Submitted:
25 May 2023
Posted:
26 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
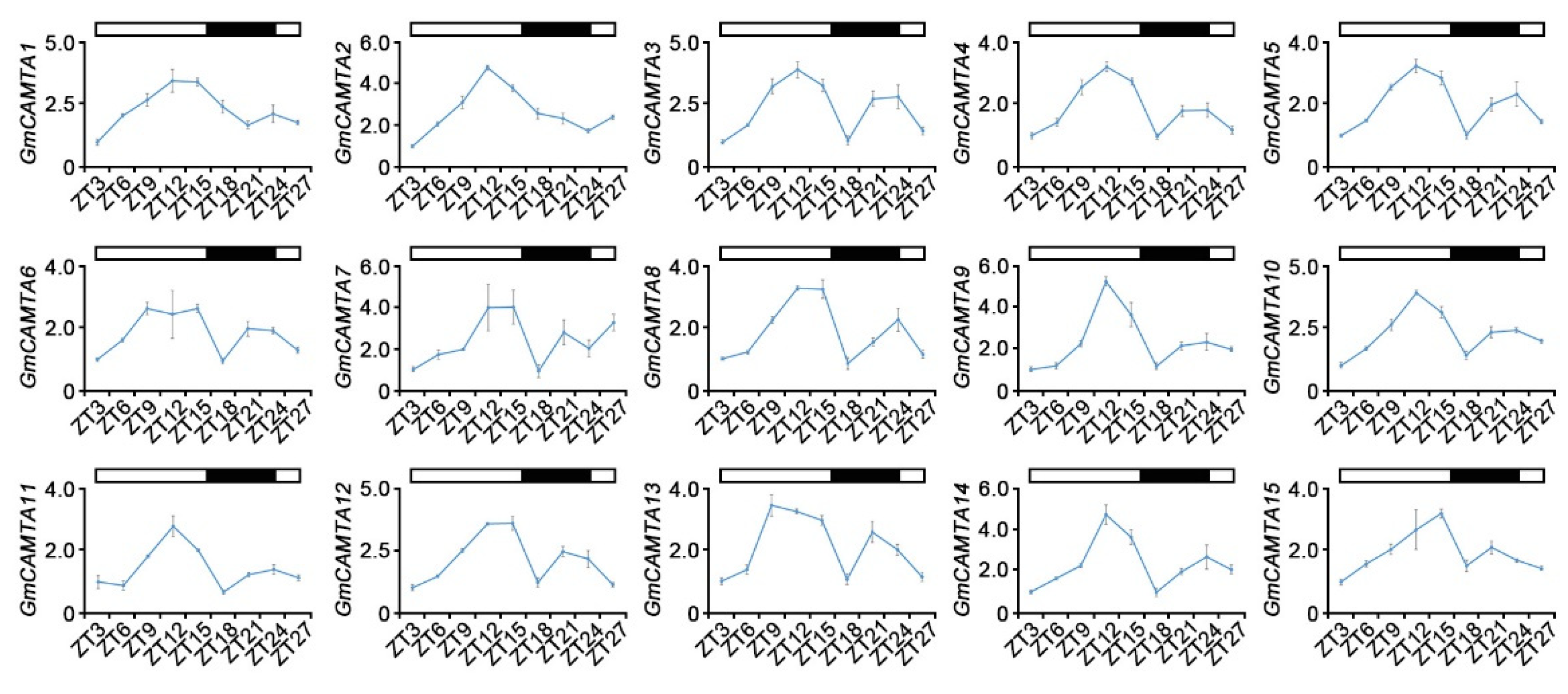
2.1. Circadian rhythms affected the expression patterns of GmCAMTAs under long-day conditions
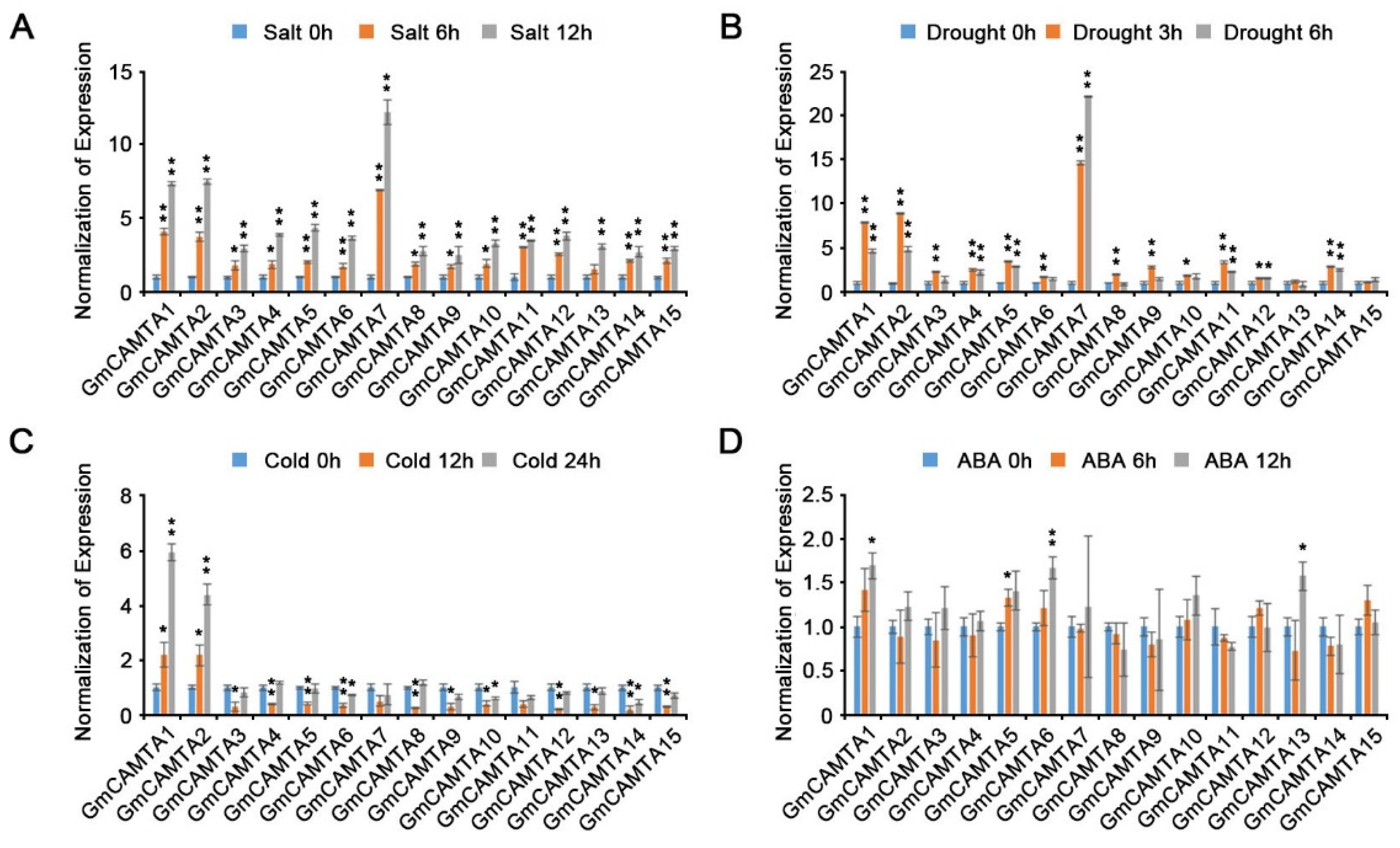
2.2. Various abiotic stresses affected the expression of GmCAMTAs
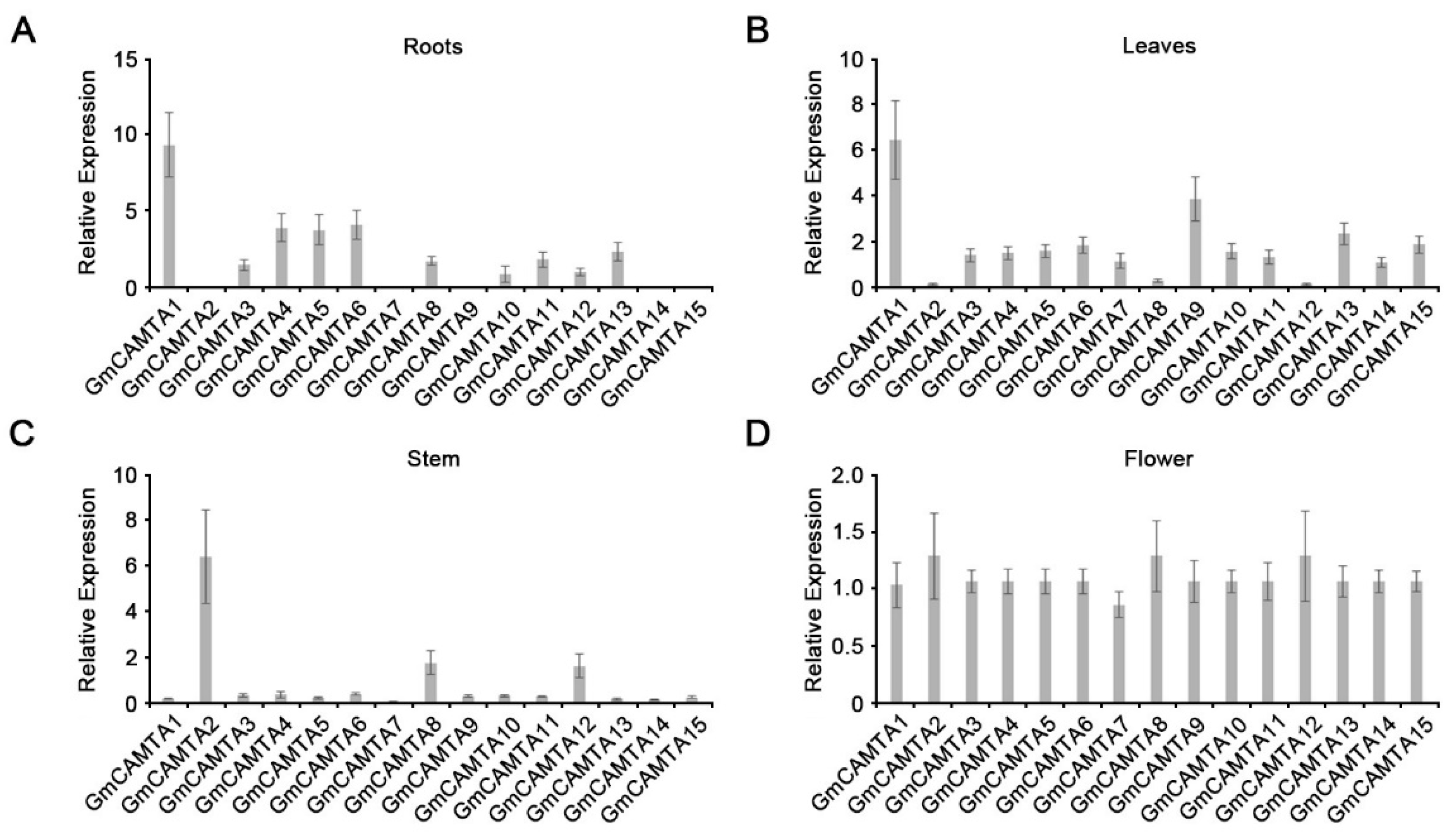
2.3. The transcripts of GmCAMTAs had substantial specificity in developmental processes
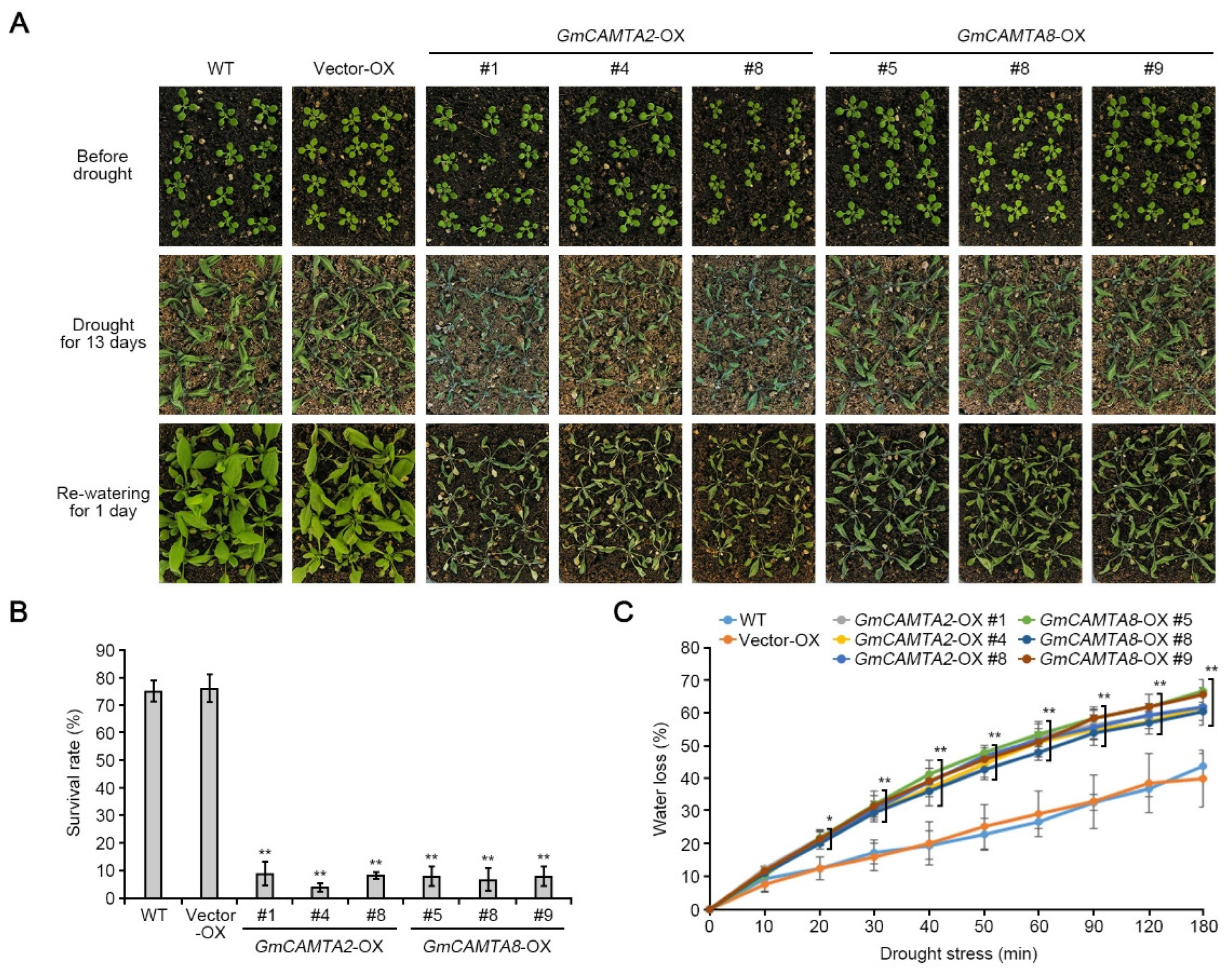
2.4. Overexpression of GmCAMTA2 and GmCAMTA8 reduced drought tolerance in Arabidopsis
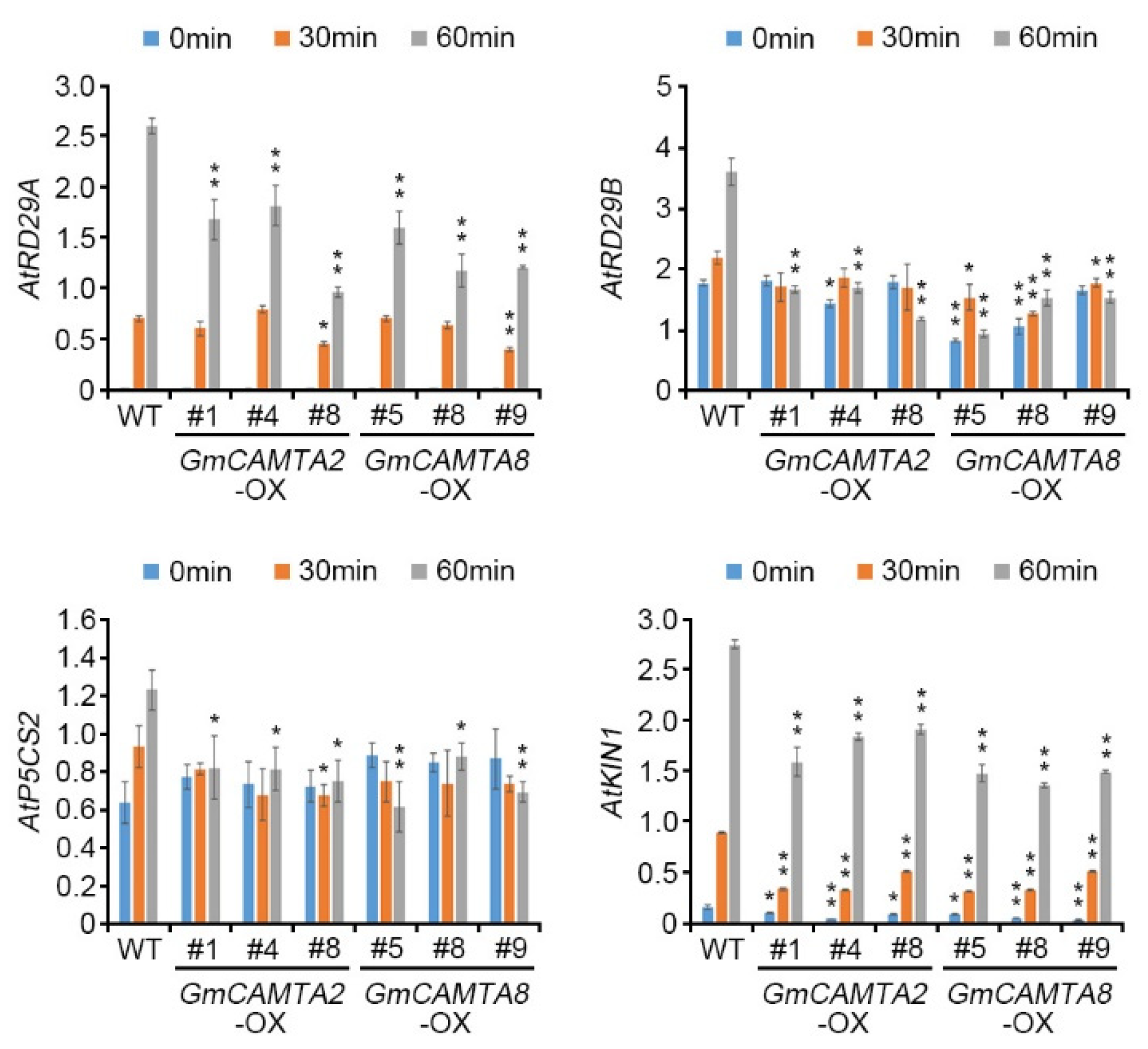
2.5. Overexpression of GmCAMTA2 and GmCAMTA8 regulated the transcripts of stress-responsive genes in drought stress responses
3. Discussion
4. Materials and Methods
4.1. Plant materials and growth conditions
4.2. Identification of GmCAMTA genes in soybean
4.3. Generation of GmCMATA-overexpressing Arabidopsis transgenic plants
4.4. Physiological assay of drought stress
4.5. Analysis of gene expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Demidchik, V.; Maathuis, F.J.M. Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 2007, 175, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Shabala, S.; Isayenkov, S.; Cuin, T.A.; Pottosin, I. Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 2018, 220, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium Signalling in Plant Biotic Interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Jin, L.; Peng, R. Crosstalk between Ca2+ and Other Regulators Assists Plants in Responding to Abiotic Stress. Plants 2022, 11, 1351. [Google Scholar] [CrossRef]
