Submitted:
30 May 2023
Posted:
30 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Phylogenetic tree analysis
2.2. Protein structural prediction and analysis
2.3. Mutational analysis
3. Results
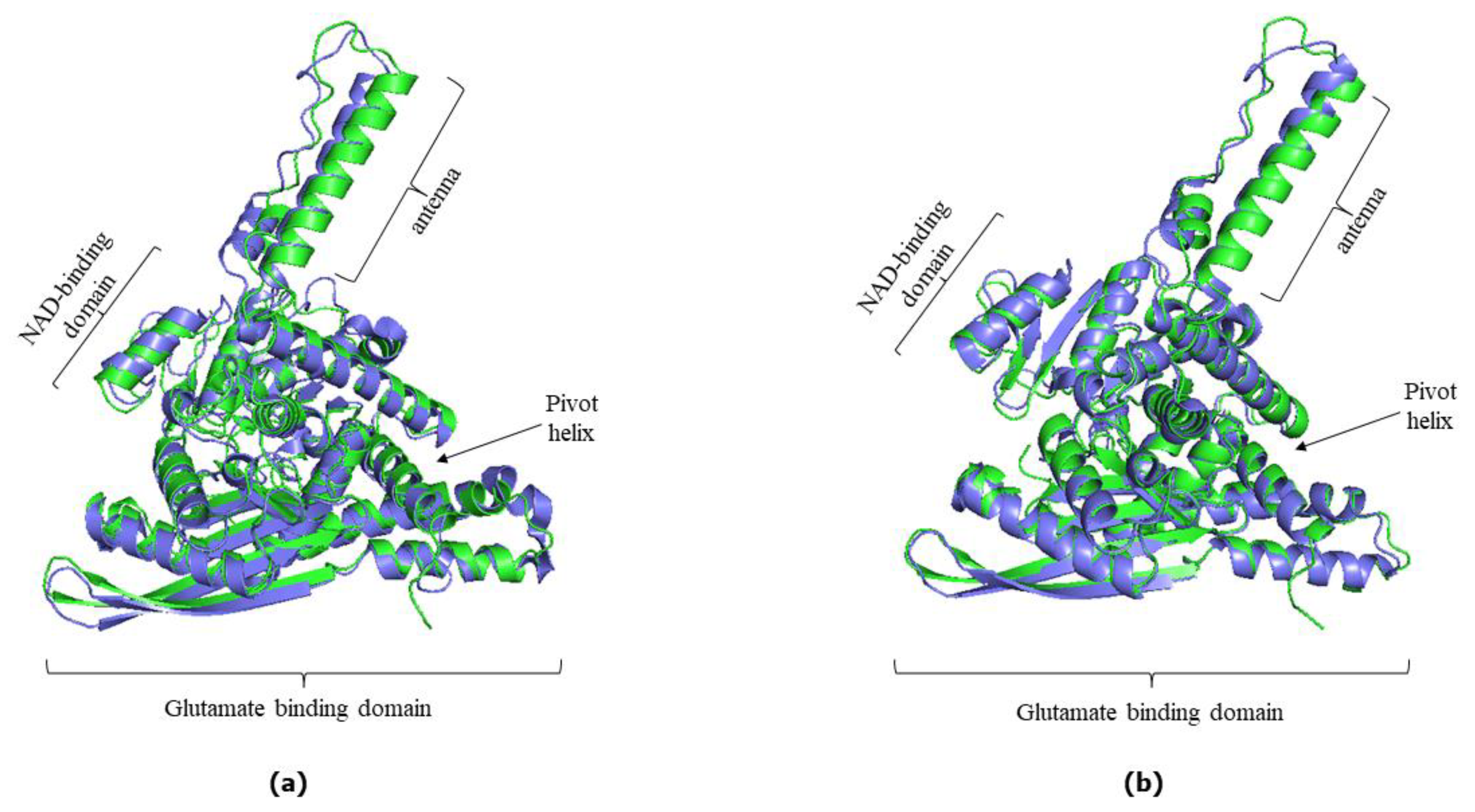
3.1. AlphaFold predicted Versus experimentally determined hGDH1 and hGDH2 structure
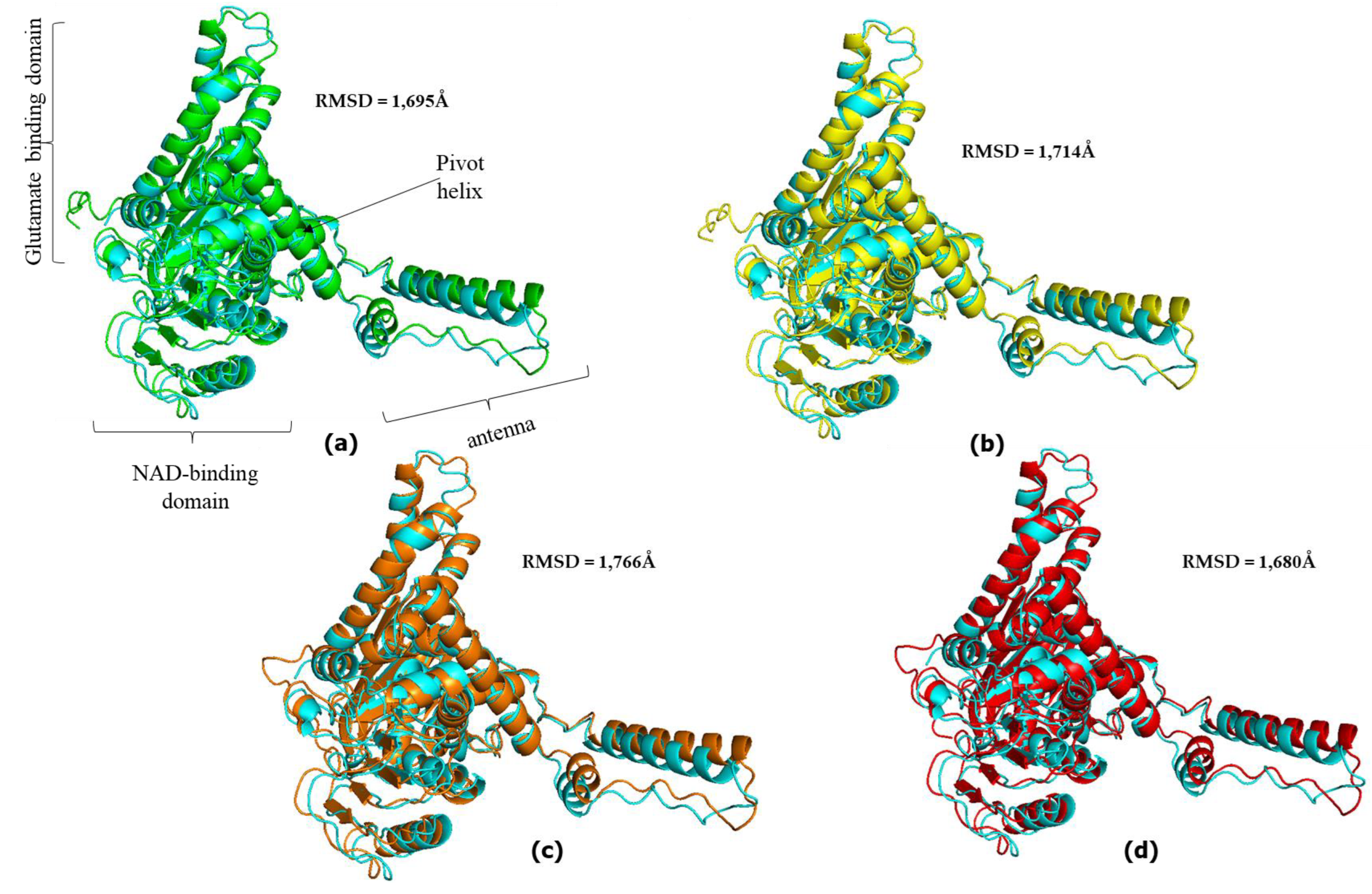
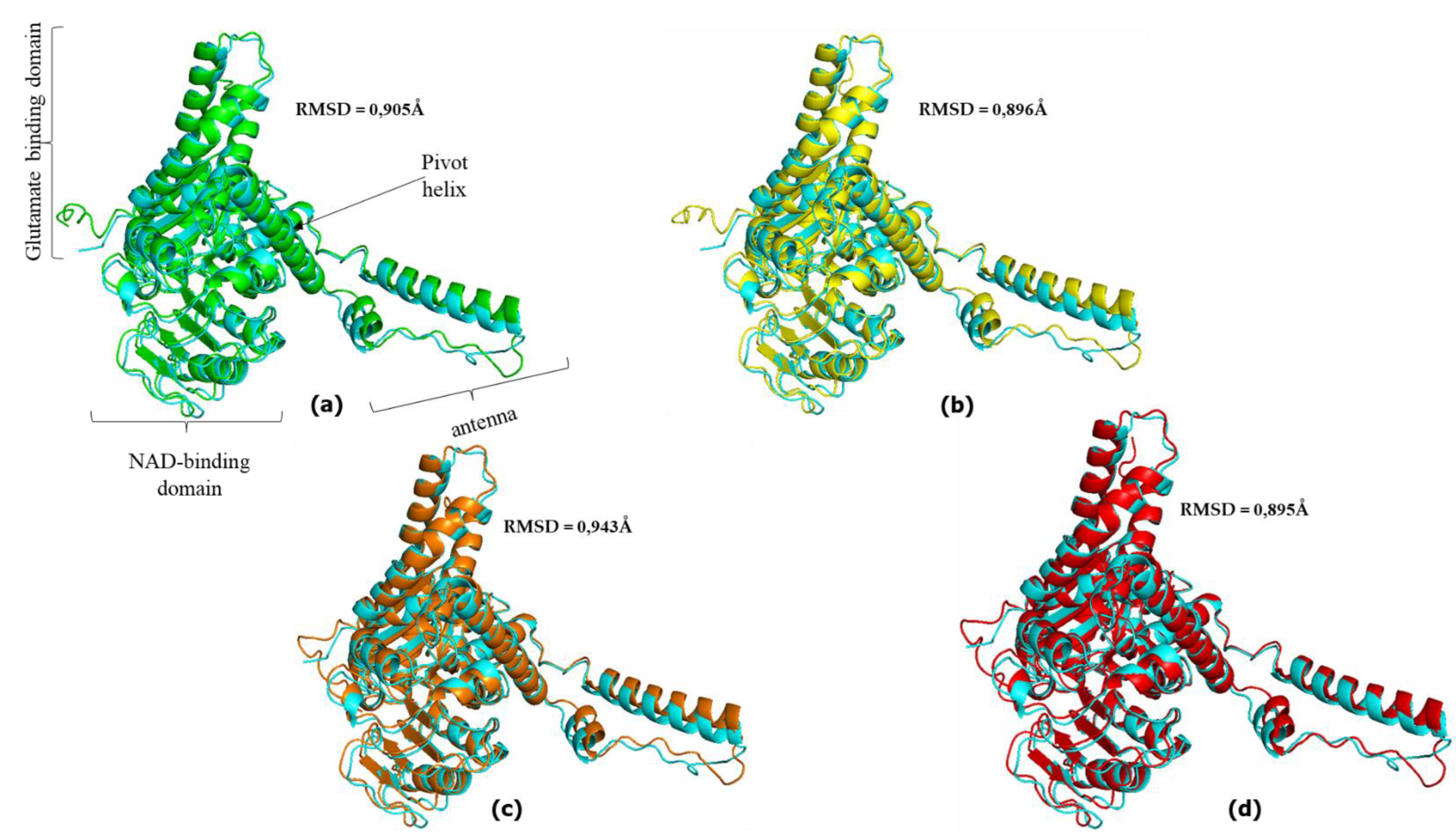
3.3. Great ape GDH2 AlphaFold Colab predicted structures and comparison with predicted hGDH2
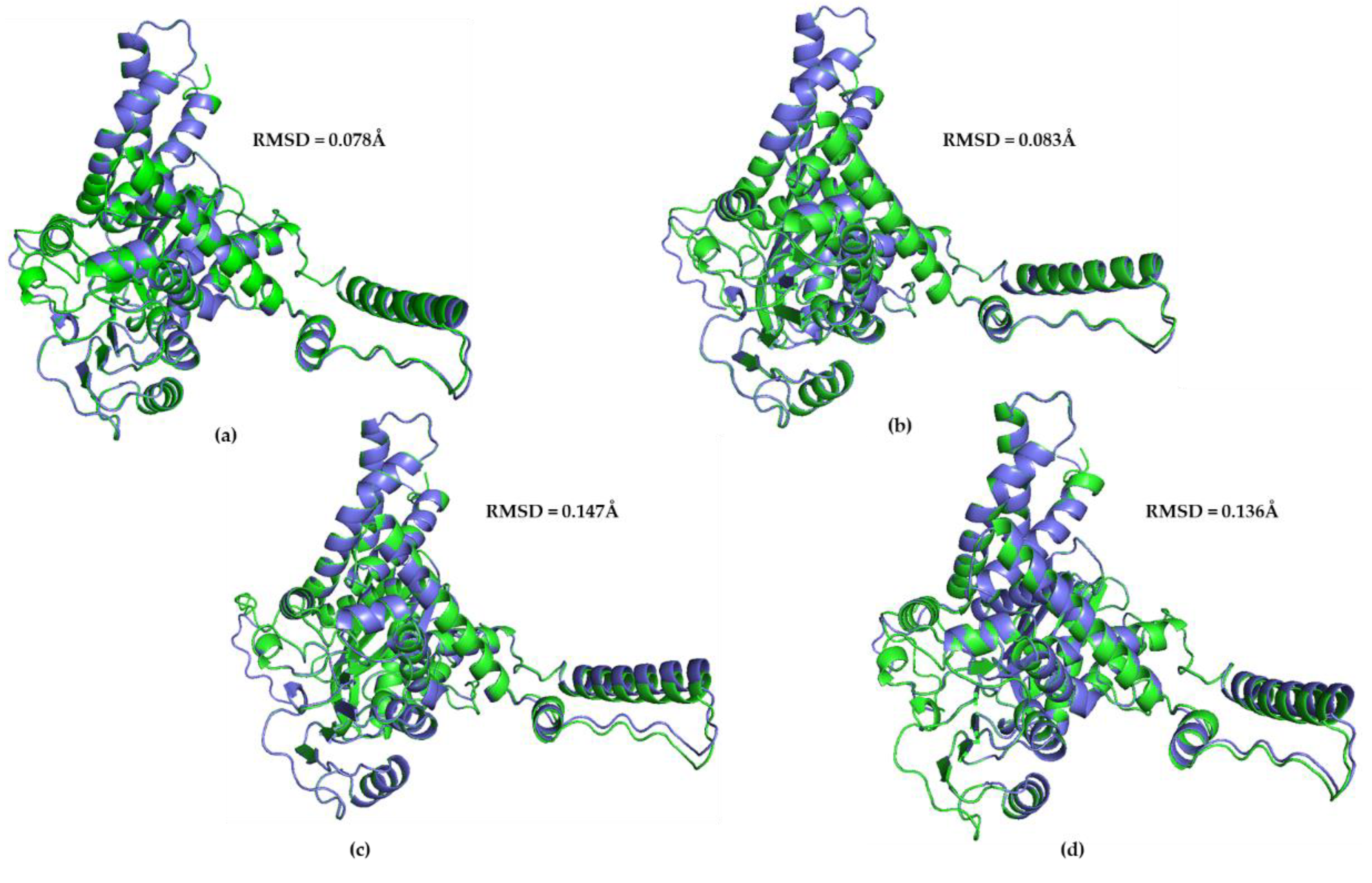
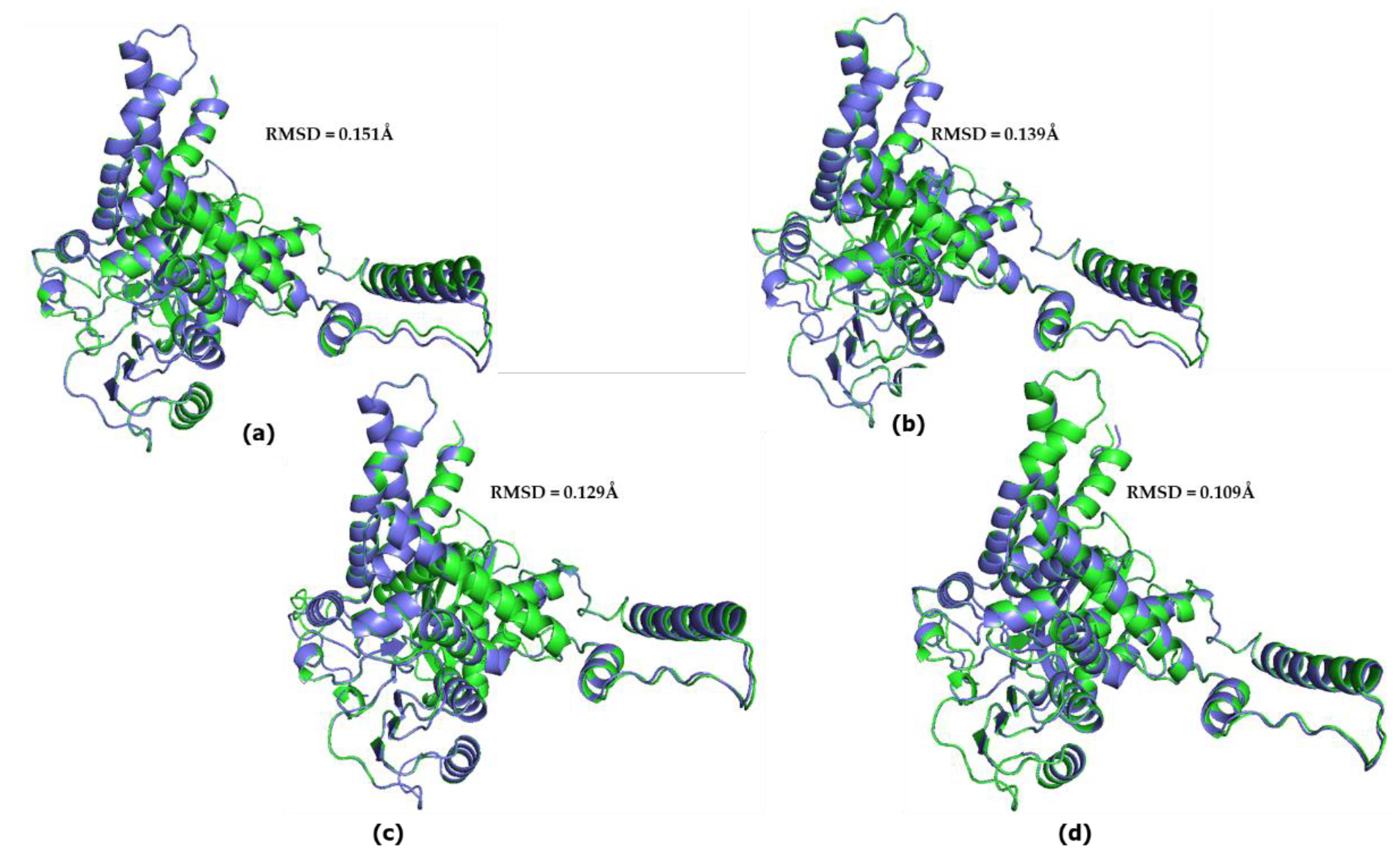
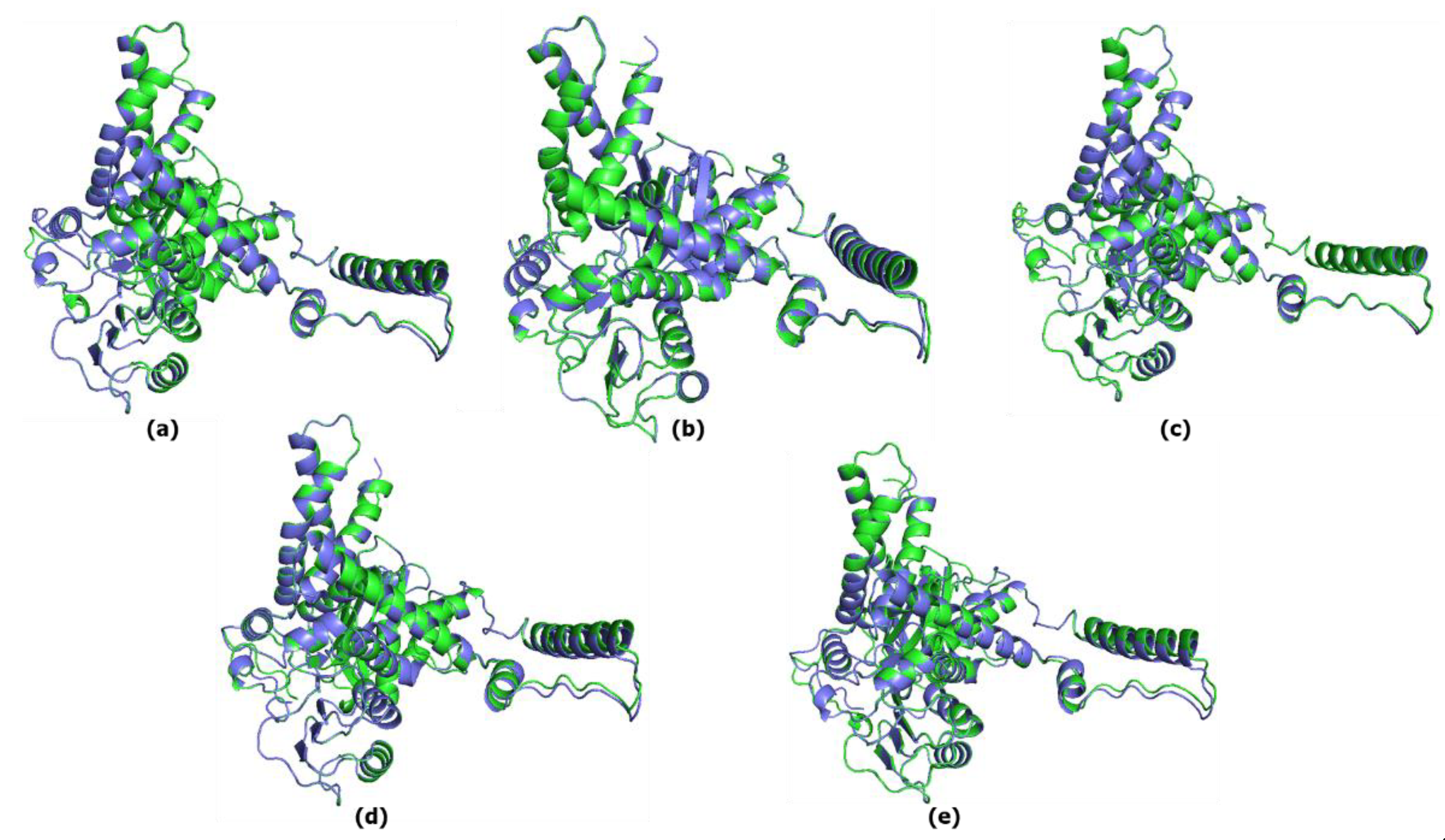
3.4. Great ape GDH2 AlphaFold Colab predicted structures during evolution
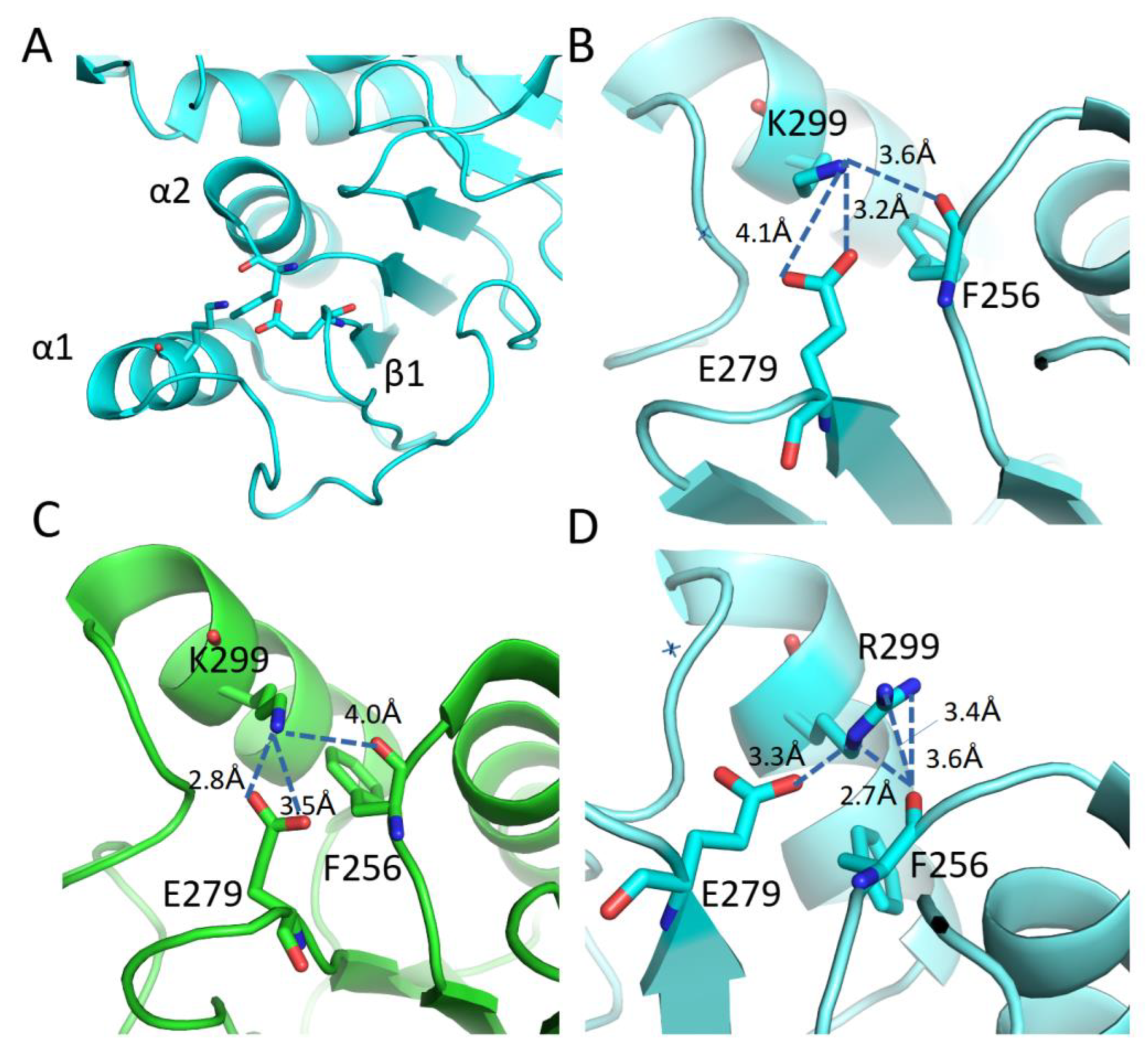
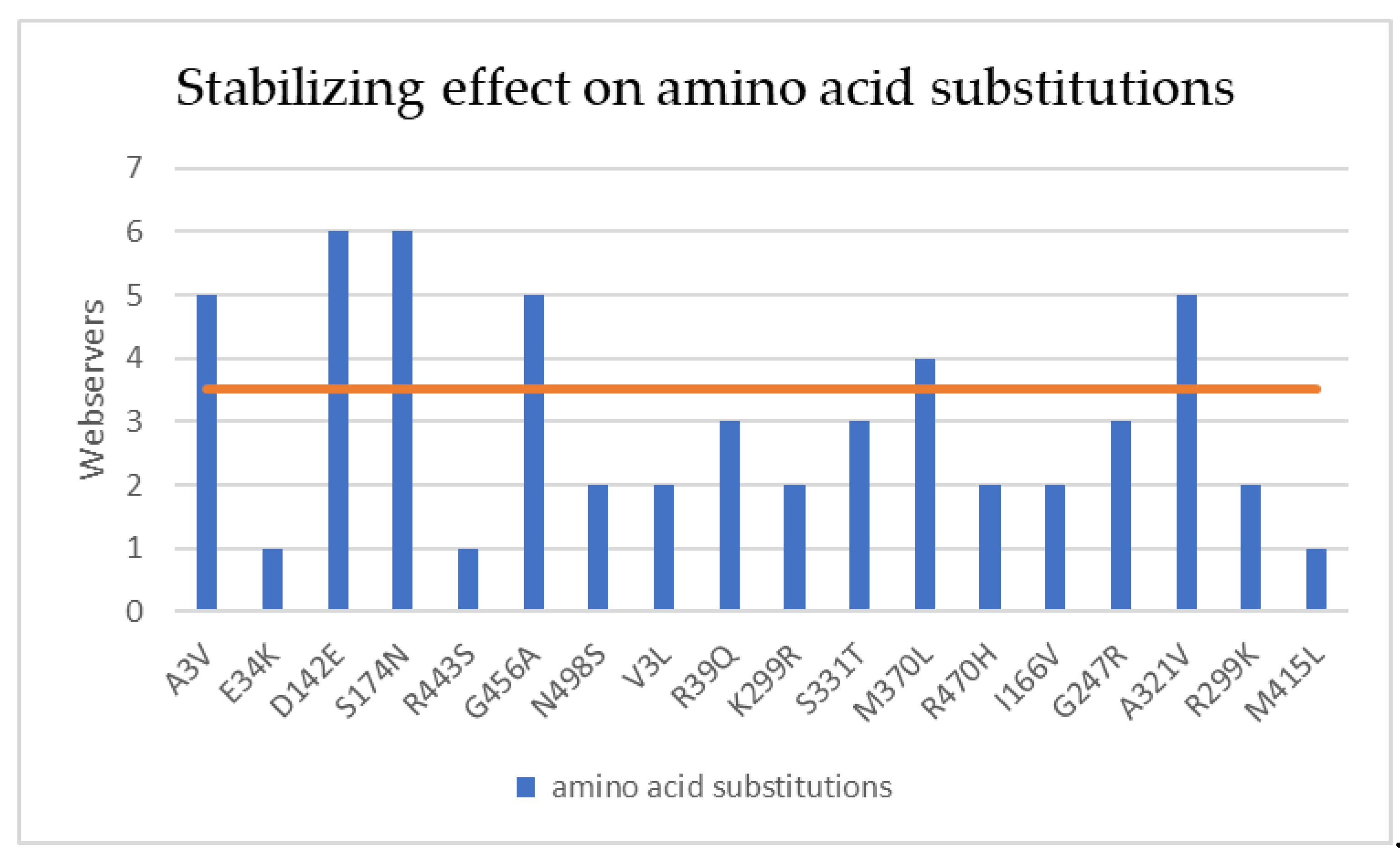
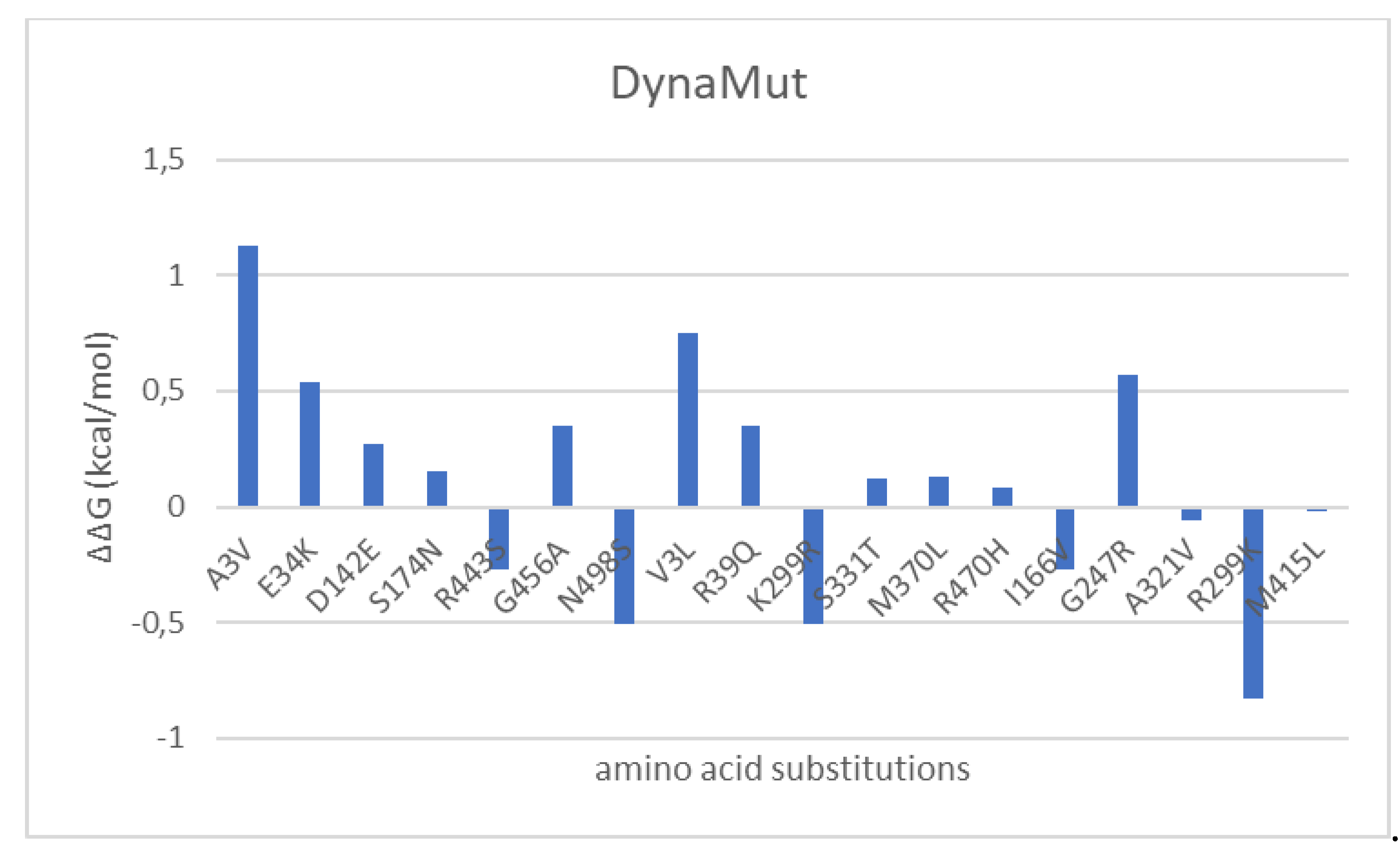
3.4. Mutational and intramolecular interactions analysis
4. Discussion
Strengths and limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hudson, R.C.; Daniel, R.M. L-Glutamate Dehydrogenases: Distribution, Properties and Mechanism. Comp. Biochem. Physiol. Part B Comp. Biochem. 1993, 106, 767–792. [Google Scholar] [CrossRef] [PubMed]
- Plaitakis, A.; Zaganas, I. Regulation of Human Glutamate Dehydrogenases: Implications for Glutamate, Ammonia and Energy Metabolism in Brain. J Neurosci Res 2001, 66, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Shashidharan, P.; Michaelidis, T.M.; Robakis, N.K.; Kresovali, A.; Papamatheakis, J.; Plaitakis, A. Novel Human Glutamate Dehydrogenase Expressed in Neural and Testicular Tissues and Encoded by an X-Linked Intronless Gene. Journal of Biological Chemistry 1994, 269, 16971–16976. [Google Scholar] [CrossRef] [PubMed]
