Submitted:
29 May 2023
Posted:
30 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Animals and sampling procedure
2.2. RNA isolation, library preparation and sequencing of the transcriptome
2.3. Transcriptome data processing, differential expression analysis and Gene Ontology classification
2.4. Reverse-transcriptase real-time qPCR validation
3. Results
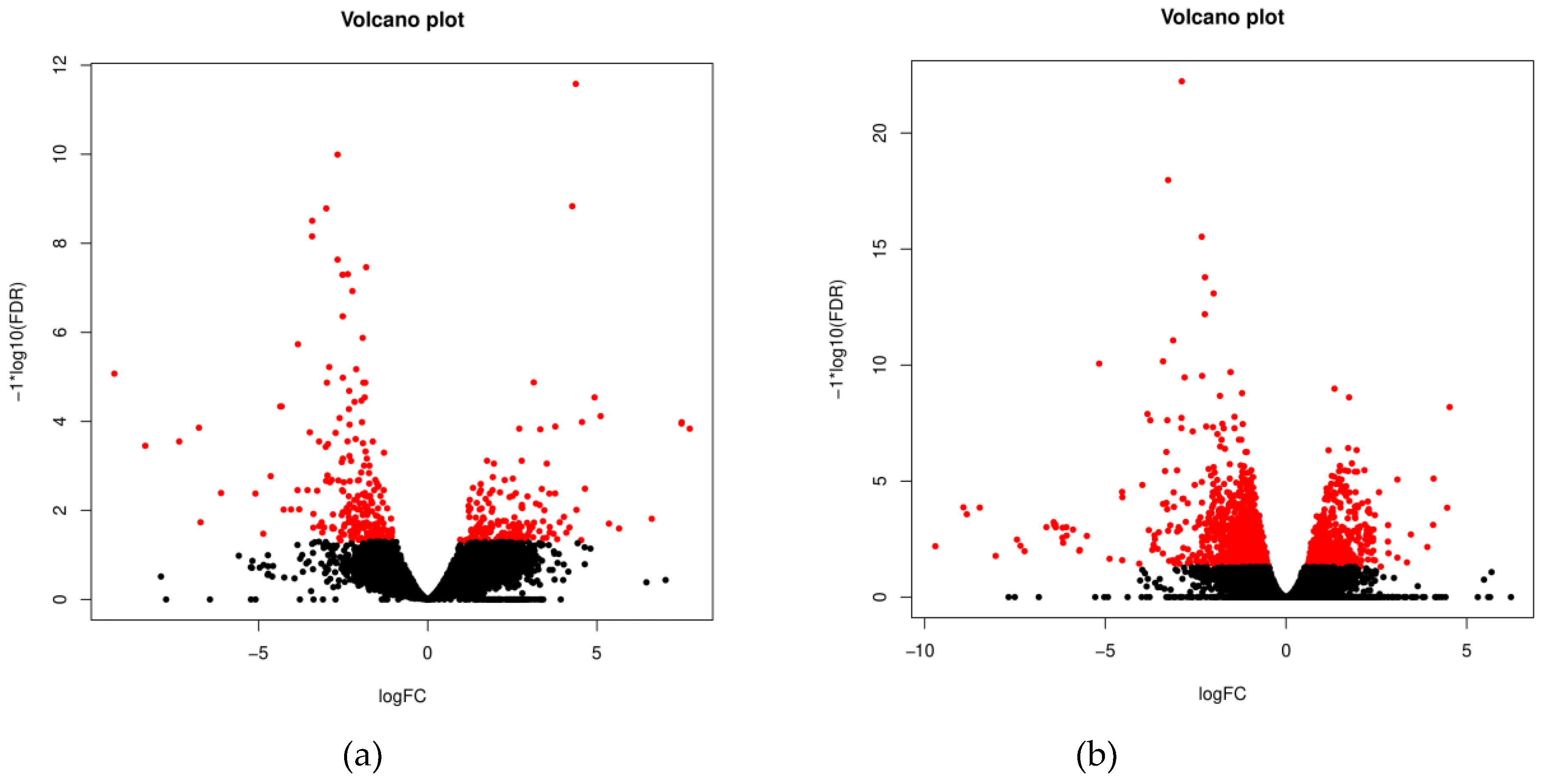
3.1. Sequencing quality overview
3.2. Characterization of the ovine whole blood transcriptome
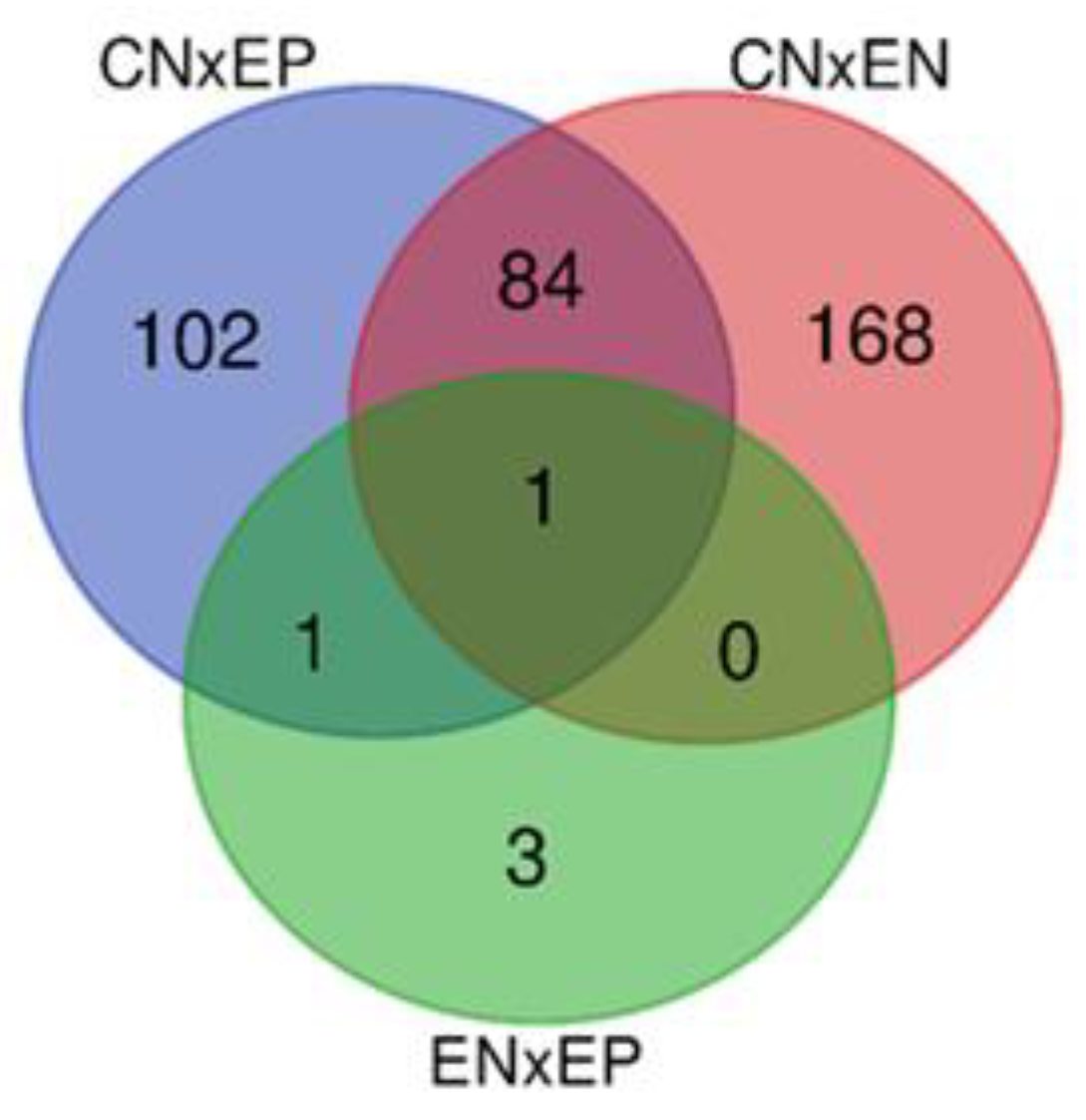
3.3. Functional significance of unique and common DEGs
3.3.1. Unique genes with the highest DE
3.3.2. Common genes with the highest DE
3.4. Enrichment of gene ontology (GO) terms and pathways in the investigated ovine groups
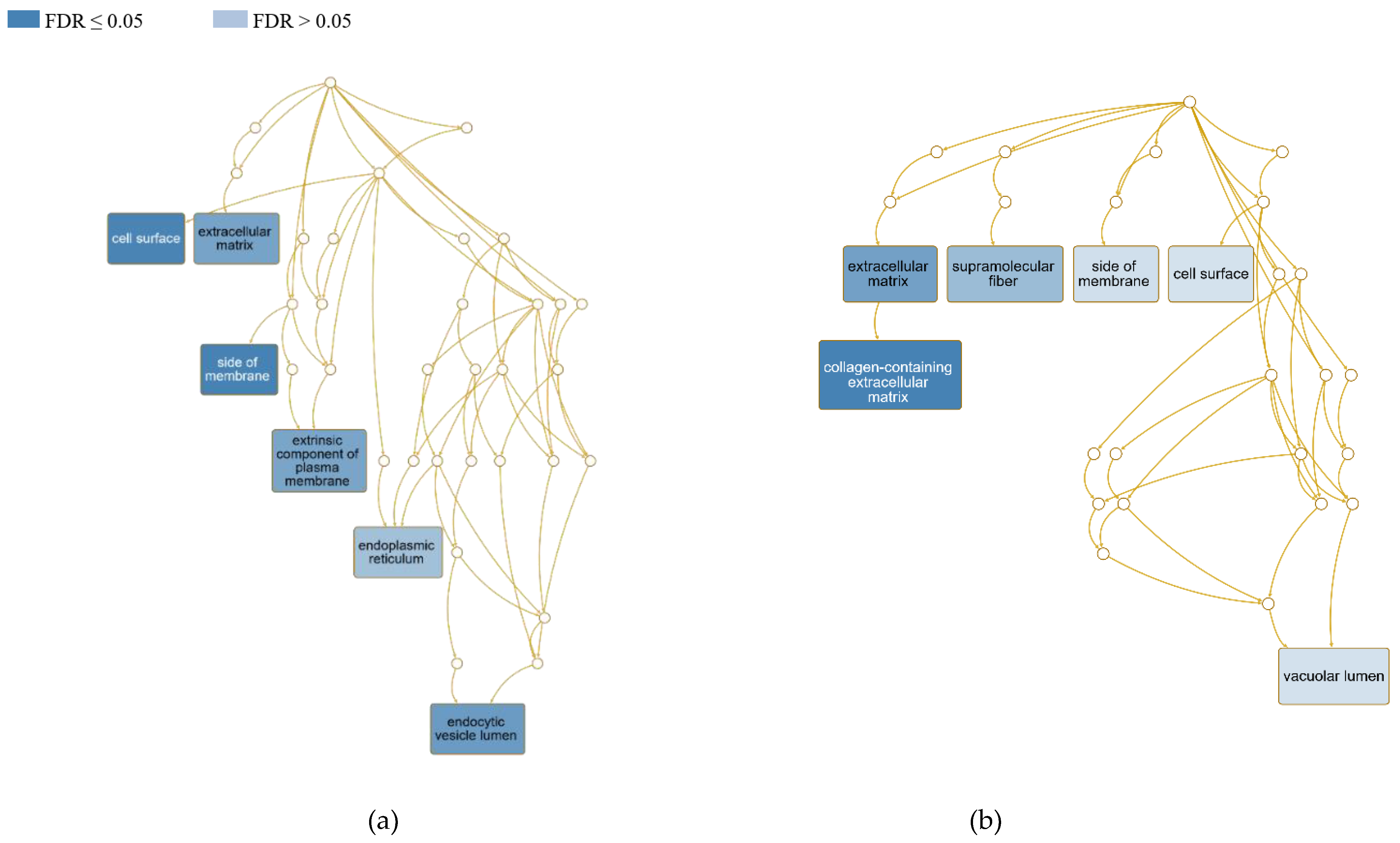
3.4.1. Cellular localization of differentially expressed genes
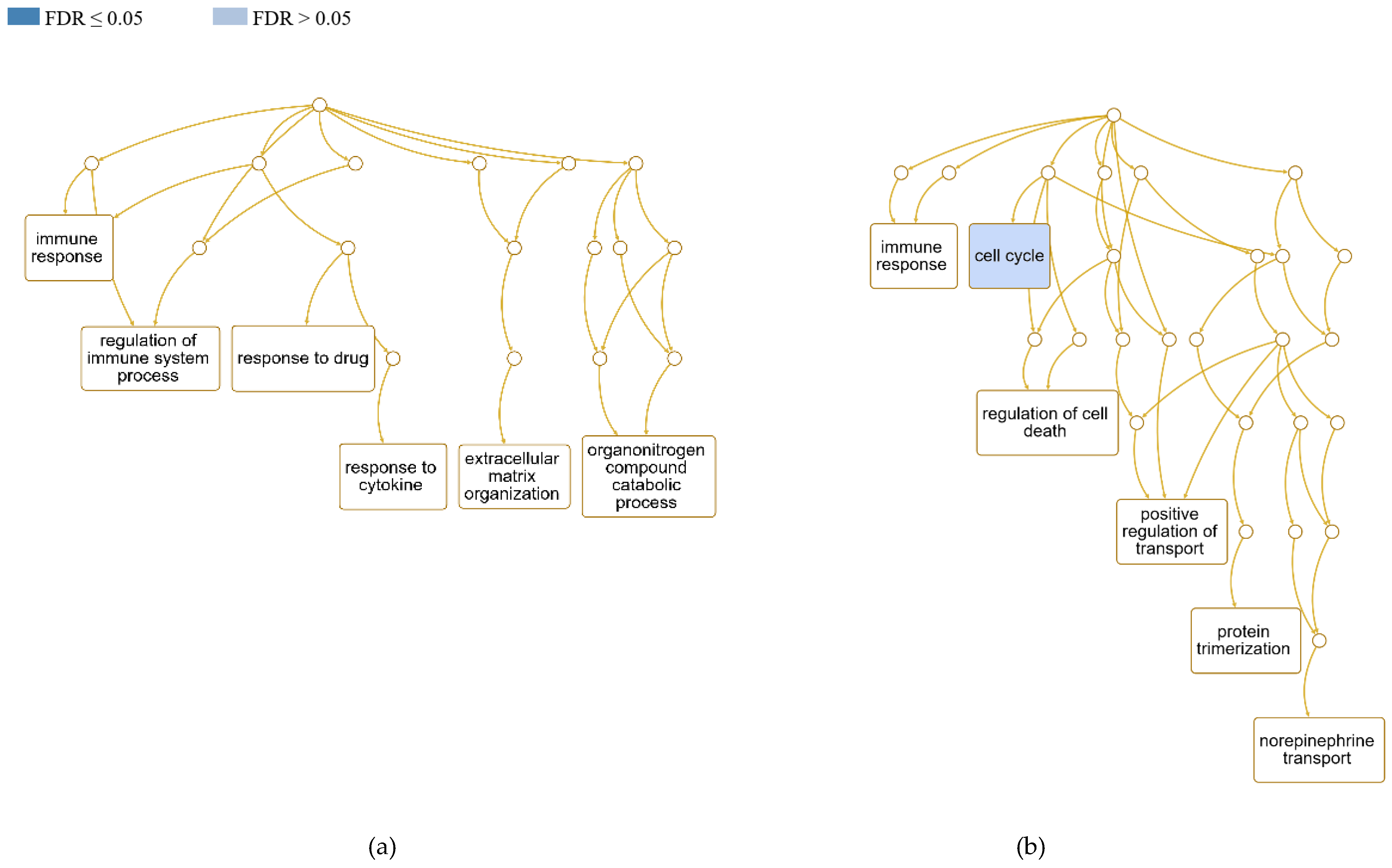
3.4.2. Influenced biological processes and role of DEGs in the host immune response
3.4.3. Differential gene expression in biological pathways influenced by C.pseudotuberculosis
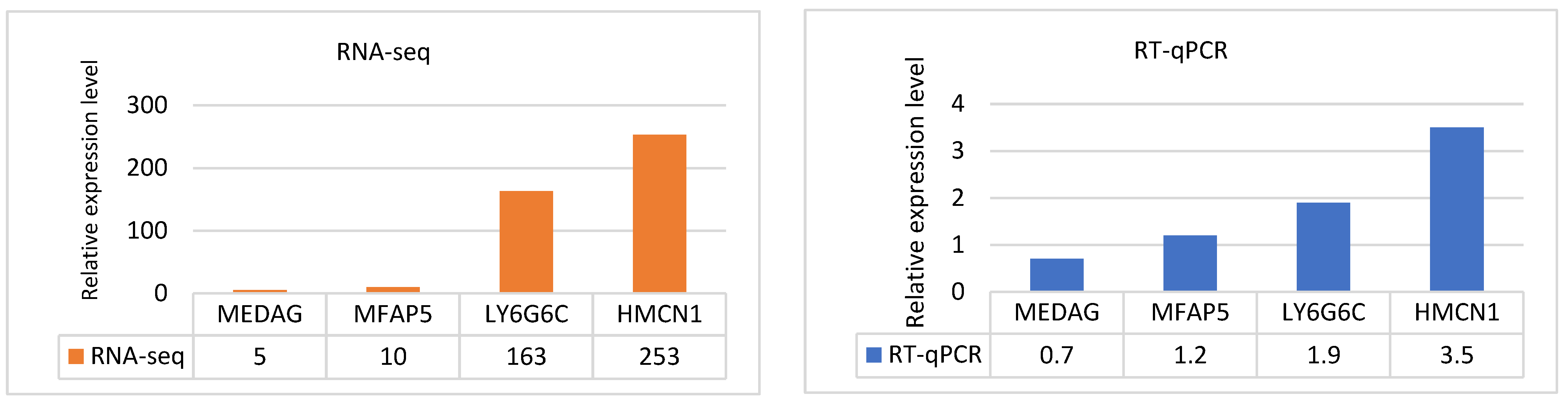
3.5. Validation of RNA-Seq data by RT‒qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Odhah, M.N.; Jesse, F.F.A.; Bura, P.; Chung, E.L.T.; Nor, N.F.M.; Norsidin, J.M.; Garba, B.; Mohd-Lila, M.-A. Current Review on Mycolic Acid Immunogen of Corynebacterium pseudotuberculosis. Journal of Advanced Veterinary Research 2022, 12, 177–186. [Google Scholar]
- Guimarães, A.S.; Borges, F.; Pauletti, R.B.; Seyffert, N.; Ribeiro, D.; Lage, A.; Heinemann, M.B.; Miyoshi, A.; Gouveia, A. Caseous Lymphadenitis: Epidemiology, Diagnosis, and Control. IIOABJ 2011, 2, 33–43. [Google Scholar]
- Zamprogna, T.D.; Ribeiro, D.; Azevedo, V.A.C.; Lara, G.H.B.; Motta, R.G.; da Silva, R.C.; Siqueira, A.K.; de Nardi, G.; Listoni, F.J.P.; Martins, L.D.A.; et al. Bacteriological, cytological, and molecular investigation of Corynebacterium pseudotuberculosis, mycobacteria, and other bacteria in caseous lymphadenitis and healthy lymph nodes of slaughtered sheep. Braz. J. Microbiol. 2021, 52, 431–438. [Google Scholar] [CrossRef]
- Baird, G.J.; Fontainet, M.C. Corynebacterium pseudotuberculosis and its role in ovine caseous lymphadenitis. J. Comp. Pathol. 2007, 137, 179–210. [Google Scholar] [CrossRef]
- Dominguez, M.C.R.; Jimenez, R.M.D.; Guerreo, J.A.V. Caseous lymphadenitis: virulence factors, pathogenesis and vaccines. Review. Rev. Mex. Cienc. Pecu. 2021, 12, 1221–1249. [Google Scholar] [CrossRef]
- Mattiello, S.; Battini, M.; Mantova, E.; Noe, L.; Grosso, L.; Barbieri, S. Evidence of poor welfare in goats with external abscesses. A preliminary study. Large Anim. Rev. 2018, 24, 113–118. [Google Scholar]
- Gascoigne, E.; Ogden, N.; Lovatt, F.; Davies, P. Update on caseous lymphadenitis in sheep. In Practice 2020, 42, 105–114. [Google Scholar] [CrossRef]
- Augustine, J.L.; Renshaw, H.W. Survival of Corynebacterium pseudotuberculosis in axenic purulent exudate on common barnyard fomites. Am. J. Vet. Res. 1986, 47, 713–715. [Google Scholar] [PubMed]
