Submitted:
26 May 2023
Posted:
29 May 2023
You are already at the latest version
Abstract
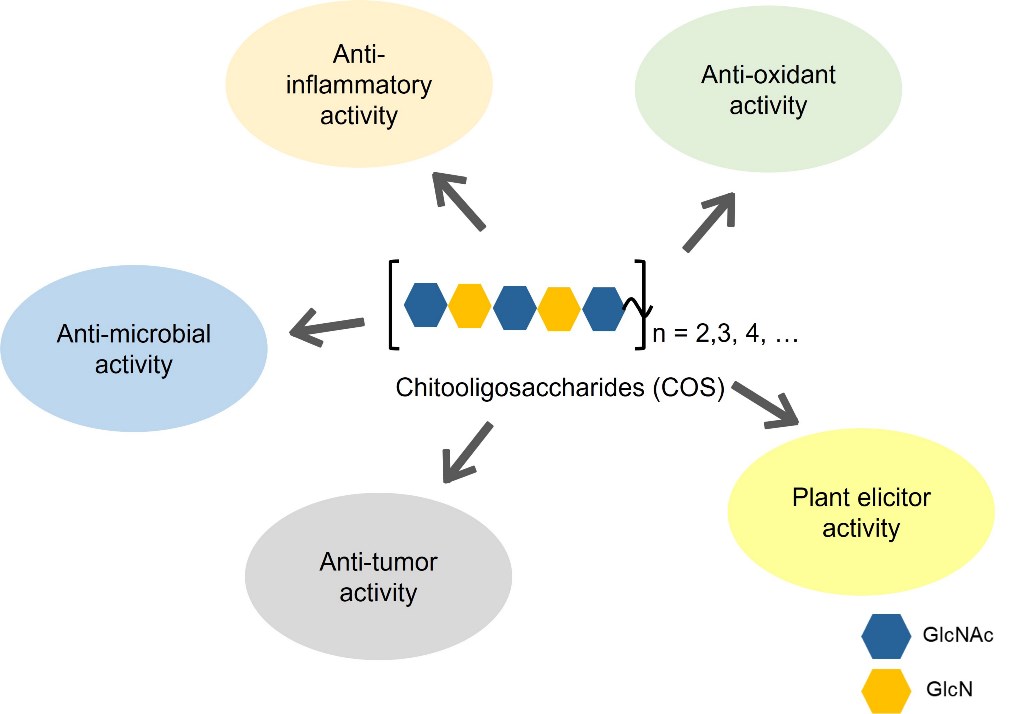
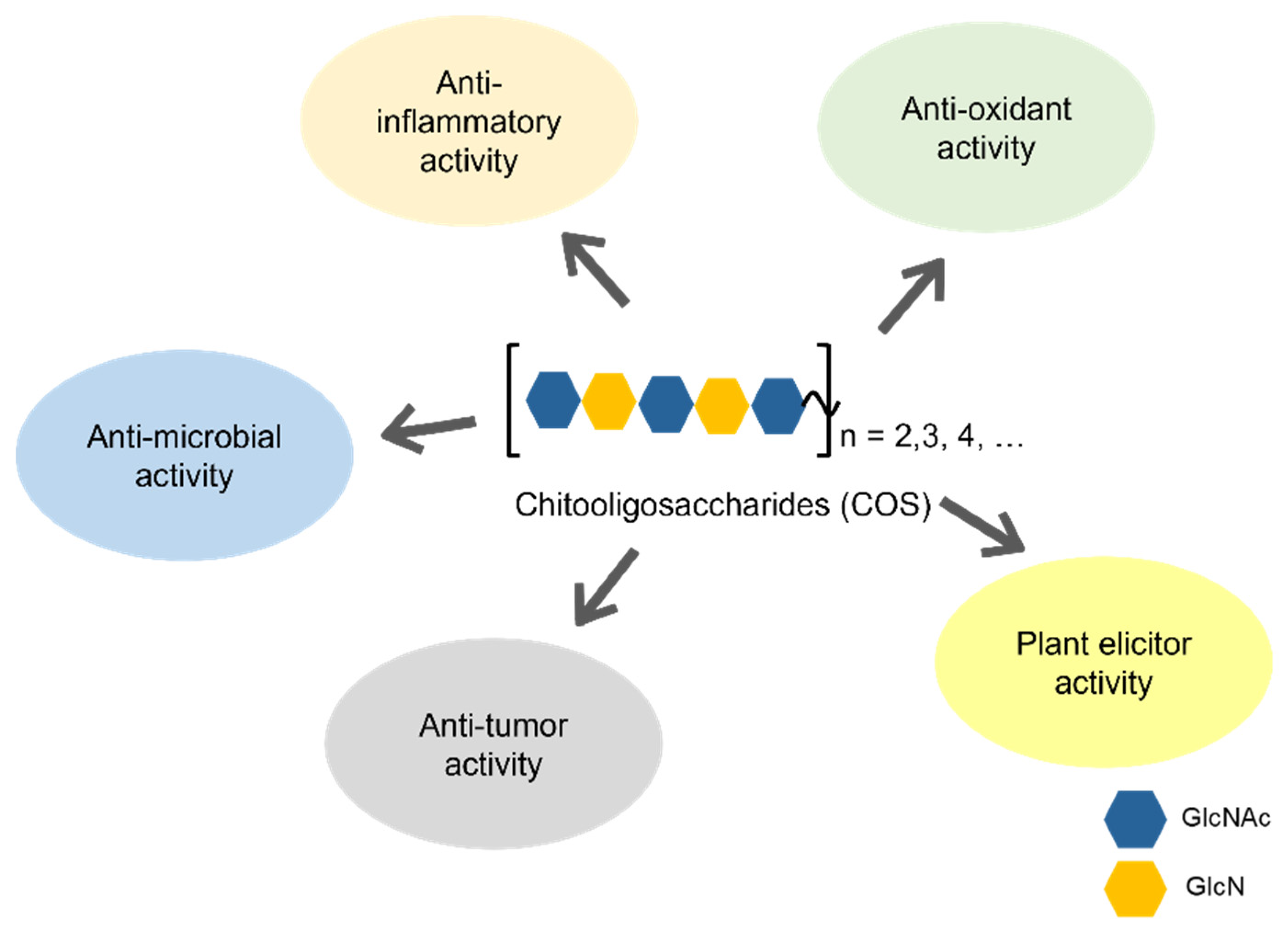
Keywords:
1. Introduction
2. Biological activities of COS
2.1. Anti-microbial activity
2.2. Anti-inflammatory activity
2.3. Anti-oxidant activity
2.4. Anti-tumor activity
2.5. Plant elicitor activity
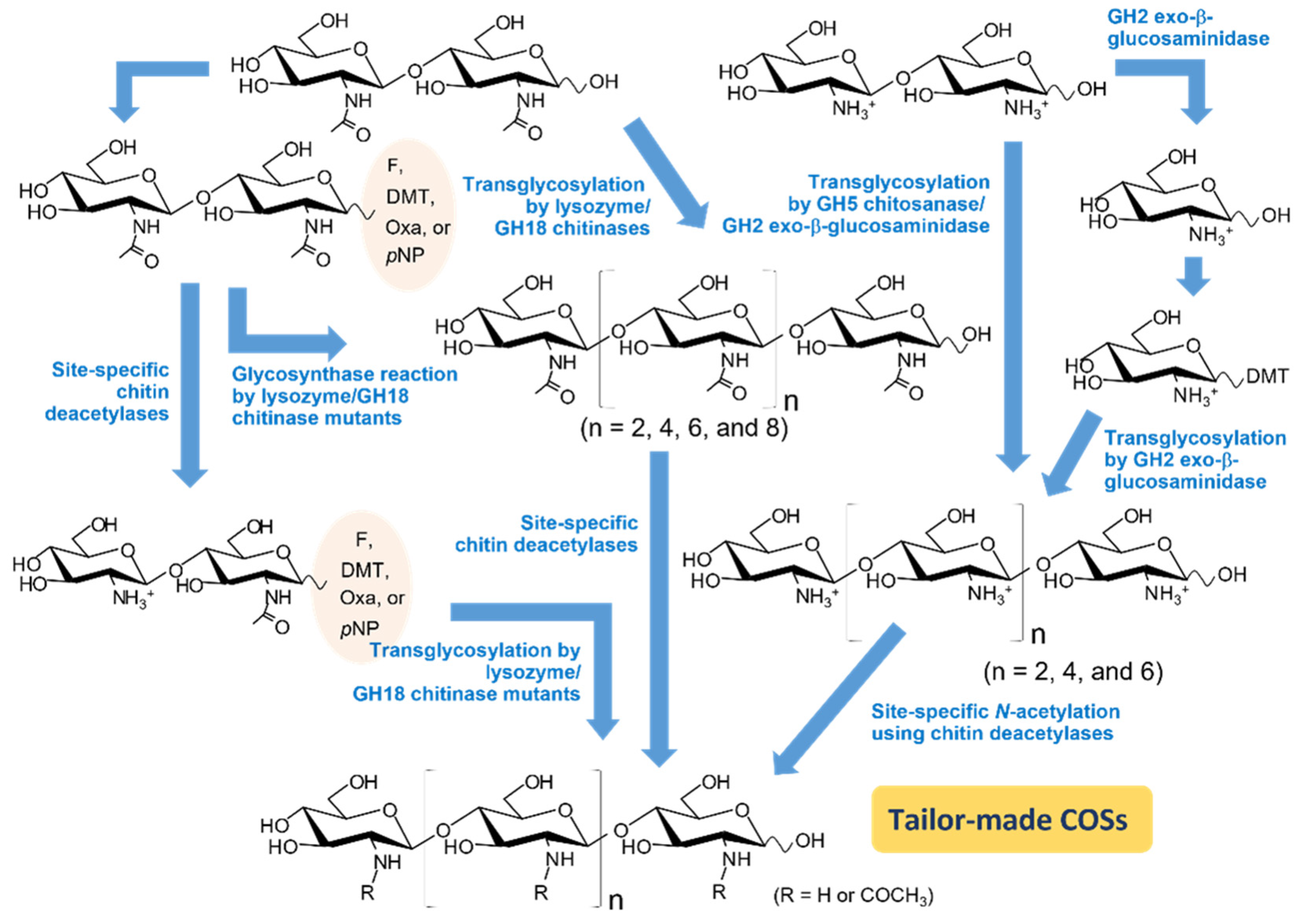
3. Synthetic strategies of COS with promising functions
3.1. Use of transglycosylation (TG) activity in chitinolytic enzymes
3.2. Mutation strategies for enhancing TG activity
3.3. Converting chitinolytic enzymes to glycosynthase
3.4. Use of activated sugars as donor substrates
3.5. Use of site-specific chitin deacetylases
3.6. Metabolic engineering approaches
4. Conclusions and future prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tharanathan, R.N.; Kittur, F.S. Chitin-the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 2003, 43, 61–87. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.J.; Park, R.D. Bioproduction of chitooligosaccharides: present and perspectives. Mar. Drugs. 2014, 2014 12, 5328–5356. [Google Scholar] [CrossRef]
- Liaqat, F.; Eltem, R. Chitooligosaccharides and their biological activities: A comprehensive review. Carbohydr. Polym. 2018, 184, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Kasaai, M.R. The use of various types of NMR and IR spectroscopy for structural characterization of chitin and chitosan. in “Chitin, Chitosan, oligosaccharides, and their derivatives: Biological activities and application”, ed. by Kim, Se-K., CRC Press (Taylor & Francis Group), 2011, pp. 149-170.