- Burki, F.; Kaessmann, H. Birth and Adaptive Evolution of a Hominoid Gene That Supports High Neurotransmitter Flux. Nat Genet 2004, 36, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Kanavouras, K.; Mastorodemos, V.; Borompokas, N.; Spanaki, C.; Plaitakis, A. Properties and Molecular Evolution of Human GLUD2 (Neural and Testicular Tissue-Specific) Glutamate Dehydrogenase. Journal of Neuroscience Research 2007, 85, 3398–3406. [Google Scholar] [CrossRef] [PubMed]
- Kalef-Ezra, E.; Kotzamani, D.; Zaganas, I.; Katrakili, N.; Plaitakis, A.; Tokatlidis, K. Import of a Major Mitochondrial Enzyme Depends on Synergy between Two Distinct Helices of Its Presequence. Biochem J 2016, 473, 2813–2829. [Google Scholar] [CrossRef] [PubMed]
- Mastorodemos, V.; Kotzamani, D.; Zaganas, I.; Arianoglou, G.; Latsoudis, H.; Plaitakis, A. Human GLUD1 and GLUD2 Glutamate Dehydrogenase Localize to Mitochondria and Endoplasmic Reticulum. Biochem. Cell Biol. 2009, 87, 505–516. [Google Scholar] [CrossRef]
- Kotzamani, D.; Plaitakis, A. Alpha Helical Structures in the Leader Sequence of Human GLUD2 Glutamate Dehydrogenase Responsible for Mitochondrial Import. Neurochemistry International 2012, 61, 463–469. [Google Scholar] [CrossRef]
- Zaganas, I.; Spanaki, C.; Karpusas, M.; Plaitakis, A. Substitution of Ser for Arg-443 in the Regulatory Domain of Human Housekeeping (GLUD1) Glutamate Dehydrogenase Virtually Abolishes Basal Activity and Markedly Alters the Activation of the Enzyme by ADP and L-Leucine. J Biol Chem 2002, 277, 46552–46558. [Google Scholar] [CrossRef]