- Williamson, L.H. Caseous Lymphadenitis in Small Ruminants. Vet. Clin. North Am.: Large Anim. Pract. 2001, 17, 359–371. [Google Scholar] [CrossRef]
- Fontaine, M.C.; Baird, G.J. Caseous lymphadenitis. Small Ruminant Res. 2008, 76, 42–48. [Google Scholar] [CrossRef]
- Pepin, M.; Paton, M. Caseous lymphadenitis in sheep and goats. In Infectious and Parasitic Diseases of Livestock, 1st ed.; Lefevre, C., Blancou, J., Chermette, r., Uilenberg, G., Ed.; CABI: 2011; pp. 1151–1163.
- Reboucas, M.F.; Loureiro, D.; Barral, T.D.; Seyffert, N.; Raynal, J.T.; Sousa, T.J.; Figueiredo, H.C.P.; Azevedo, V.; Meyer, R.; Portela, R.W. Cell wall glycolipids from Corynebacterium pseudotuberculosis strains with different virulences differ in terms of composition and immune recognition. Braz. J. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Souček, A.; Michalec, Č.; Součková, A. Identification and characterization of a new enzyme of the group “phospholipase D” isolated from Corynebacterium ovis. Biochimica et Biophysica Acta (BBA) - Enzymology 1971, 227, 116–128. [Google Scholar] [CrossRef]
- Pepin, M.; Sanchis, R.; Paton, M. Caseous lymphadenitis in sheep and goats. Point Veterinaire 1999, 30, 33–40. [Google Scholar]
- de Pinho, R.B.; Silva, M.T.D.; Bezerra, F.S.B.; Borsuk, S. Vaccines for caseous lymphadenitis: up-to-date and forward-looking strategies. Appl. Microbiol. Biotechnol. 2021, 105, 2287–2296. [Google Scholar] [CrossRef]
- Franco, E.F.; Rana, P.; Cavalcante, A.L.Q.; da Silva, A.L.; Gomide, A.C.P.; Folador, A.R.C.; Azevedo, V.; Ghosh, P.; Ramos, R.T.J. Co-Expression Networks for Causal Gene Identification Based on RNA-Seq Data of Corynebacterium pseudotuberculosis. Genes 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.C.; Silva, A.; Trost, E.; Blom, J.; Ramos, R.; Carneiro, A.; Ali, A.; Santos, A.R.; Pinto, A.C.; Diniz, C.; et al. The Pan-Genome of the Animal Pathogen Corynebacterium pseudotuberculosis Reveals Differences in Genome Plasticity between the Biovar ovis and equi Strains. PLoS One 2013, 8. [Google Scholar] [CrossRef]
- Bastos, B.L.; Portela, R.W.D.; Dorella, F.A.; Ribeiro, D.; Seyffert, N.; de Paula Castro, T.L.; Miyoshi, A.; Oliveira, S.C.; Meyer, R.; Azevedo, V. Corynebacterium pseudotuberculosis: Immunological Responses in Animal Models and Zoonotic Potential. J. Clin. Cell. Immunol. 2012, 01. [Google Scholar] [CrossRef]
- Vale, V.L.C.; Silva, M.d.C.; de Souza, A.P.; Trindade, S.C.; de Moura-Costa, L.F.; Dos Santos-Lima, E.K.N.; Nascimento, I.L.d.O.; Cardoso, H.S.P.; Marques, E.d.J.; Paule, B.J.A.; et al. Humoral and cellular immune responses in mice against secreted and somatic antigens from a Corynebacterium pseudotuberculosis attenuated strain: Immune response against a C. pseudotuberculosis strain. BMC Vet. Res. 2016, 12, 195–195. [Google Scholar] [CrossRef]
- Paule, B.J.A.; Azevedo, V.; Regis, L.F.; Carminati, R.; Bahia, C.R.; Vale, V.L.C.; Moura-Costa, L.F.; Freire, S.M.; Nascimento, I.; Schaer, R.; et al. Experimental Corynebacterium pseudotuberculosis primary infection in goats: kinetics of IgG and interferon-gamma production, IgG avidity and antigen recognition by Western blotting. Vet. Immunol. Immunopathol. 2003, 96, 129–139. [Google Scholar] [CrossRef]