- Peter, M.G.; Eberlin, M.N. Application of Mass spectrometry to analyze the structure and Bioactivity of chitooligosaccharides. in “Chitin, Chitosan, oligosaccharides, and their derivatives: Biological activities and application”, ed. by Kim, Se-K., CRC Press (Talor & Francis Group), 2011, pp. 127-148.
- Mengíbar, M.; Mateos-Aparicio, I.; Miralles, B.; Heras, A. Influence of the physico-chemical characteristics of chitooligosaccharides (COS) on antioxidant activity. Carbohydr. Polym. 2013, 97, 776–782. [Google Scholar] [CrossRef]
- Hao, W.; Li, K.; Ge, X.; Yang, H.; Xu, C.; Liu, S. Yu, H.; Li, P.; Xing, R. The effect of N-acetylation on the anti-inflammatory activity of chitooligosaccharides and its potential for relieving endotoxemia. Int. J. Mol. Sci. 2022, 23, 8205. [Google Scholar] [CrossRef]
- Cord-Landwehr, S.; Moerschbacher, B.M. Deciphering the ChitoCode: fungal chitins and chitosans as functional biopolymers. Fungal Biol. Biotechnol. 2021, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Park, P.J.; Kim, S.K. Antimicrobial effect of chitooligosaccharides produced by bioreactor. Carbohydr. Polym. 2001, 44, 71–76. [Google Scholar] [CrossRef]
- Li, K.; Xing, R.; Liu, S.; Qin, Y.; Yu, H.; Li, P. Size and pH effects of chitooligomers on antibacterial activity against Staphylococcus aureus. Int. J. Biol. Macromol. 2014, 64, 302–305. [Google Scholar] [CrossRef]
- No, H.K.; Park, N.Y.; Lee, S.H.; Meyers, S.P. Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int. J. Food Microbiol. 2002, 74, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Aktuganov, GE.; Safina, V.R.; Galimzianova, N.F.; Elena A Gilvanova, Lyudmila Yu Kuzmina, Alexander I Melentiev, Andrei H Baymiev, Sergey A Lopatin.. Constitutive chitosanase from and its potential for preparation of antimicrobial chitooligomers World J Microbiol Biotechnol 2022 Jul 22;38(10):167.