- Zaganas, I.V.; Kanavouras, K.; Borompokas, N.; Arianoglou, G.; Dimovasili, C.; Latsoudis, H.; Vlassi, M.; Mastorodemos, V. The Odyssey of a Young Gene: Structure-Function Studies in Human Glutamate Dehydrogenases Reveal Evolutionary-Acquired Complex Allosteric Regulation Mechanisms. Neurochem Res 2014, 39, 471–486. [Google Scholar] [CrossRef]
- Zaganas, I.; Plaitakis, A. Single Amino Acid Substitution (G456A) in the Vicinity of the GTP Binding Domain of Human Housekeeping Glutamate Dehydrogenase Markedly Attenuates GTP Inhibition and Abolishes the Cooperative Behavior of the Enzyme *. Journal of Biological Chemistry 2002, 277, 26422–26428. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.A.; Lieu, Y.K.; Hsu, B.Y.; Burlina, A.B.; Greenberg, C.R.; Hopwood, N.J.; Perlman, K.; Rich, B.H.; Zammarchi, E.; Poncz, M. Hyperinsulinism and Hyperammonemia in Infants with Regulatory Mutations of the Glutamate Dehydrogenase Gene. N Engl J Med 1998, 338, 1352–1357. [Google Scholar] [CrossRef] [PubMed]
- Spanaki, C.; Zaganas, I.; Kleopa, K.A.; Plaitakis, A. Human GLUD2 Glutamate Dehydrogenase Is Expressed in Neural and Testicular Supporting Cells. J Biol Chem 2010, 285, 16748–16756. [Google Scholar] [CrossRef] [PubMed]
- Mathioudakis, L.; Dimovasili, C.; Bourbouli, M.; Latsoudis, H.; Kokosali, E.; Gouna, G.; Vogiatzi, E.; Basta, M.; Kapetanaki, S.; Panagiotakis, S.; et al. Study of Alzheimer’s Disease- and Frontotemporal Dementia-Associated Genes in the Cretan Aging Cohort. Neurobiol Aging 2022, S0197-4580(22)00146-4. [CrossRef]