- Fu, M.Z.; Su, H.; Su, Z.Q.; Yin, Z.; Jin, J.; Wang, L.X.; Zhang, Q.; Xu, X.G. Transcriptome analysis of Corynebacterium pseudotuberculosis-infected spleen of dairy goats. Microb. Pathog. 2020, 147, 9. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.Z.; Yan, Y.C.; Su, H.; Wang, J.J.; Shi, X.J.; Zhou, H.C.; Zhang, Q.; Xu, X.G. Spleen proteome profiling of dairy goats infected with C. pseudotuberculosis by TMT-based quantitative proteomics approach. J. Proteomics 2021, 248, 11. [Google Scholar] [CrossRef]
- Fu, M.; Xu, X.; Cheng, Z.; Zhu, J.; Sun, A.; Xu, G.; An, X. Combined Transcriptomic and Proteomic of Corynebacterium pseudotuberculosis Infection in the Spleen of Dairy Goats. Animals 2022, 12, 3270. [Google Scholar] [CrossRef]
- Hu, G.Y.; Do, D.N.; Gray, J.; Miar, Y. Selection for Favorable Health Traits: A Potential Approach to Cope with Diseases in Farm Animals. Animals 2020, 10, 28. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.X.; Wang, J.; Jaehnig, E.J.; Shi, Z.A.; Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2021, 50, D687–D692. [Google Scholar] [CrossRef]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; R, A.M.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: connecting communities. Nucleic Acids Res 2021, 49, D613–D621. [Google Scholar] [CrossRef] [PubMed]
- El-Enbaawy, M.I.; Saad, M.M.; Selim, S.A. Humoral and cellular immune responses of a murine model against Corynebacterium pseudotuberculosis antigens. Egypt. J. Immunol. 2005, 12, 13–19. [Google Scholar] [PubMed]
- Hoelzle, L.E.; Scherrer, T.; Muntwyler, J.; Wittenbrink, M.M.; Philipp, W.; Hoelzle, K. Differences in the antigen structures of Corynebacterium pseudotuberculosis and the induced humoral immune response in sheep and goats. Vet. Microbiol. 2013, 164, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.; Regis, L.; Vale, V.; Paule, B.; Carminati, R.; Bahia, R.; Moura-Costa, L.; Schaer, R.; Nascimento, I.; Freire, S. In vitro IFN-gamma production by goat blood cells after stimulation with somatic and secreted Corynebacterium pseudotuberculosis antigens. Vet. Immunol. Immunopathol. 2005, 107, 249–254. [Google Scholar] [CrossRef]
- Pepin, M.; Seow, H.F.; Corner, L.; Rothel, J.S.; Hodgson, A.L.M.; Wood, P.R. Cytokine gene expression in sheep following experimental infection with various strains of Corynebacterium pseudotuberculosis differing in virulence. Vet. Res. 1997, 28, 149–163. [Google Scholar] [PubMed]
- Sandri, M.; Stefanon, B.; Loor, J.J. Transcriptome profiles of whole blood in Italian Holstein and Italian Simmental lactating cows diverging for genetic merit for milk protein. J. Dairy Sci. 2015, 98, 6119–6127. [Google Scholar] [CrossRef] [PubMed]