- Iqbal, Z.; Shariq Iqbal, M.; Singh, S.P.; Buaboocha, T. Ca2+/Calmodulin Complex Triggers CAMTA Transcriptional Machinery Under Stress in Plants: Signaling Cascade and Molecular Regulation. Front. Plant Sci. 2020, 11, 598327. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Day, I.S.; Reddy, V.S.; Shad Ali, G.; Reddy, A.S. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 2002, 3, RESEARCH0056. [Google Scholar] [CrossRef]
- Sun, Q.P.; Guo, Y.; Sun, Y.; Sun, D.Y.; Wang, X.J. Influx of extracellular Ca2+ involved in jasmonic-acid-induced elevation of [Ca2+]cyt and JR1 expression in Arabidopsis thaliana. J Plant Res. 2006, 119, 343–350. [Google Scholar] [CrossRef]
- Poovaiah, B.W.; Du, L. Calcium signaling: Decoding mechanism of calcium signatures. New Phytol. 2018, 217, 1394–1396. [Google Scholar] [CrossRef]
- Xiao, P.; Feng, J.W.; Zhu, X.T.; Gao, J. Evolution Analyses of CAMTA Transcription Factor in Plants and Its Enhancing Effect on Cold-tolerance. Front Plant Sci. 2021, 12, 758187. [Google Scholar] [CrossRef]
- Wang, G.; Zeng, H.; Hu, X.; Zhu, Y.; Chen, Y.; Shen, C.; Wang, H.; Poovaiah, B.W.; Du, L. Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil. 2015, 386, 205–221. [Google Scholar] [CrossRef]