- Takeuchi, Y.; Nakayama, Y.; Fukusaki, E.; Irino, Y. Glutamate Production from Ammonia via Glutamate Dehydrogenase 2 Activity Supports Cancer Cell Proliferation under Glutamine Depletion. Biochem Biophys Res Commun 2018, 495, 761–767. [Google Scholar] [CrossRef]
- Smith, T.J.; Schmidt, T.; Fang, J.; Wu, J.; Siuzdak, G.; Stanley, C.A. The Structure of Apo Human Glutamate Dehydrogenase Details Subunit Communication and Allostery. J Mol Biol 2002, 318, 765–777. [Google Scholar] [CrossRef]
- Smith, T.J.; Peterson, P.E.; Schmidt, T.; Fang, J.; Stanley, C.A. Structures of Bovine Glutamate Dehydrogenase Complexes Elucidate the Mechanism of Purine Regulation. J Mol Biol 2001, 307, 707–720. [Google Scholar] [CrossRef]
- Dimovasili, C.; Fadouloglou, V.E.; Kefala, A.; Providaki, M.; Kotsifaki, D.; Kanavouras, K.; Sarrou, I.; Plaitakis, A.; Zaganas, I.; Kokkinidis, M. Crystal Structure of Glutamate Dehydrogenase 2, a Positively Selected Novel Human Enzyme Involved in Brain Biology and Cancer Pathophysiology. J Neurochem 2021, 157, 802–815. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved Protein Structure Prediction Using Potentials from Deep Learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Nei, M. MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinform 2004, 5, 150–163. [Google Scholar] [CrossRef]
