular surveillance of HPV is needed for the detection of new strains or types emerging among symptomatic and asymptomatic populations in these remote islands. This will help policymakers to implement preventive measures against HPV-associated cervical cancer. Study on specificity of HPV variants will be helpful to develop broad-spectrum vaccines aiming HR-HPV variants.
5. Conclusion:
This is the first-ever community-based cross-sectional study conducted in Andaman and Nicobar Islands which interestingly revealed prevalence of wide range of the genotype distribution of HPV among women in this small island. The knowledge about the regional circulating genotypes of HPV is essential to formulate an effective vaccine. These findings will emphasize and help to initiate stronger public health awareness programs and prevention strategies for the women of Andaman and Nicobar Islands. The findings in the current study provide sufficient data to highlight the important of screening for cervical cancer and promote vaccination and vaccine awareness in women living in remote rural geographical location.
Author Contributions
The study concept and study design were contributed by NM and RP. Data collection and sample collection was contributed by RP. Data analysis, interpretation, and critical evaluation were all contributed by RP, NM, PV, and HK. Manuscript writing and correction of manuscript were by all the authors. The final article was approved by all the authors. All authors read and approved the final manuscript.
Funding
This study was supported by the extramural funds of the Indian Council of Medical Research. [File no. Tribal/117/2020-ECD-II (2020-5180)].
Institutional Review Board Statement
This research was approved by the Institutional Human Ethical Committee (IHEC), ICMR- Regional Medical Research Centre, Port Blair, Andaman and Nicobar Islands (RMRC/2017/06/29/3).
Data Availability Statement
The datasets of the current study are available from the corresponding author upon reasonable request.
Acknowledgments
We acknowledge the excellent support of Directorate of public health services, Andaman and Nicobar Islands and extend our gratefulness to Director General and secretory, Department of Health Research, Indian Council of Medical Research (ICMR), Ministry of Health and Family welfare (MOHFW), India for conducting this research.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- World Health Organisation Team. Cervical cancer. World Health Organisation (WHO). 2023. https://www.who.int/news-room/fact-sheets/detail/cervical-cancer. Last accessed:. 20 April.
- International Agency for Research on Cancer (IARC). Global cancer observatory (GCO). World Health Organisation-International Agency for Research on Cancer. 2020. Available at http://gco.iarcfr/today/online-analysis. Last accessed:. 20 April.
- International Institute for Population Sciences (IIPS). National Family Health Survey (NFHS-4), India, 2015-2016. IIPS. 2017. Available at http://rchiips.org/nfhs/nfhs-4Reports/India.pdf Last accessed:. 12 November.
- 4. Thobias AR, Patel KA, Gokani R, Parekh C, Desai A, Patel JB. Prevalence of Human Papilloma Virus Infection in Cervical Cancer Patients from Western Region of India. Indian J Gynecol Oncol.
- Press, D. Epidemiology of cervical cancer with special focus on India. 2015;405–14.
- Graham, SV. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 1493. [Google Scholar] [CrossRef]
- INTERNATIONAL AGENCY FOR RESEARCH ON CANCER IARC Monographs on the Evaluation of Carcinogenic Risks to Humans VOLUME 90 Human Papillomaviruses.
- 8. Muñoz N, Bosch FX, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. [CrossRef]
- Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445(1-2):232-243. [CrossRef]
- Arbyn M, Tommasino M, Depuydt C, Dillner J. Are 20 human papillomavirus types causing cervical cancer?. J Pathol. 2014;234(4):431-435. [CrossRef]
- Office of the Registrar General & Census Commissioner, India. Census 2011. Ministry of Home affairs, Government of India. 2011. Available at https:// censusindia.gov.in/census.website/data/data-visualizations/PopulationSearch_ PCA_ Indicators. Last accessed:. 20 April.