- Park, H.E.; Park, H.T.; Jung, Y.H.; Yoo, H.S. Gene expression profiles of immune-regulatory genes in whole blood of cattle with a subclinical infection of Mycobacterium avium subsp paratuberculosis. PLoS One 2018, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Malvisi, M.; Curti, N.; Remondini, D.; De Iorio, M.G.; Palazzo, F.; Gandini, G.; Vitali, S.; Polli, M.; Williams, J.L.; Minozzi, G. Combinatorial Discriminant Analysis Applied to RNAseq Data Reveals a Set of 10 Transcripts as Signatures of Exposure of Cattle to Mycobacterium avium subsp. paratuberculosis. Animals 2020, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.; Huerta, B.; Galan-Relano, A.; Gomez-Gascon, L.; Almeida, A.; Viegas, I.; Maldonado, A. Utility assessment of an Enzyme-linked immunosorbent assay for detection of subclinical cases of caseous lymphadenitis in small ruminant flocks. Vet. Med. Sci. 2020, 6, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Kuria, J.K.N.; Mbuthia, P.G.; Kang'ethe, E.K.; Wahome, R.G. Caseous lymphadenitis in goats: The pathogenesis, incubation period and serological response after experimental infection. Vet. Res. Commun. 2001, 25, 89–97. [Google Scholar] [CrossRef]
- Langenegger, C.H.; Langenegger, J. Serologic and allergic monitoring of experimental infections in goats by Corynebacterium-pseudotuberculosis. Pesqui. Vet. Bras. 1991, 11, 1–7. [Google Scholar]
- Malvisi, M.; Palazzo, F.; Morandi, N.; Lazzari, B.; Williams, J.L.; Pagnacco, G.; Minozzi, G. Responses of Bovine Innate Immunity to Mycobacterium avium subsp paratuberculosis Infection Revealed by Changes in Gene Expression and Levels of MicroRNA. PLoS One 2016, 11, 23. [Google Scholar] [CrossRef]
- David, J.; Barkema, H.W.; Guan, L.L.; De Buck, J. Gene-expression profiling of calves 6 and 9 months after inoculation with Mycobacterium avium subspecies paratuberculosis. Vet. Res. 2014, 45, 12. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.H.; Kim, Y.K.; Kim, M.R.; Jang, J.H.; Lee, S. Emerging Roles of Vascular Cell Adhesion Molecule-1 (VCAM-1) in Immunological Disorders and Cancer. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Park, H.E.; Shin, M.K.; Park, H.T.; Jung, M.; Il Cho, Y.; Yoo, H.S. Gene expression profiles of putative biomarker candidates in Mycobacterium avium subsp paratuberculosis-infected cattle. Pathog. Dis. 2016, 74, 4. [Google Scholar] [CrossRef]
- Mecham, R.P.; Gibson, M.A. The microfibril-associated glycoproteins (MAGPs) and the microfibrillar niche. Matrix Biol. 2015, 47, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Vogel, B.E.; Hedgecock, E.M. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development (Cambridge, England) 2001, 128, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, X.; Sairam, M.R. Novel genes of visceral adiposity: identification of mouse and human mesenteric estrogen-dependent adipose (MEDA)-4 gene and its adipogenic function. Endocrinology 2012, 153, 2665–2676. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Schmidt, T.A.; Krawetz, R.J.; Dufour, A. Proteoglycan 4: From Mere Lubricant to Regulator of Tissue Homeostasis and Inflammation: Does proteoglycan 4 have the ability to buffer the inflammatory response? BioEssays 2019, 41, e1800166. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, A.; Kamishikiryo, J.; Mori, D.; Toyonaga, K.; Okabe, Y.; Toji, A.; Kanda, R.; Miyake, Y.; Ose, T.; Yamasaki, S.; et al. Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL. Proc. Natl. Acad. Sci. USA 2013, 110, 17438–17443. [Google Scholar] [CrossRef]