- Chen, C.-W.; Lin, J.; Chu, Y.-W. IStable: Off-the-Shelf Predictor Integration for Predicting Protein Stability Changes. BMC Bioinformatics 2013, 14 Suppl 2, S5. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, H.; Zhang, N.; Zhu, Z.; Wang, S.; Li, M. PremPS: Predicting the Impact of Missense Mutations on Protein Stability. PLoS Comput Biol 2020, 16, e1008543. [Google Scholar] [CrossRef] [PubMed]
- Laimer, J.; Hiebl-Flach, J.; Lengauer, D.; Lackner, P. MAESTROweb: A Web Server for Structure-Based Protein Stability Prediction. Bioinformatics 2016, 32, 1414–1416. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A Server for Predicting Effects of Mutations on Protein Stability. Nucleic Acids Res 2017, 45, W229–W235. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the Impact of Mutations on Protein Conformation, Flexibility and Stability. Nucleic Acids Research 2018, 46, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Arap, W.; Pasqualini, R. Predicting Proteome-Scale Protein Structure with Artificial Intelligence. N Engl J Med 2021, 385, 2191–2194. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly Accurate Protein Structure Prediction for the Human Proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Carvalho, L.S.; Pessoa, D.M.A.; Mountford, J.K.; Davies, W.I.L.; Hunt, D.M. The Genetic and Evolutionary Drives behind Primate Color Vision. Front. Ecol. Evol. 2017, 5. [Google Scholar] [CrossRef]