- Bouché, N.; Scharlat, A.; Snedden, W.; Bouchez, D.; Fromm, H. A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem. 2002, 277, 21851–21861. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Han, J.; Reddig, K.; Li, H.S. A potential dimerization region of dCAMTA is critical for termination of fly visual response. J. Biol. Chem. 2007, 282, 21253–21258. [Google Scholar] [CrossRef] [PubMed]
- Finkler, A.; Ashery-Padan, R.; Fromm, H. CAMTAs: Calmodulin-binding transcription activators from plants to human. FEBS Lett. 2007, 581, 3893–3898. [Google Scholar] [CrossRef]
- Rhoads, A.R.; Friedberg, F. Sequence motifs for calmodulin recognition. FASEB J. 1997, 11, 331–340. [Google Scholar] [CrossRef]
- Yang, T.; Peng, H.; Whitaker, B.D.; Conway, W.S. Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 2012, 12, 19. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhao, M.; Xing, F.; Mao, G.; Wang, Y.; Dai, Y.; Niu, M.; Yuan, H. Identification and Expression Analysis of CAMTA Genes in Tea Plant Reveal Their Complex Regulatory Role in Stress Responses. Front Plant Sci. 2022, 13, 910768. [Google Scholar] [CrossRef]
- Wei, M.; Xu, X.; Li, C. Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress. Sci. Rep. 2017, 7, 17910. [Google Scholar] [CrossRef]
- Meer, L.; Mumtaz, S.; Labbo, A.M.; Khan, M.J.; Sadiq, I. Genome-wide identification and expression analysis of calmodulin-binding transcription activator genes in banana under drought stress. Sci. Hortic. 2019, 244, 10–14. [Google Scholar] [CrossRef]