- Ganan, M.; Silje B Lorentzen, S.B.; Jane W Agger, J.W.; Heyward, C.A.; Bakke, O.; Knutsen, S.H.; Aam, B.B.; Eijsink, V.G.H.; Gaustad, P.; Sørlie, M. Antifungal activity of well-defined chitooligosaccharide preparations against medically relevant yeasts. PLoS One 2019, 14, e0210208. [Google Scholar] [CrossRef]
- Kulikov, S.N.; Lisovskaya, S.A.; Zelenikhin, P.V.; Bezrodnykh, E.A.; Shakirova, D.R.; Blagodatskikh, I.V.; Tikhonov, V.E. Antifungal activity of oligochitosans (short-chain chitosans) against some Candida species and clinical isolates of Candida albicans: Molecular weight-activity relationship. Eur. J. Med. Chem. 2014, 74, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Li, K.; Ge, X.; Yang, H.; Xu, C.; Liu, S.; Yu, H.; Li, P.; Xing, R. The Effect of N-acetylation on the anti-inflammatory activity of chitooligosaccharides and its potential for relieving endotoxemia. Int. J. Mol. Sci. 2022, 23, 8205. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.J.; Park, J.K.; Il Park, Y. Anti-inflammatory effects of low-molecular weight chitosan oligosaccharides in IgE–antigen complex-stimulated RBL-2H3 cells and asthma model mice. Int. Immunopharmacol. 2012, 12, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Pangestuti, R.; Bak, S.-S.; Kim, S.-K. Attenuation of pro-inflammatory mediators in LPS-stimulated BV2 microglia by chitooligosaccharides via the MAPK signaling pathway. Int. J. Biol. Macromol. 2011, 49, 599–606. [Google Scholar] [CrossRef]
- Wei, P.; Ma, P.; Xu, Q.-S.; Bai, Q.-H.; Gu, J.-G.; Xi, H.; Du, Y.-G.; Yu, C. Chitosan oligosaccharides suppress production of nitric oxide in lipopolysaccharide-induced N9 murine microglial cells in Vitro. Glycoconj.. J. 2012, 29, 285–295. [Google Scholar] [CrossRef]
- Sánchez, Á.; Mengíbar, M.; Fernández, M.; Alemany, S.; Heras, A.; Acosta, N. Influence of preparation methods of chitooligosaccharides on their physicochemical properties and their anti-inflammatory effects in mice and in RAW264.7 Macrophages. Mar. Drugs 2018, 16, 430. [Google Scholar] [CrossRef]
- Ngo, D.-N.; Kim, M.-M.; Kim, S.-K. Chitin oligosaccharides inhibit oxidative stress in live cells. Carbohydr. Polym. 2008, 74, 228–234. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Ji, L.; Du, X.; Sang, Q.; Chen, F. Enzymatic single-step preparation and antioxidant activity of hetero-chitooligosaccharides using non-pretreated housefly larvae powder. Carbohydr. Polym. 2017, 172, 113–119. [Google Scholar] [CrossRef]
- Liu, H.T.; Li, W.M.; Xu, G.; Li, X.Y.; Bai, X.F.; Wei, P.; Yu, C.; Du, Y.G. Chitosan oligosaccharides attenuate hydrogen peroxide-induced stress injury in human umbilical vein endothelial cells. Pharmacol. Res. 2009, 59, 167–175. [Google Scholar] [CrossRef]
- Liu, H.T.; He, J.L.; Li, W.M.; Yang, Z.; Wang, Y.X.; Bai, X.F.; Yu, C.; Du, Y.G. Chitosan oligosaccharides protect human umbilical vein endothelial cells from hydrogen peroxide-induced apoptosis. Carbohydr. Polym. 2010, 80, 1062–1071. [Google Scholar] [CrossRef]
- Jia, Y.; Ma, Y.; Zou, P.; Cheng, G.; Zhou, J.; Cai, S. Effects of different oligochitosans on isoflavone metabolites, antioxidant activity, and isoflavone biosynthetic genes in soybean (Glycine max) seeds during germination. J. Agric. Food Chem. 2019, 67, 4652–4661. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Li, K.; Ma, Y.; Li, R.; Xing, R.; Yu, H.; Li., P. Preparation and antioxidant activity of chitosan dimers with different sequences. Mar. Drugs 2021, 19, 366. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, K.; Matsumoto, T.; Aizawa, K.; Tokoro, A.; Naruse, R.; Suzuki, S.; Suzuki, M. Antimetastatic and growth-inhibitory effects of N-acetylchitohexaose in mice bearing Lewis lung Carcinoma. Jap. J. Cancer Res. 1990, 81, 259–265. [Google Scholar] [CrossRef]
- Salah, R.; Michaud, P.; Mati, F.; Harrat, Z.; Lounici, H.; Abdi, N.; Drouiche, N.; Mameri, N. Anticancer activity of chemically prepared shrimp low molecular weight chitin evaluation with the human monocyte leukaemia cell line, THP-1. Int. J. Biol. Macromol. 2013, 52, 333–339. [Google Scholar] [CrossRef]
- Park, J.K.; Chung, M.J.; Choi, H.N.; Park, Y.I. Effects of the molecular weight and the degree of deacetylation of chitosan oligosaccharides on antitumor activity. Int. J. Mol. Sci. 2011, 12, 266–277. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Chen, X.; Tian, J.; Li, L.; Zhao, M.; Jiao, Y.; Zhou, C. Effect of chitooligosaccharides on cyclin D1, bcl-xl and bcl-2 mRNA expression in A549 cells using quantitative PCR. Chinese Sci. Bull. 2011, 56, 1629–1632. [Google Scholar] [CrossRef]
- Zhai, X.; Li, C.; Ren, D.; Wang, J.; Ma, C.; Abd El-Aty, A.M. The impact of chitooligosaccharides and their derivatives on the in vitro and in vivo antitumor activity: A comprehensive review. Carbohydr. Polym. 2021, 266, 118132. [Google Scholar] [CrossRef]