- Glazko, G.; Veeramachaneni, V.; Nei, M.; Makałowski, W. Eighty Percent of Proteins Are Different between Humans and Chimpanzees. Gene 2005, 346, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Janiak, M.C. Digestive Enzymes of Human and Nonhuman Primates: Digestive Enzymes of Human and Nonhuman Primates. Evol. Anthropol. 2016, 25, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Gröhs Ferrareze, P.A.; Zimerman, R.A.; Franceschi, V.B.; Caldana, G.D.; Netz, P.A.; Thompson, C.E. Molecular Evolution and Structural Analyses of the Spike Glycoprotein from Brazilian SARS-CoV-2 Genomes: The Impact of Selected Mutations. J Biomol Struct Dyn 2022, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Beniashvili, D.Sh. An Overview of the World Literature on Spontaneous Tumors in Nonhuman Primates. J. Med. Primatol. 1989, 18, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Toledano, A.; Álvarez, M.I.; López-Rodríguez, A.B.; Toledano-Díaz, A.; Fernández-Verdecia, C.I. [Does Alzheimer’s disease exist in all primates? Alzheimer pathology in non-human primates and its pathophysiological implications (II)]. Neurologia 2014, 29, 42–55. [Google Scholar] [CrossRef]
- Mastorodemos, V.; Zaganas, I.; Spanaki, C.; Bessa, M.; Plaitakis, A. Molecular Basis of Human Glutamate Dehydrogenase Regulation under Changing Energy Demands. J. Neurosci. Res. 2005, 79, 65–73. [Google Scholar] [CrossRef]
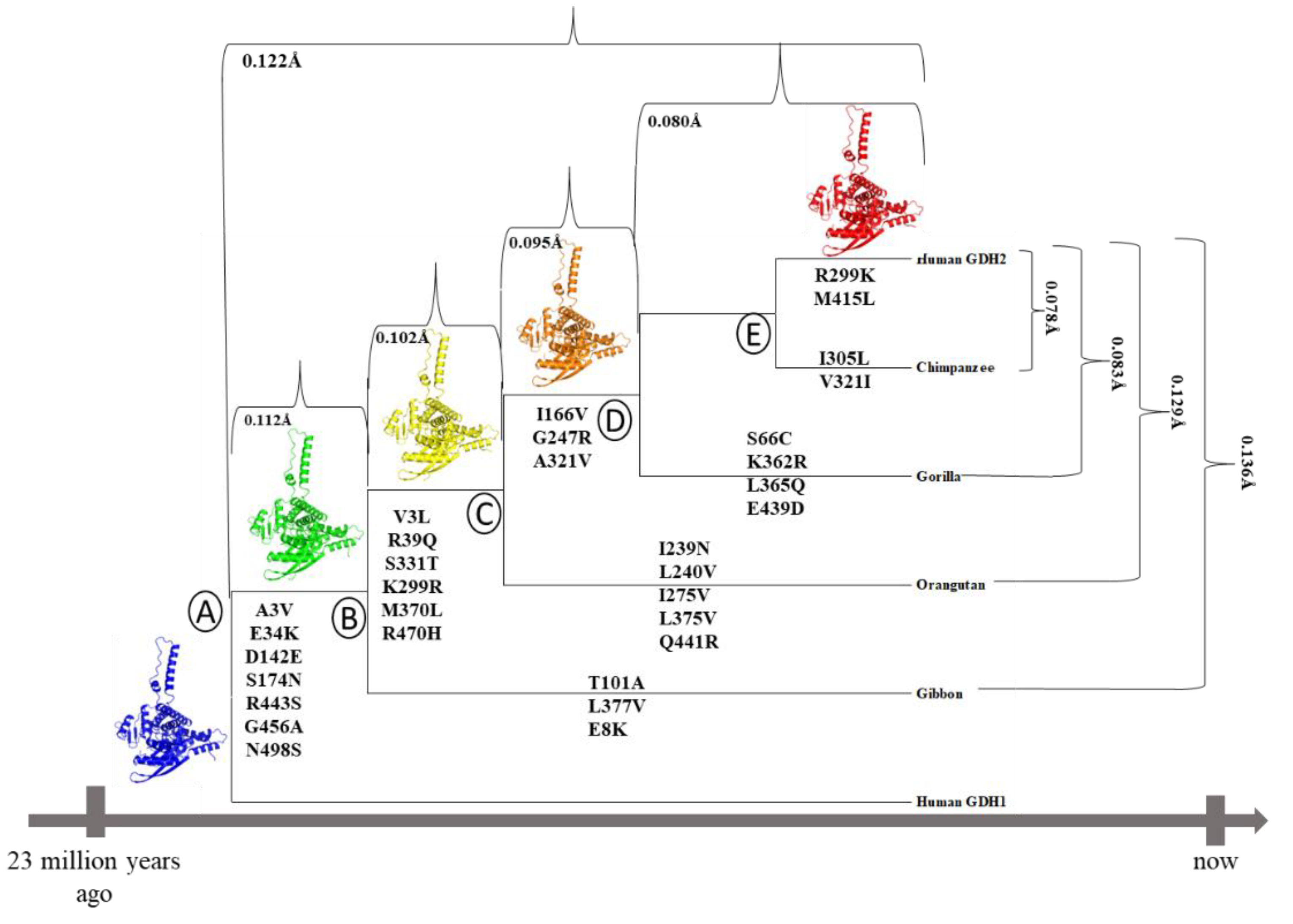
| Protein | AlphaFold pLDDT |
|---|---|
| Node A (=hGDH1) | 93.79 |
| Node B | 93.38 |
| Node C | 93.80 |
| Node D | 93.36 |
| Node E (=hGDH2) | 93.85 |
| Chimpanzee (Node E) | 93.67 |
| Gorilla (Node D) | 93.52 |
| Orangutan (Node C) | 93.86 |
| Gibbon (Node B) | 93.44 |