- Schick, J.; Etschel, P.; Bailo, R.; Ott, L.; Bhatt, A.; Lepenies, B.; Kirschning, C.; Burkovski, A.; Lang, R. Toll-Like Receptor 2 and Mincle Cooperatively Sense Corynebacterial Cell Wall Glycolipids. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef]
- Lai, J.J.; Cruz, F.M.; Rock, K.L. Immune Sensing of Cell Death through Recognition of Histone Sequences by C-Type Lectin-Receptor-2d Causes Inflammation and Tissue Injury. Immunity 2020, 52, 123–135 e126. [Google Scholar] [CrossRef]
- Al-Moussawy, M.; Abdelsamed, H.A.; Lakkis, F.G. Immunoglobulin-like receptors and the generation of innate immune memory. Immunogenetics 2022, 74, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Marino, R.; Capoferri, R.; Panelli, S.; Minozzi, G.; Strozzi, F.; Trevisi, E.; Snel, G.G.M.; Ajmone-Marsan, P.; Williams, J.L. Johne's disease in cattle: an in vitro model to study early response to infection of Mycobacterium avium subsp paratuberculosis using RNA-seq. Mol. Immunol. 2017, 91, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.; Plain, K.; Purdie, A.; Saunders, B.M.; de Silva, K. Biomarkers for Detecting Resilience against Mycobacterial Disease in Animals. Infect. Immun. 2019, 88, 23. [Google Scholar] [CrossRef]
- Kaczmarek, R.; Pasciak, M.; Szymczak-Kulus, K.; Czerwinski, M. CD1: A Singed Cat of the Three Antigen Presentation Systems. Arch. Immunol. Ther. Exp. (Warsz) 2017, 65, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Zhao, T.; Zhang, H.; Lu, H.; Zhang, Q.; Sun, L.; Fan, Z. Granzyme H induces apoptosis of target tumor cells characterized by DNA fragmentation and Bid-dependent mitochondrial damage. Mol. Immunol. 2008, 45, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Guggino, G.; Orlando, V.; Cutrera, S.; La Manna, M.P.; Di Liberto, D.; Vanini, V.; Petruccioli, E.; Dieli, F.; Goletti, D.; Caccamo, N. Granzyme A as a potential biomarker of Mycobacterium tuberculosis infection and disease. Immunol. Lett. 2015, 166, 87–91. [Google Scholar] [CrossRef]
- Rasi, V.; Wood, D.C.; Eickhoff, C.S.; Xia, M.; Pozzi, N.; Edwards, R.L.; Walch, M.; Bovenschen, N.; Hoft, D.F. Granzyme A Produced by γ(9)δ(2) T Cells Activates ER Stress Responses and ATP Production, and Protects Against Intracellular Mycobacterial Replication Independent of Enzymatic Activity. Front. Immunol. 2021, 12, 712678. [Google Scholar] [CrossRef]
- Ciucci, T.; Bosselut, R. Gimap and T cells: A matter of life or death. Eur. J. Immunol. 2014, 44, 348–351. [Google Scholar] [CrossRef]
- Datta, P.; Webb, L.M.; Avdo, I.; Pascall, J.; Butcher, G.W. Survival of mature T cells in the periphery is intrinsically dependent on GIMAP1 in mice. Eur. J. Immunol. 2017, 47, 84–93. [Google Scholar] [CrossRef]