- Vander, P.; Km, V.R.; Domard, A.; Eddine, E.G.N.; Moerschbacher, B.M. Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 1998, 118, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, B.; Sarma, P.V.S.R.N.; Ankati, S.; Bhuvanachandra, B.; Podile, A.R. Elicitation of defense response by transglycosylated chitooligosaccharides in rice seedlings. Carbohydr. Res. 2021 510, 108459. [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; Zhou, J.M.; Chai, J. Chitin-induced dimerization activates a plant immune receptor. Science. 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife. 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, J.; Han, Z.; Gong, X.; Zhang, H.; Chai, J. Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure. 2016, 24, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Hayafune, M.; Berisio, R.; Marchetti, R.; Silipo, A.; Kayama, M.; Desaki, Y.; Arima, S.; Squeglia, F.; Ruggiero, A.; Tokuyasu, K.; Molinaro, A.; Kaku, H.; Shibuya, N. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, E404–E413. [Google Scholar] [CrossRef]
- Gubaeva, E.; Gubaev, A.; Melcher, R.L.J.; Cord-Landwehr, S.; Singh, R.; El Gueddari, N.E.; Moerschbacher, B.M. ‘Slipped Sandwich’ model for chitin and chitosan perception in Arabidopsis. Mol. Plant Microbe Interact. 2018, 31, 1145–1153. [Google Scholar] [CrossRef]
- Mészáros, Z.; Nekvasilová, P.; Bojarová, P.; Křen, V.; Slámová, K. Advanced glycosidases as ingenious biosynthetic instruments. Biotechnol Adv. 2021, 49, 107733. [Google Scholar] [CrossRef]
- Chipman, D.M.; Pollock, J.J.; Sharon, N. Lysozyme-catalyzed hydrolysis and transglycosylation reactions of bacterial cell wall oligosaccharides. J. Biol. Chem. 1968, 243, 487–496. [Google Scholar] [CrossRef]
- Masaki, A.; Fukamizo, T.; Otakara, A.; Torikata, T.; Hayashi, K.; Imoto, T. Lysozyme-catalyzed reaction of chitooligosaccharides. J. Biochem. 1981, 90, 527–533. [Google Scholar] [CrossRef]
- Masaki, A.; Fukamizo, T.; Otakara, A.; Torikata, T.; Hayashi, K.; Imoto, T. Estimation of rate constants in lysozyme-catalyzed reaction of chitooligosaccharides. J. Biochem. 1981, 90, 1167–1175. [Google Scholar] [CrossRef]
- Fukamizo, T.; Torikata, T.; Nagayama, T.; Minematsu, T.; Hayashi, K. Enzymatic activity of avian egg-white lysozymes. J. Biochem. 1983, 94, 115–122. [Google Scholar] [CrossRef]
- Fukamizo, T.; Minematsu, T.; Yanase, Y.; Hayashi, K.; Goto., S. Substrate size dependence of lysozyme-catalyzed reaction. Arch. Biochem. Biophys. 1986, 250, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Sakabe, Y.; Ogata, M.; Michishita, K.; Dohra, H.; Kawagishi, H.; Totani, K.; Nikaido, M.; Nakamura, T.; Koshino, H.; Usui, T. Enzymatic synthesis of an α-chitin-like substance via lysozyme-mediated transglycosylation. Carbohydr. Res. 2012, 347, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Umemoto, N.; Ohnuma, T.; Numata, T.; Suzuki, A.; Usui, T.; Fukamizo, T. A novel transition-state analogue for lysozyme, 4-O-β-tri-N-acetylchitotriosyl moranoline, provided evidence supporting the covalent glycosyl-enzyme intermediate. J. Biol. Chem. 2013, 288, 6072–6082. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Matsui, M.; Kono, H.; Matsuzaki, Y.; Kato, Y.; Usui, T. A novel analytical procedure for assaying lysozyme activity using an end-blocked chitotetraose derivative as substrate. Anal. Biochem. 2017, 538, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Madhuprakash, J.; Singh, A.; Kumar, S.; Sinha, M.; Kaur, P.; Sharma, S.; Podile, A.R.; Singh, T.P. Structure of chitinase D from Serratia proteamaculans reveals the structural basis of its dual action of hydrolysis and transglycosylation. Int. J. Biochem. Mol. Biol. 2013, 4, 166–178. [Google Scholar]
- Bhuvanachandra, B.; Podile, A.R. A transglycosylating chitinase from Chitiniphilus shinanonensis (CsChiL) hydrolyzes chitin in a processive manner. Int. J. Biol. Macromol. 2020, 145, 1–10. [Google Scholar] [CrossRef]
- Fukamizo, T.; Goto, S.; Torikata, T.; Araki, T. State of binding subsites in Asp 101-modified lysozymes. J. Biochem. 1990, 107, 445–451. [Google Scholar] [CrossRef]
- Fukamizo, T.; Goto, S.; Torikata, T.; Araki, T. Enhancement of transglycosylation of lysozyme by chemical modification. Agric. Biol. Chem. 1989, 53, 2641–2651. [Google Scholar] [CrossRef]
- Fukamizo, T.; Fleury, A.; Côté, N.; Mitsutomi, M.; Brzezinski, R. Exo-b-D-glucosaminidase from Amycolatopsis orientalis: catalytic residues, sugar recognition specificity, kinetics, and synergism. Glycobiol. 2006, 16, 1064–1072. [Google Scholar] [CrossRef]