| Dynamut | iMutant | MUpro | iSTABLE | PremPS | MaestroWeb | SDM | ||
|---|---|---|---|---|---|---|---|---|
| Human Node A-B | A3V | S | D | S | D | S | S | S |
| Human Node A-B | E34K | S | D | D | D | D | D | D |
| Human Node A-B | D142E | S | D | S | S | S | S | S |
| Human Node A-B | S174N | S | S | S | S | D | S | S |
| Human Node A-B | R443S | D | D | D | D | D | S | D |
| Human Node A-B | G456A | S | D | S | S | D | S | S |
| Human Node A-B | N498S | D | S | D | D | D | S | D |
| Human Node B-C | V3L | S | D | D | D | S | D | D |
| Human Node B-C | R39Q | S | D | S | D | D | S | D |
| Human Node B-C | K299R | D | D | D | D | D | S | S |
| Human Node B-C | S331T | S | S | D | D | D | D | S |
| Human Node B-C | M370L | S | D | D | D | S | S | S |
| Human Node B-C | R470H | S | D | D | D | D | S | D |
| Human Node C-D | I166V | D | D | D | D | D | S | S |
| Human Node C-D | G247R | S | S | D | S | D | D | D |
| Human Node C-D | A321V | D | S | S | S | S | S | D |
| Human Node D-E | R299K | D | D | D | D | S | S | D |
| Human Node D-E | M415L | D | D | D | D | D | D | S |
| Chimpanzee Node E | I305L | D | D | D | D | S | D | S |
| Chimpanzee Node E | V321I | S | D | D | D | D | S | D |
| Gorilla Node D | S66C | D | D | D | D | D | S | S |
| Gorilla Node D | K362R | D | D | S | S | D | S | S |
| Gorilla Node D | L365Q | D | D | D | D | D | S | S |
| Gorilla Node D | E439D | D | S | D | S | D | S | D |
| Orangutan Node C | I239N | D | D | D | D | D | S | D |
| Orangutan Node C | L240V | D | D | D | D | S | S | D |