- Saeidi, K.; Zare, N.; Baghizadeh, A.; Asghari-Zakaria, R. Phaseolus vulgaris genome possesses CAMTA genes, and phavuCAMTA1 contributes to the drought tolerance. J Genet. 2019, 98, 31. [Google Scholar] [CrossRef]
- Yue, R.; Lu, C.; Sun, T.; Peng, T.; Han, X.; Qi, J.; Yan, S.; Tie, S. Identification and expression profiling analysis of calmodulin-binding transcription activator genes in maize (Zea mays L.) under abiotic and biotic stresses. Front. Plant Sci. 2015, 6, 576. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, T.; Xu, L.; Pi, E.; Wang, S.; Wang, H.; Shen, C. Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments. Front. Plant Sci. 2015, 6, 459. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef] [PubMed]
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009, 21, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.; Aysha, J.; Ketehouli, T.; Yang, J.; Du, L.; Wang, F.; Li, H. Calmodulin binding transcription activators: An interplay between calcium signalling and plant stress tolerance. J Plant Physiol. 2021, 256, 153327. [Google Scholar] [CrossRef]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef]
- Mitsuda, N.; Isono, T.; Sato, M.H. Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell Physiol. 2003, 44, 975–981. [Google Scholar] [CrossRef]
- Shkolnik, D.; Finkler, A.; Pasmanik-Chor, M.; Fromm, H. CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: A Key Regulator of Na+ Homeostasis during Germination. Plant Physiol. 2019, 180, 1101–1118. [Google Scholar] [CrossRef]
- Koo, S.C.; Choi, M.S.; Chun, H.J.; Shin, D.B.; Park, B.S.; Kim, Y.H.; Park, H.M.; Seo, H.S.; Song, J.T.; Kang, K.Y.; Yun, D.J.; Chung, W.S.; Cho, M.J.; Kim, M.C. The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Mol. Cells. 2009, 27, 563–570. [Google Scholar] [CrossRef]
- Noman, M.; Jameel, A.; Qiang, W.D.; Ahmad, N.; Liu, W.C.; Wang, F.W.; Li, H.Y. Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean. Int. J. Mol. Sci. 2019, 20, 4849. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Z.; Cao, X.; Duan, W.; Wei, C.; Zhang, C.; Jiang, D.; Li, M.; Chen, K.; Qiao, Y.; Liu, H.; Zhang, B. Genome-Wide Analysis of Calmodulin Binding Transcription Activator (CAMTA) Gene Family in Peach (Prunus persica L. Batsch) and Ectopic Expression of PpCAMTA1 in Arabidopsis camta2,3 Mutant Restore Plant Development. Int. J. Mol. Sci. 2022, 23, 10500. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.; Khare, D. Molecular approaches for genetic improvement of seed quality and characterization of genetic diversity in soybean: A critical review. Biotechnol. Lett. 2016, 38, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Haun, W.J.; Hyten, D.L.; Xu, W.W.; Gerhardt, D.J.; Albert, T.J.; Richmond, T.; Jeddeloh, J.A.; Jia, G.; Springer, N.M.; Vance, C.P.; Stupar, R.M. The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol. 