- Tanabe, T.; Morinaga, K.; Fukamizo, T.; Mitsutomi, M. Novel chitosanase from Streptomyces griseus HUT 6037 with transglycosylation activity. Biosci. Biotechnol. Biochem. 2003, 67, 354–364. [Google Scholar] [CrossRef]
- Aronson, N.N., Jr.; Halloran, B.A.; Alexeyev, M.F.; Zhou, X.E.; Wang, Y.; Meehan, E.J.; Chen, L. Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia marcescens chitinase A enhances transglycosylation. Biosci. Biotechnol. Biochem. 2006, 70, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Zakariassen, H.; Hansen, M.C.; Jøranli, M.; Eijsink, V.G.H.; Sørlie, M. Mutational effects on transglycosylating activity of family 18 chitinases and construction of a hypertransglycosylating mutant. Biochemistry 2011, 50, 5693–5703. [Google Scholar] [CrossRef]
- Sirimontree, P.; Suginta, W.; Sritho, N.; Kanda, Y.; Shinya, S.; Ohnuma, T.; Fukamizo, T. Mutation strategies for obtaining chitooligosaccharides with longer chains by transglycosylation reaction of family GH18 chitinase. Biosci. Biotechnol. Biochem. 2014, 78, 2014–2021. [Google Scholar] [CrossRef] [PubMed]
- Madhuprakash, J.; Dalhus, B.; Rani, T.S.; Podile, A.R.; Eijsink, V.G.H.; Sørlie, M. Key residues affecting transglycosylation activity in family 18 chitinases: Insights into donor and acceptor subsites. Biochemistry. 2018, 57, 4325–4337. [Google Scholar] [CrossRef] [PubMed]
- Madhuprakash, J.; Tanneeru, K.; Purushotham, P.; Guruprasad, L.; Podile, A.R. Transglycosylation by chitinase D from Serratia proteamaculans improved through altered substrate interactions. J. Biol. Chem. 2012, 287, 44619–44627. [Google Scholar] [CrossRef]
- Umemoto, N.; Kanda, Y.; Ohnuma, T.; Osawa, T.; Numata, T.; Sakuda, S.; Taira, T.; Fukamizo, T. Crystal structures and inhibitor binding properties of plant class V chitinases: the cycad enzyme exhibits unique structural and functional features. Plant J. 2015, 82, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Umemoto, N.; Ohnuma, T.; Mizuhara, M.; Sato, H.; Skriver, K.; Fukamizo, T. Introduction of a tryptophan side chain into subsite +1 enhances transglycosylation activity of a GH-18 chitinase from Arabidopsis thaliana, AtChiC. Glycobiology. 2013, 23, 81–90. [Google Scholar] [CrossRef]
- Umemoto, N.; Ohnuma, T.; Osawa, T.; Numata, T.; Fukamizo, T. Modulation of the transglycosylation activity of plant family GH18 chitinase by removing or introducing a tryptophan side chain. FEBS Lett. 2015, 589, 2327–2333. [Google Scholar] [CrossRef]
- Faijes, M.; Planas, A. In vitro synthesis of artificial polysaccharides by glycosidases and glycosynthases. Carbohydr. Res. 2007, 342, 1581–1594. [Google Scholar] [CrossRef]
- Danby, P.M.; Withers, S.G. Advances in enzymatic glycoside synthesis. ACS Chem. Biol. 2016, 11, 1784–1794. [Google Scholar] [CrossRef]
- Honda, Y.; Kitaoka, M. The first glycosynthase derived from an inverting glycoside hydrolase. J. Biol. Chem. 2006, 281, 1426–1431. [Google Scholar] [CrossRef]
- van Aalten, D.M.; Komander, D.; Synstad, B.; Gåseidnes, S.; Peter, M.G.; Eijsink, V.G. Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 8979–8984. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Fukuda, T.; Dozen, S.; Honda, Y.; Kitaoka, M.; Fukamizo, T. A glycosynthase derived from an inverting GH19 chitinase from the moss Bryum coronatum. Biochem. J. 2012, 444, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Dozen, S.; Honda, Y.; Kitaoka, M.; Fukamizo, T. A glycosynthase derived from an inverting chitinase with an extended binding cleft. J. Biochem. 2016, 160, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Umemoto, N.; Nagata, T.; Shinya, S.; Numata, T.; Taira, T.; Fukamizo, T. Crystal structure of a “loopless” GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center. Biochim. Biophys. Acta. 2014, 1844, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Umemoto, N.; Kondo, K.; Numata, T.; Fukamizo, T. Complete subsite mapping of a “loopful” GH19 chitinase from rye seeds based on its crystal structure. FEBS Lett. 2013, 587, 2691–2697. [Google Scholar] [CrossRef]
- Rousseau, A.; Armand, S.; Cottaz, S.; Fort, S. Size-controlled synthesis of β(1→4)-GlcNAc oligosaccharides using an endo-glycosynthase. Chemistry. 2021, 27, 17637–17646. [Google Scholar] [CrossRef] [PubMed]
- Rastelli, R.A.; Bucke, C. Enzymatic synthesis of oligosaccharides, Biotechnol. Genet. Eng. Rev., 1992, 10, 253–282. [Google Scholar] [CrossRef]
- Harmsen, R.A.G.; Aam, B.B.; Madhuprakash, J.; Hamre, A.G.; Goddard-Borger, E.D.; Withers, S.G.; Eijsink, V.G.H.; Sørlie, M. Chemoenzymatic synthesis of chito-oligosaccharides with alternating N-D-acetylglucosamine and D-glucosamine. Biochemistry, 2020, 59, 4581–4590. [Google Scholar] [CrossRef]