| Orangutan Node C | I275V | D | D | D | D | D | D | D |
| Orangutan Node C | L375V | D | D | D | D | D | S | D |
| Orangutan Node C | Q441R | D | S | D | S | D | D | D |
| Gibbon Node B | E8K | S | D | D | D | S | D | D |
| Gibbon Node B | T101A | D | D | D | D | D | D | S |
| Gibbon Node B | L377V | D | D | D | D | D | S | D |
| Evolutionary step | Amino acid substitutions | Bonds lost | Bonds gained | Interactions lost | Interactions gained |
|---|---|---|---|---|---|
| 1st | A3V | Ser1 | Ser1 Ala5 |
||
| 1st | E34K | Lys31 |
Lys31, Asp30 |
Leu32 | |
| 1st | D142E | Gln144, Glu146 | Arg178, Gln146, Arg178 | Trp182 | |
| 1st | S174N | Tyr99 | Tyr99 | Pro137 | |
| 1st | R443S | Ala447, Phe440, Glu439 | Phe440, Ala447 | Gln441, Ser445 |
|
| 1st | G456A | His454, Tyr459, Thr460 |
Val453, His454, Tys459, Ile452 |
Phe387 | |
| 1st | N498S | Gly501, Ala500, Phe494 | Val496, Phe494, Ile52 |
||
| 2nd | V3L | Ser1 | |||
| 2nd | R39Q | ||||
| 2nd | K299R | Glu296, His302, Gln301, Glu296 | Leu295, Glu296, | Phe256 |
Phe256, Leu295, Ile305 |
| 2nd | S331T | Lost: Gln334 | |||
| 2nd | M370L | Ile347 | Ile347 |
Ile347, Phe230, Met237 |
Tyr236, Leu479, Leu481 |
| 2nd | R470H | Met473, Ala472 | |||
| 3rd | I166V | Pro92 | Gly163, Gly160, Ile162 |
||
| 3rd | G247R | Lys249 | |||
| 3rd | A321V | Ile318 | Cys323, Lys344 | Tyr314, Ile318, Val252, Cys323 |
|
| 4th | R299K | His302, Glu296, His302, Gln301, Asp297 | Glu279, Ile305 |
Gln301, Phe256 | |
| 4th | M415L | Gln418, His412, Val417 | Val417 |
Leu413 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