- Limoges, M.A.; Cloutier, M.; Nandi, M.; Ilangumaran, S.; Ramanathan, S. The GIMAP Family Proteins: An Incomplete Puzzle. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Oliveira, A.; Oliveira, L.C.; Aburjaile, F.; Benevides, L.; Tiwari, S.; Jamal, S.B.; Silva, A.; Figueiredo, H.C.P.; Ghosh, P.; Portela, R.W.; et al. Insight of Genus Corynebacterium: Ascertaining the Role of Pathogenic and Non-pathogenic Species. Front. Microbiol. 2017, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, L.; Egan, R.; Baquero, M.; Mansz, A.; Plattner, B.L. WC1(+) and WC1(neg) gammadelta T lymphocytes in intestinal mucosa of healthy and Mycobacterium avium subspecies paratuberculosis-infected calves. Vet. Immunol. Immunopathol. 2019, 216, 109919. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, A.; Yirsaw, A.; Kim, S.; Wilson, K.; McLaughlin, J.; Madigan, M.; Loonie, K.; Britton, E.; Zhang, F.Q.; Damani-Yokota, P.; et al. Gene characterization and expression of the gamma delta T cell co-receptor WC1 in sheep. Dev. Comp. Immunol. 2021, 116. [Google Scholar] [CrossRef]
- Yirsaw, A.W.; Gillespie, A.; Britton, E.; Doerle, A.; Johnson, L.; Marston, S.; Telfer, J.; Baldwin, C.L. Goat gamma delta T cell subpopulations defined by WC1 expression, responses to pathogens and cytokine production. Dev. Comp. Immunol. 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, N.; Gendron, D.; Ariel, O.; Dudemaine, P.; Ibeagha-Awemu, E. Transcriptome profiling of primary bovine macrophages from cows with Johne's disease suggests a tolerance state induced by convergent signalling via NF-kappa B and its synergistic tolerized genes. J. Anim. Sci. 2018, 96, 22–22. [Google Scholar] [CrossRef]
- da Silva, W.M.; Seyffert, N.; Silva, A.; Azevedo, V. A journey through the Corynebacterium pseudotuberculosis proteome promotes insights into its functional genome. PeerJ 2021, 9, 31. [Google Scholar] [CrossRef]
- Yozwiak, M.L.; Songer, J.G. Effects of Corynebacterium-pseudotuberculosis phospholipase-D on viability and chemotactic responses of ovine neutrophils. Am. J. Vet. Res. 1993, 54, 392–397. [Google Scholar]
- Schweighoffer, E.; Tybulewicz, V.L. BAFF signaling in health and disease. Curr. Opin. Immunol. 2021, 71, 124–131. [Google Scholar] [CrossRef]
- Bastos, B.L.; Loureiro, D.; Raynal, J.T.; Guedes, M.T.; Vale, V.L.C.; Moura-Costa, L.F.; Guimaraes, J.E.; Azevedo, V.; Portela, R.W.; Meyer, R. Association between haptoglobin and IgM levels and the clinical progression of caseous lymphadenitis in sheep. BMC Vet. Res. 2013, 9, 254. [Google Scholar] [CrossRef]
- Areschoug, T.; Gordon, S. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell. Microbiol. 2009, 11, 1160–1169. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell. Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed]
- Hempel, R.J.; Bannantine, J.P.; Stabel, J.R. Transcriptional Profiling of Ileocecal Valve of Holstein Dairy Cows Infected with Mycobacterium avium subsp Paratuberculosis. PLoS One 2016, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Kelley, J.; Walter, L.; Trowsdale, J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet 2005, 1, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Cemerski, S.; Shaw, A. Immune synapses in T-cell activation. Curr. Opin. Immunol. 2006, 18, 298–304. [Google Scholar] [CrossRef]