- Tanaka, T.; Noguchi, M.; Kobayashi, A.; Shoda, S. A novel glycosyl donor for chemo-enzymatic oligosaccharide synthesis: 4,6-dimethoxy-1,3,5-triazin-2-yl glycoside. Chem. Commun. (Camb). 2008, 17, 2016–2018. [Google Scholar] [CrossRef]
- Tanaka, T.; Wada, T.; Noguchi, M.; Ishihara, M.; Kobayashi, A.; Ohnuma, T.; Fukamizo, T.; Brzezinski, R.; Shoda, S.-I. 4,6-Dimethoxy-1,3,5-triazin-2-yl β-D-glycosaminides: novel substrates for transglycosylation reaction catalyzed by exo-β-D-glucosaminidase from Amycolatopsis orientalis. J. Carbohydr. Chem., 2012, 31, 634–646. [Google Scholar] [CrossRef]
- Ohnuma, T.; Tanaka, T.; Urasaki, A.; Dozen, S.; Fukamizo, T. A novel method for chemo-enzymatic synthesis of chitin oligosaccharide catalyzed by the mutant of inverting family GH19 chitinase using 4,6-dimethoxy-1,3,5-triazin-2-yl α-chitobioside as a glycosyl donor. J. Biochem. 2019, 165, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, M.; Tanaka, T.; Gyakushi, H.; Kobayashi, A.; Shoda, S. Efficient synthesis of sugar oxazolines from unprotected N-acetyl-2-amino sugars by using chloroformamidinium reagent in water. J. Org. Chem. 2009, 74, 2210–2212. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Tanaka, T.; Noguchi, M.; Kobayashi, A.; Ishikura, K.; Ikenuma, T.; Seno, H.; Watanabe, T.; Kohri, M.; Shoda, S.-I. One-pot chemoenzymatic route to chitoheptaose via specific transglycosylation of chitopentaose-oxazoline on chitinase-template. Chem. Lett. 2012, 41, 689–690. [Google Scholar] [CrossRef]
- Alsina, C.; Sancho-Vaello, E.; Aranda-Martínez, A.; Faijes, M.; Planas, A. Auxiliary active site mutations enhance the glycosynthase activity of a GH18 chitinase for polymerization of chitooligosaccharides. Carbohydr. Polym. 2021, 252, 117121. [Google Scholar] [CrossRef]
- Umemoto, N.; Saito, N.; Noguchi, M.; Shoda, S.-I.; Ohnuma, T.; Watanabe, T.; Sakuda, S.; Fukamizo, T. Plant chitinase mutants as the catalysts for chitooligosaccharide synthesis using the sugar oxazoline derivatives. J. Agric. Food Chem. 2022, 70, 12897–12906. [Google Scholar] [CrossRef]
- Hembach, L.; Cord-Landwehr, S.; Moerschbacher, B.M. Enzymatic production of all fourteen partially acetylated chitosan tetramers using different chitin deacetylases acting in forward or reverse mode. Sci. Rep. 2017, 7, 17692. [Google Scholar] [CrossRef]
- Cord-Landwehr, S.; Ihmor, P.; Niehues, A.; Luftmann, H.; Moerschbacher, B.M.; Mormann, M. Quantitative mass-spectrometric sequencing of chitosan oligomers revealing cleavage sites of chitosan hydrolases. Anal. Chem. 2017, 89, 2893–2900. [Google Scholar] [CrossRef]
- Hamer, S.N.; Cord-Landwehr, S.; Biarnés, X.; Planas, A.; Waegeman, H.; Moerschbacher, B.M.; Kolkenbrock, S. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci.Rep. 2015, 5, 8716. [Google Scholar] [CrossRef]
- Sreekumar, S.; Wattjes, J.; Niehues, A.; Mengoni, T.; Mendes, A.C.; Morris, E.R.; Goycoolea, F.M.; Moerschbacher, B.M. Biotechnologically produced chitosans with nonrandom acetylation patterns differ from conventional chitosans in properties and activities. Nat. Commun. 2022, 13, 7125. [Google Scholar] [CrossRef]
- Ling, M.; Li, J.; Du, G.; Liu, L. Metabolic engineering for the production of chitooligosaccharides: advances and perspectives. Emerg. Top Life Sci. 2018, 2, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.; Wu, Y.; Tian, R.; Liu, Y.; Yu, W.; Tao, G.; Lv, X.; Li, J.; Du, G.; Amaro, R.L.; Liu, L. Combinatorial pathway engineering of Bacillus subtilis for production of structurally defined and homogeneous chitooligosaccharides. Metab. Eng. 2022, 70, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Weyer, R.; Hellmann, M.J.; Hamer-Timmermann, S.N.; Singh, R.; Moerschbacher, B.M. Customized chitooligosaccharide production-controlling their length via engineering of rhizobial chitin synthases and the choice of expression system. Front. Bioeng. Biotechnol. 2022, 10, 1073447. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Fukamizo, T.; Suginta, W. Bioeconomic production of high-quality chitobiose from chitin food wastes using an in-house chitinase from Vibrio campbellii. Biores. Bioprocess. 2022, 9, 86. [Google Scholar] [CrossRef]
- Chen, T.; Cheng, G.; Jiao, S.; Ren, L.; Zhao, C.; Wei, J.; Han, J.; Pei, M.; Du, Y.; Li, J.J. Expression and biochemical characterization of a novel marine chitosanase from Streptomyces niveus suitable for preparation of chitobiose. Mar. Drugs. 2021, 19, 300. [Google Scholar] [CrossRef]
- Li, Y.; Gou, Y.; Liu, Z.; Xie, T.; Wang, G. Structure-based rational design of chitosanase CsnMY002 for high yields of chitobiose. Colloids Surf. B Biointerfaces. 2021, 202, 111692. [Google Scholar] [CrossRef]
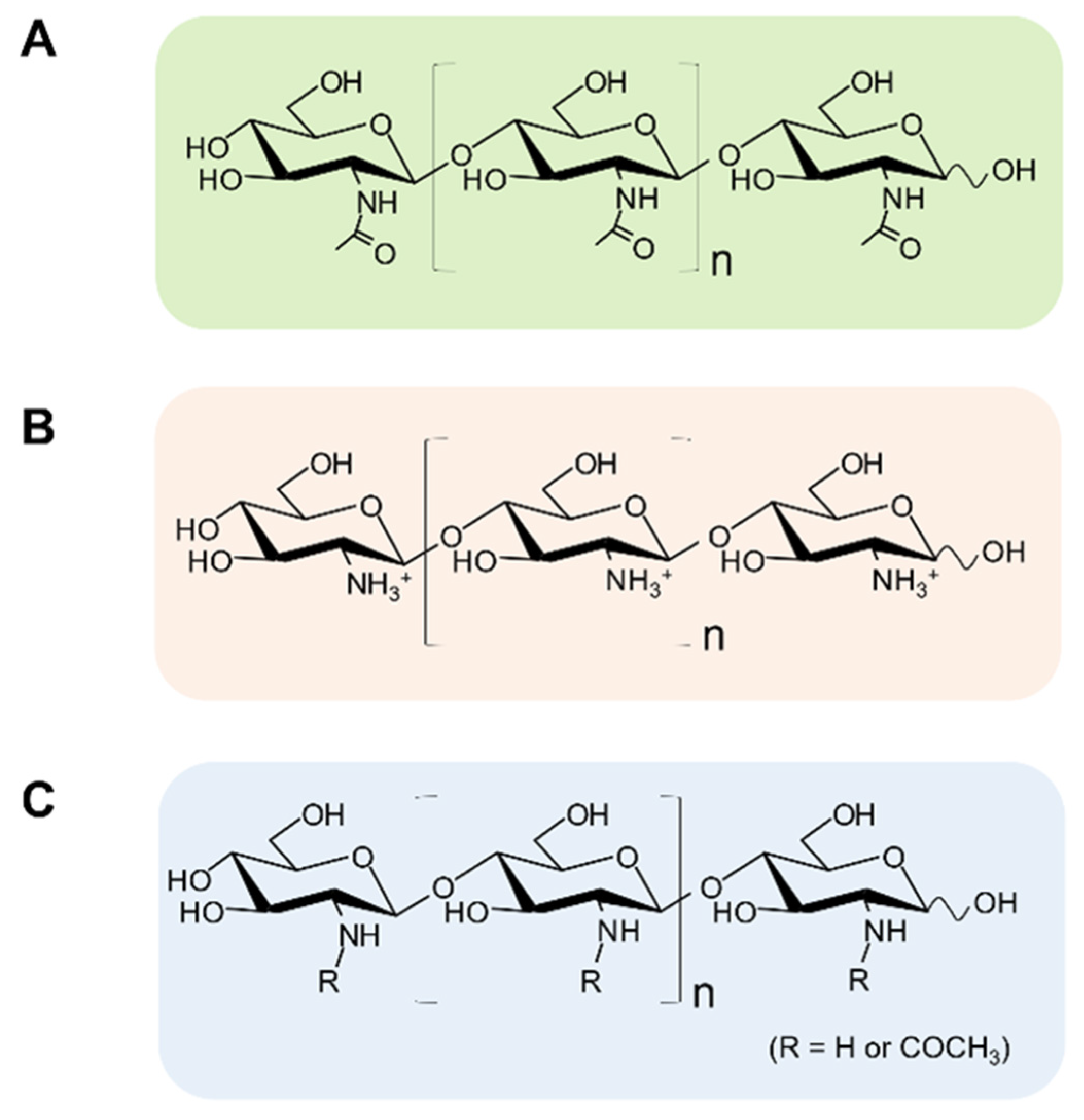
| MW or DP | Source, DA or sequence | Activity | Assay | Ref | |
|---|---|---|---|---|---|