- Parvez R, Hedau S, Bhattacharya D, et al. High-risk HPV infection among the tribal and non-tribal women of the Andaman and Nicobar Islands, India. Public Health. 2012;126(1):67-69. [CrossRef]
- Gopalkrishna V, Aggarwal N, Malhotra VL, Koranne RV, Mohan VP, Mittal A and Das BC. Chlamydia trachomatis and human papillomavirus infection in Indian women with sexually transmitted diseases and cervical precancerous and canerous lesions. European Society of Clinical Microbiology and Infectious Diseases 2000, 6, 88–93. [Google Scholar]
- Das BC, Sharma JK, Gopalkrishna V et al. A high frequency of human papillomavirus DNA sequences in cervical carcinomas of Indian women as revealed by southern blot hybridization and polymerase chain reaction. Journal of Medical Virology 1992, 36, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Pande S, Jain N, Prusty BK, et al. Human papillomavirus type 16 variant analysis of E6, E7, and L1 genes and long control region in biopsy samples from cervical cancer patients in north India. J Clin Microbiol. 2008;46(3):1060-1066. [CrossRef]
- Tamura, K. , Stecher G., and Kumar S. (2021). MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution. [CrossRef]
- 17. Li M, Du X, Lu M, Zhang W, Sun Z, Li L, Ye M, Fan W, Jiang S, Liu A, Wang M, Meng Y, Li Y. Prevalence characteristics of single and multiple HPV infections in women with cervical cancer and precancerous lesions in Beijing, China. J Med Virol.
- Bhatla N, Lal N, Bao YP, Ng T, Qiao YL. A meta-analysis of human papillomavirus type-distribution in women from South Asia: implications for vaccination. Vaccine. 2008;26(23):2811-2817. [CrossRef]
- Kranti Suresh Vora, Shahin Saiyed, Rajendra Joshi, Senthilkumar Natesan. Prevalence of high-risk HPV among marginalized urban women in India and its implications on vaccination: A cross sectional study. 01 September. [CrossRef]
- National Cancer Institute. HPV and cancer. National Institute of Health. 2023. Available at: https://www.cancer.gov/about-cancer/causes-prevention/risk/infectious-agents/hpv-and-cancer. Last accessed:. 20 April.
- Bruni L, Albero G, Serrano B, Mena M, Collado JJ, Gómez D, Muñoz J, Bosch FX, de Sanjosé S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in Asia. Summary Report . Last accessed: 20 April 2023. 10 March.
- Bruni L, Albero G, Serrano B, Mena M, Collado JJ, Gómez D, Muñoz J, Bosch FX, de Sanjosé S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in India. Summary Report . Last accessed: 20 April 2023. 10 March.
- Torres-Poveda K, Ruiz-Fraga I, Madrid-Marina V, Chavez M, Richardson V. High risk HPV infection prevalence and associated cofactors: a population-based study in female ISSSTE beneficiaries attending the HPV screening and early detection of cervical cancer program. BMC Cancer. 2019;19(1):1205. Published 2019 Dec 10. [CrossRef]
- Kovacevic G, Milosevic V, Knezevic P, Knezevic A, Knezevic I, Radovanov J, Nikolic N, Patic A, Petrovic V, Cvjetkovic IH, Stanisic L. Prevalence of oncogenic Human papillomavirus and genetic diversity in the L1 gene of HPV16 HPV 18 HPV31 and HPV33 found in women from Vojvodina Province Serbia. Biologicals. 2019 Mar 1;58:57-63.
- 25. Gheit T, Vaccarella S, Schmitt M, et al. Prevalence of human papillomavirus types in cervical and oral cancers in central India. Vaccine. [CrossRef]
- Munjal K, Adamson CS, Rajendran V, Nandedkar S, Cooper K, Evans MF. Human papillomavirus type distribution in invasive cervical cancers from Madhya Pradesh: implications for vaccination programs in central India. Int J Gynecol Pathol. 2014;33(5):531-536. [CrossRef]
- Clifford GM, Tenet V, Georges D, et al. Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: Whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res. 2019;7:67-74. [CrossRef]
- Zu Y, Ou Z, Wu D, et al. Genetic characteristics of human papillomavirus type 16, 18, 52 and 58 in southern China. Genomics. 2021;113(6):3895-3906. [CrossRef]
-
29. Xiuli Wang1†, Shuizhong Han2†, Xingwei Li1, Xiaochuan Wang1et.al.Prevalence and distribution of humanpapillomavirus (HPV) in Luoyang city of Henan province during 2015–2021 and the genetic variability of HPV16 and 52. Virology Journal (2022) 19:37. [CrossRef]
- Salavatiha Z, Shoja Z, Heydari N, et al. Lineage analysis of human papillomavirus type 18 based on E6 region in cervical samples of Iranian women [published online ahead of print, 2020 Jul 9]. J Med Virol. 2020;10.1002/jmv.26283. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).