| Group | Trimmed reads | Mapped reads | Mapped reads % | Total paired reads | Unique reads | Unique reads % | Transcripts | Genes |
| EP | 34 454 888 | 26 542 080 | 77.02 | 25 825 876 | 14 965 299 | 56.34 | 27 430 | 12 230 |
| EN | 30 881 461 | 24 175 800 | 78.29 | 23 691 710 | 13 300 932 | 55.02 | 26 730 | 12 080 |
| CN | 30 495 644 | 19 935 851 | 63.64 | 19 501 543 | 11 341 457 | 57.03 | 26 753 | 11 932 |
| Gene Name | Gene Symbol | FC | padj |
|---|---|---|---|
| CNxEP | |||
| killer cell lectin-like receptor subfamily B member 1 | KLRB1 | 7.74 | 1.46E-04 |
| kelch like family memeber 18 | KLHL18 | 6.62 | 1.53E-02 |
| chloride voltage-gated channel Ka | CLCNKA | 5.35 | 1.98E-02 |
| immunoglobulin superfamily DCC subclass member 3 | IGDCC3 | 5.11 | 7.60E-05 |
| protein-glucosyl galactosyl hydroxylysine glucosidase | PGGHG | 4.64 | 3.25E-03 |
| asporin | ASPN | -6.71 | 1.84E-02 |
| secreted protein acidic and cysteine rich | SPARC | -4.36 | 4.60E-05 |
| secreted phosphoprotein 1 | SPP1 | -4.32 | 4.62E-05 |
| solute carrier family 7 member 11 | SLC7A11 | -3.21 | 2.24E-02 |
| twist family bHLH transcription Factor 1 | TWIST1 | -3.14 | 1.84E-02 |
| CNxEN | |||
| zinc finger protein 227 | LOC101109384 | 3.45 | 1.99E-03 |
| ankyrin repeat domain 7 | ANKRD7 | 2.82 | 7.95E-04 |
| crystallin gamma S | CRYGS | 2.62 | 4.84E-02 |
| antigen WC1.1 | WC1.1 | 2.42 | 2.12E-03 |
| tripartite motif containing 45 | TRIM45 | 2.42 | 2.50E-02 |
| proteoglycan 4 | PRG4 | -7.44 | 3.33E-03 |
| secreted frizzled related protein 1 | SFRP1 | -7.35 | 6.14E-03 |
| tubulointerstitial nephritis antigen like 1 | TINAGL1 | -4.53 | 4.98E-05 |
| collagen type VI alpha 1 chain | COL6A1 | -4.06 | 3.62E-02 |
| myeloperoxidase | MPO | -3.80 | 1.30E-03 |
| ENxEP | |||
| vascular cell adhesion protein 1 | VCAM1 | 7.50 | 2.68E-02 |
| phospholipid phosphatase related 5 | PLPPR5 | 6.12 | 2.57E-02 |
| Group | EP | EN | |||
|---|---|---|---|---|---|
| Gene Name | Gene Symbol | FC | padj | FC | padj |
| Same direction of expression | |||||
| hemicentin 1 | HMCN1 | 4.93 | 2.89E-05 | 4.07 | 7.72E-04 |
| MHC class I polypeptide-related sequence B | MICB | 4.37 | 2.63E-12 | 4.52 | 6.42E-09 |
| lymphocyte antigen 6 family member G6C | LY6G6C | 3.77 | 1.30E-04 | 4.08 | 7.84E-06 |
| tyrosine-protein phosphatase nonreceptor type substrate 1 | PTPN1 | 3.59 | 4.17E-03 | 3.91 | 6.92E-03 |
| microfibril associated protein 5 | MFAP5 | -9.26 | 8.45E-06 | -6.63 | 9.73E-04 |
| syndecan 2 | SDC2 | -8.35 | 3.52E-04 | -5.72 | 1.00E-02 |
| mesenteric estrogen dependent adipogenesis | MEDAG | -7.34 | 2.83E-04 | -8.93 | 1.36E-04 |
| decorin | DCN | -6.76 | 1.39E-04 | -5.51 | 2.31E-03 |
| Opposite direction of expression | |||||
| leukocyte immunoglobulin-like receptor subfamily A member 6 | LILRA6 | -1.72 | 9.82E-04 | 3.08 | 8.59E-06 |
| GTPase IMAP family member 7 | GIMAP7 | -1.50 | 2.43E-03 | 2.57 | 3.02E-05 |