| Anti-microbial activity | Higher MW (10 kDa), Medium MW (5 kDa), and Lower MW (1 kDa) |
from chitosan | Higher activity (10 kDa) Lower activity (1 and 5 kDa) |
Growth inhibition toward Gram-positive, Gram-negative, and Lactic acid bacteria |
[9] |
| Chitosan polymers and Chitosan oligomers |
from chitosan | Higher activity (polymer) Lower activity (oligomer) |
Growth inhibition toward Staphyllococcus aureus Gram-positive and Gram-negative bacteria |
[10,11] | |
| DPs of 2, 3, and 4, and Lower MW Initial polymer |
DA, 0.15-0.5 chitosan |
Higher activity (DP of 2-4 and Lower MW) Lower activity (polymer) |
Growth inhibition toward fungi and bacteria |
[12] | |
| Average DPs of 17, 31, 54, and 62 | from chitosan (DA, 0.15) |
Higher activity (DP of 31 and 54) Lower activity (DP of 17 and 62) |
Growth inhibition toward yeast, fungi, including Candida |
[13] | |
| Chitosans (70 and 600 kDa) Chitosan oligomers |
from chitosan | Higher activity (oligomers) |
Growth inhibition toward Candida | [14] |
| MW or DP | Source, DA or sequence | Activity | Assay | Ref. | |
|---|---|---|---|---|---|
| Anti-inflammatory activity | COS with similar DP distribution | Different DAs of 0,0.12,0.50, and 0.85 | COS with DA of 0.12 has the highest activity | Inhibition of LPS-induced inflammatory cytokines burst, down-regulating its mRNA expression, and reducing phosphorylation of IκBα. | [15] |
| Chitosan oligomer mixture (DPs of 3-5) |
DA of 0.0 | Active toward allergic asthma inflammation in vivo | Inhibition of degranulation, cytokine generation in RBL-2H3 cells, and lung inflammation in allergic asthma model mice | [16] | |
| Chitosan oligomers with MW <1 kDa, 1-3 kDa, 3-5 kDa, and 5-10 kDa | from chitosan |
The lower the MW, the higher the activity. | Inhibition of iNOS and cyclooxygenase expressions | [17] | |
| MW<1 kDa (DP2-6; 4, 16, 29,37, and 14%, respectively) | DA <0.05 | Active toward N9 microglia cells | Inhibition of NO production by suppressing iNOS expression | [18] | |
| 42 %fully de-N-acetylated/54% mono-N-acetylated oligomers (42/54) and 50% fully de-N—acetylated/27% mono-N-acetylated oligomers (50/27) | from chitosan |
42/54 attenuated the inflammation both in vivo and in vitro, but 50/27 promoted the inflammatory response | Effects of COS preparations on the inflammation in lipopolysaccharide-induced mice and in RAW264.7 macrophages | [19] |
| MW or DP | Source, DA or sequence | Activity | Assay | Ref | |
|---|---|---|---|---|---|
| Anti-oxidant activity | COS with DP of 1,2, and 3 | fully N-acetylated crab chitin (DA about 1.0) | Attenuated peroxidase activity and oxidation of DNA/membrane proteins, and exhibited free-radical scavenging effects | Experiments using human myeloid cells, HL-60 and mouse macrophages RAW 264.7 | [20] |
| Hetero-COS with DP of 2-6 | from housefly larvae powder | The COS with major DP of 5 exhibited the highest activity | Hydroxyl-scavenging effects | [21] | |
| COS | from chitosan (DA = 5%) |
Protective effects on H2O2-induced apoptosis or oxidative damage | Experiments using human umbilical vein endothelial cell | [22,23] | |
| COS with different DPs (3-7) | from chitosan | COS with DP of 6 has the highest activity | Effects on isoflavone contents and antioxidant activity in soyabean seeds during germination | [24] | |
| COS dimers, AA, DA, AD, and DD (A, GlcNAc; D, GlcN) |
Site-specific chitin deacetylase treatments of the purified (GlcNAc)2 |
DD>AD>>DA, AA toward superoxide radicals AA>DA>>AD, DD toward hydroxyl radicals DD>>AD, DA, AA toward DPPH |
All scavenging effects were examined in vitro | [25] |
| MW or DP | Source, DA or sequence | Activity | Assay | Ref | |
|---|---|---|---|---|---|
| Anti-tumor activity | (GlcNAc)6 | from crab chitin | Decreased the pulmonary nodules | Experiments using Lewis lung carcinoma mice | [26] |
| Chitin, chitosan 2.5-338 kDa |
from shrimp shell chitin | Activity of chitin (2.5 kDa) was higher than those of chitin (338 kDa) and chitosan (12 kDa) | Experiments using human leukemia cells | [27] | |
| COS from chitosan hydrolysis (DPs of 2,3, and 4 are the major components) | Enzymatic hydrolysis of high molecular weight chitosan | COS with lower MW> COS with higher MW | Experiments using prostate and lung cancer cells and hepatoma cells | [28] | |
| Chitosan oligomers DP 2-6 | Chemical and enzymatic hydrolysis of chitosan | For effective inhibition of the cell proliferation, DP should be at least 6 | Inhibitory effect on A549 cell proliferation | [29] |
| MW or DP | Source, DA or sequence | Activity | Assay | Ref | |
|---|---|---|---|---|---|
| Plant elicitor | (GlcNAc)n (n=4,5,6,7,8,9, and 10), (GlcN)n (n=5,6, and 7) and Partially N-acetylated chitosans |
Chemical hydrolysis of chitin (fluorolysis) deacetylation of the high MW chitin |