2011, 155, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yuan, L.; Xie, Q. The circadian clock ticks in plant stress responses. Stress Biol. 2022, 2, 15. [Google Scholar] [CrossRef]
- Ma, X.; Su, Z.; Ma, H. Molecular genetic analyses of abiotic stress responses during plant reproductive development. J. Exp. Bot. 2020, 71, 2870–2885. [Google Scholar] [CrossRef]
- Sabbioni, G.; Funck, D.; Forlani, G. Enzymology and Regulation of δ1-Pyrroline-5-Carboxylate Synthetase 2 From Rice. Front. Plant Sci. 2021, 12, 672702. [Google Scholar] [CrossRef]
- Zhou, L.; Zhou, J.; Xiong, Y.; Liu, C.; Wang, J.; Wang, G.; Cai, Y. Overexpression of a maize plasma membrane intrinsic protein ZmPIP1;1 confers drought and salt tolerance in Arabidopsis. PLoS ONE 2018, 13, e0198639. [Google Scholar] [CrossRef]
- Más, P. Circadian clock function in Arabidopsis thaliana: Time beyond transcription. Trends Cell Biol. 2008, 18, 273–281. [Google Scholar] [CrossRef]
- Martí Ruiz, M.C.; Hubbard, K.E.; Gardner, M.J.; Jung, H.J.; Aubry, S.; Hotta, C.T.; Mohd-Noh, N.I.; Robertson, F.C.; Hearn, T.J.; Tsai, Y.C.; Dodd, A.N.; Hannah, M.; Carré, I.A.; Davies, J.M.; Braam, J.; Webb, A.A.R. Circadian oscillations of cytosolic free calcium regulate the Arabidopsis circadian clock. Nat. Plants 2018, 4, 690–698. [Google Scholar] [CrossRef]