| DEGs ordered according to their FC | Biological processes | |||||
|---|---|---|---|---|---|---|
| EP | Upregulated | Downregulated | ||||
| KLRB1; DCN; IGDCC3; HMCN1; PRG3 | antigen-receptor mediated SP | humoral IR | ||||
| ADAMTS13; MICB; SPARC; SPP1; IL13 | B-cell receptor signalling pathway | cytokine production | ||||
| LY6G6C; PTPN1; OLA-I; COL1A1; BTN2A2 | response to other organism | calcium ion homeostasis | ||||
| GZMH; LAMB1; TNIP2; GATA6; SPIB | adaptive IR-somatic recomb. of IRCs built from Ig domains | inflammatory response | ||||
| GIMAP8; NFKB2; C4BPA; GZMA; IBSP | response to cytokine | complement activation | ||||
| TNFRSF13C; SH2B2; PAX5; BLK; MT2A | cell adhesion | |||||
| CXCR6; JCHAIN; IFITM1; CXCL10; UBD | lymphocyte activation | |||||
| C3AR1; FCRL3; CDA; GHR; UBA7; CLECL1 | innate IR | |||||
| ADGRE3; CD1A; HSP90B1; LILRA6; OVAR | ||||||
| BPI; ULBP1; CD1E; GIMAP7 | ||||||
| EN | ||||||
| PRG4; DCN; MICB; LY6G6C; HMCN1; PTPN1 | positive regulation of NK cell mediated cytotoxicity | innate IR | ||||
| MPO; COL1A1; IL13; GZMB; GIMAP8 | gamma-delta T-cell activation | regulation of response to stress | ||||
| C4BPA; SERPINB4; BMPR1A; LAMB1 | I-kappaB kinase/NF-kappaB signalling | regulation of protein metabolic process | ||||
| GIMAP1; LILRA6; DQB; CLEC4E; IL1RN | response to stress | apoptotic process | ||||
| GIMAP7; TRIM45; DNM1; GZMH; HMGB2 | apoptotic process | locomotion | ||||
| PRG3; SOX13; C3AR1; GZMA; ULBP2 | calcium ion homeostasis | |||||
| WC1.1; CCL14; DNM3; NLRP2; TNFSF10 | ||||||
| GCA; KIR3DP1; LILRA5; BLK; UBD; PILRB | ||||||
| SERPINB9; CD226; FCER1A; ADGRE1; OLA-I | ||||||
| CLEC2D; TNFRSF25; GHSR; PSPC1; BPI | ||||||
| Gene Set | Description | Size | Overlap | ER | p value | FDR |
|---|---|---|---|---|---|---|
| EP | ||||||
| R-HSA-2173782 | Binding and Uptake of Ligands by Scavenger Receptors | 42 | 6 | 14.22 | 3.48E-06 | 0.01 |
| R-HSA-3000178 | ECM proteoglycans | 76 | 6 | 7.86 | 1.10E-04 | 0.06 |
| R-HSA-1474244 | Extracellular matrix organization | 301 | 11 | 3.64 | 2.11E-04 | 0.06 |
| R-HSA-1480926 | O2/CO2 exchange in erythrocytes | 13 | 3 | 22.98 | 2.62E-04 | 0.06 |
| R-HSA-198933 | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | 132 | 8 | 5.28 | 3.56E-04 | 0.08 |
| R-HSA-1280215 | Cytokine Signalling in Immune system | 688 | 16 | 2.32 | 1.33E-03 | 0.25 |
| R-HSA-168256 | Immune System | 1997 | 32 | 1.60 | 3.35E-03 | 0.40 |
| EN | ||||||
| hsa04115 | p53 signall pathway | 72 | 7 | 6.21 | 1.23E-04 | 0.04 |
| R-HSA-198933 | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | 132 | 7 | 3.73 | 2.73E-03 | 0.11 |
| R-HSA-1793185 | Chondroitin sulfate/dermatan sulfate metabolism | 50 | 4 | 9.28 | 8.80E-04 | 0.19 |
| R-HSA-1480926 | O2/CO2 exchange in erythrocytes | 13 | 3 | 18.45 | 4.99E-04 | 0.29 |
| hsa04110 | Cell cycle | 124 | 7 | 3.60 | 3.22E-03 | 0.34 |
| R-HSA-5633008 | TP53 Regulates Transcription of Cell Death Genes | 44 | 4 | 7.27 | 2.15E-03 | 0.41 |
| R-HSA-3000170 | Syndecan interactions | 27 | 3 | 8.88 | 4.49E-03 | 0.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