(GlcN)n were not active as elicitors. (GlcNAc)n with a DP ≥7 strongly elicited POD activities but not PAL activities. Partially N-acetylated chitosans elicited both PAL and POD activities. |
Induction of phenylalanine ammonia-lyase (PAL), peroxidase (POD) in healthy wheat leaves | [31] |
| (GlcNAc)n (n=5,6, and 7) |
Enzymatic transglycosylation | (GlcNAc)7 strongly induced oxidative burst response as well as peroxidase and PAL activities. | Induction of phenylalanine ammonia-lyase (PAL), peroxidase (POD), PR protein gene expression in rice seedlings | [32] | |
| (GlcNAc)8 and hetero-COS with a DP of 8 (A, GlcNAc; D, GlcN) |
Enzymatic synthesis | AAAAAAAA was elicitor active, but DADADADA inactive | Inhibition of CEBiP-dimerization and Reactive oxygen generation | [37] | |
| (GlcNAc)6 and two hetero-COS with DP of 6 (A, GlcNAc; D, GlcN) |
Enzymatic deacetylation of (GlcNAc)6 |
AAAAAA > ADAAAA > ADAADA |
Inhibition of reactive oxygen generation | [38] |
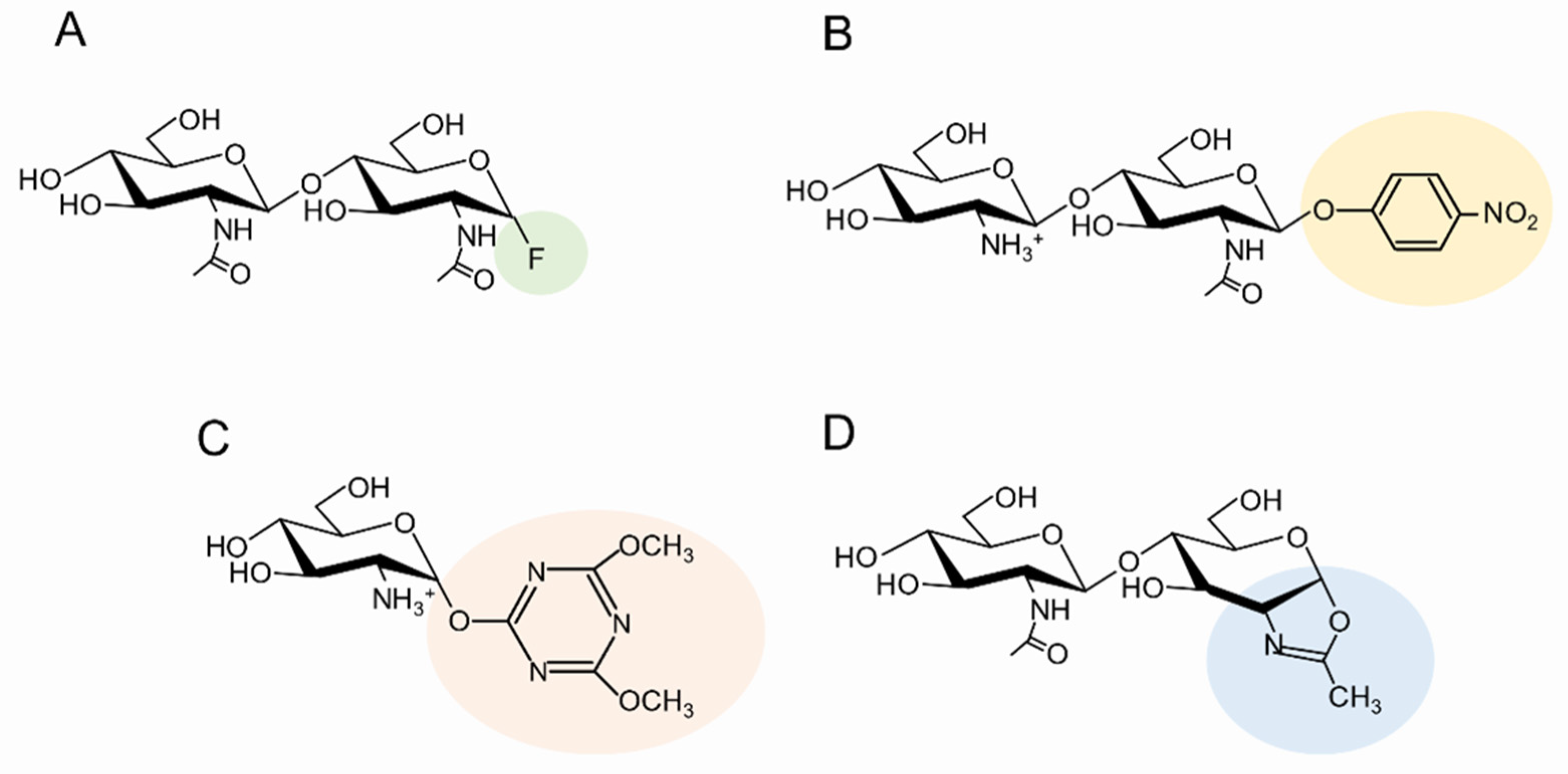
| Catalysts | Substrates | Products | Ref. | |
|---|---|---|---|---|
| Donor | acceptor | |||
| Hen egg white lysozyme Wild type |
(GlcNAc)3 | (GlcNAc)n (n = 3–15) | [45] | |
| Hen egg white lysozyme Wild type |
(GlcNAc)4 | Moranoline (1-deoxynojirimycin) | 4-O-b-di(tri)-N-acetylchitobi(tri)osyl moranoline | [46] |
| Hen egg white lysozyme Wild type |
(GlcNAc)4 | 2-acetamide-2,3-dideoxy-glucopyranose | 44-O-β-d-galactosyl-β-tri-N-acetylchitotriosyl 2-acetamide-2,3-dideoxy-glucopyranose | [47] |
| Hen egg white lysozyme Asp101, Trp62-modified |
(GlcNAc)5 | (GlcNAc)9 | [51] | |
| Amycolatopsis orientalis GH2 exo-b-D-glucosaminidase | (GlcN)4 | (GlcN)5 and (GlcN)6 | [52] | |
| Streptomyces griseus HUT6037 GH5 endo-chitosanase | (GlcN)5 | (GlcNAc)3 | (GlcN)2-(GlcNAc)3 (GlcN)3-(GlcNAc)3 |
[53] |
|
Serratia marcescens GH18 chitinase A mutated at Trp at site -3. |
(GlcNAc)4 or (GlcNAc)5 | (GlcNAc)6 or (GlcNAc)7 | [54] | |
|
Serratia marcescens GH18 chitinases A and B mutated at the middle Asp in the DxDxE motif. |
(GlcNAc)4 | (GlcNAc)3 produced through the transglycosylation product (GlcNAc)6 | [55] | |
| Vibrio harveyi GH18 chitinase A mutated at the middle Asp in the DxDxE motif. | (GlcNAc)4 (GlcNAc)6 |
(GlcNAc)6 (GlcNAc)8 |
[56] | |
|
Serratia proteamaculans GH18 chitinase D triple-mutated at the glycon- or aglycon-binding aromatic residues as well as at the middle Asp in the DxDxE motif. |
(GlcNAc)4 | (GlcNAc)5 or (GlcNAc)6 | [57] | |
|
Serratia proteamaculans GH18 chitinase D single-mutated at the catalytic center and the binding groove. |
(GlcNAc)4 | (GlcNAc)5 or (GlcNAc)6 | [58] | |
| Arabidopsis thaliana GH18 chitinase C mutant, in which tryptophan side chain was introduced into the upper portion of the catalytic center. | (GlcNAc)4 | (GlcNAc)3 produced through the transglycosylation product (GlcNAc)6 | [60] | |
| Cycas revoluta GH18 chitinase A mutant, in which tryptophan side chain was introduced into the upper portion of the catalytic center. | (GlcNAc)4 | (GlcNAc)3 produced through the transglycosylation product (GlcNAc)6 | [61] | |
| A glycosynthase derived from Bryum coronatum GH19 chitinase A | (GlcNAc)2-fluoride | (GlcNAc)2 | (GlcNAc)4 | [66] |
| A glycosynthase derived from Secale cereale GH19 chitinase C | (GlcNAc)3-fluoride | (GlcNAc)4 | (GlcNAc)7 | [67] |
| A chitin-oligosaccharide N-deacetylase (NodB) and a glycosynthase derived from hen egg white lysozyme (Asp52→Ser) |
GlcN-(GlcNAc)2-fluoride | (GlcNAc)3 (GlcNAc)4 (GlcNAc)5 |
GlcN-(GlcNAc)5 GlcN-(GlcNAc)6 GlcN-(GlcNAc)7 |
[70] |
| Hyper-transglycosylating mutants from Serratia marcescens GH18 chitinases A and Serratia proteamaculans GH18 chitinase D | GlcN-GlcNAc-pNP (p-nitorophenylated) obtained by enzymatic de-N-acetylation of (GlcNAc)2-pNP | (GlcN-GlcNAc)2 (GlcN-GlcNAc)3 (GlcN-GlcNAc)4 (GlcN-GlcNAc)5 |
[72] | |
| Amycolatopsis orientalis GH2 exo-b-glucosaminidase | GlcN-DMT (4,6-dimethoxy-1,3,5-triazin-2-yl) | (GlcNAc)2 | GlcN-(GlcNAc)2 | [74] |
| A glycosynthase derived from Bryum coronatum GH19 chitinase A | (GlcNAc)2-DMT | (GlcNAc)2 | (GlcNAc)4 | [75] |
| An activity-suppressed mutant from Bacillus circulans GH18 chitinase A1 | (GlcNAc)2-oxa | (GlcNAc)5 | (GlcNAc)7 | [77] |
| Catalytic-site mutants from Serratia proteamaculans GH18 chitinase D | (GlcNAc)5-oxa | (GlcNAc)10 | [78] | |
| Hyper-transglycosylating mutants from Nicotiana tobaccum GH18 chitinase C and Cycas revoluta GH18 chitinase A | (GlcNAc)2-oxa (GlcNAc)3-oxa (GlcNAc)4-oxa (GlcNAc)5-oxa |
(GlcNAc)5 (GlcNAc)4 (GlcNAc)3 (GlcNAc)2 |
(GlcNAc)7 | [79] |
| Site-specific chitin dectylases from fungi | (GlcNAc)4 | A full lineup of partially N-acetylated chitotetraoses | [80] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