- Love, J.; Dodd, A.N.; Webb, A.A. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell 2004, 16, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Tataroglu, O.; Zhao, X.; Busza, A.; Ling, J.; O'Neill, J.S.; Emery, P. Calcium and SOL Protease Mediate Temperature Resetting of Circadian Clocks. Cell 2015, 163, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, Y.; Xie, Q.; Wu, Y. Loss of CDKC;2 increases both cell division and drought tolerance in Arabidopsis thaliana. Plant J. 2017, 91, 816–828. [Google Scholar] [CrossRef]
- Shangguan, L.; Wang, X.; Leng, X.; Liu, D.; Ren, G.; Tao, R.; Zhang, C.; Fang, J. Identification and bioinformatic analysis of signal responsive/calmodulin-binding transcription activators gene models in Vitis vinifera. Mol. Biol. Rep. 2014, 41, 2937–2949. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Jaleel, C.A.; Manivannan, P.; Wahid, A.; Farooq, M.; Al-Juburi, H.J.; Somasundararam, R.; Panneerselvam, R. Drought stress in plants: A review on morphological characteristics and pigments composition. Int. J. Agric. Biol. 2009, 11, 100–105. [Google Scholar]
- Mangena, P. Water stress: Morphological and anatomic changes in soybean (Glycine max L.) plants. In Plant, Abiotic Stress and Responses to Climate Change, ed V. Andjelkovic; Intech Open: London, UK, 2018; pp. 9–31. [Google Scholar] [CrossRef]
- Lee, H. Stem Cell Maintenance and Abiotic Stress Response in Shoot Apical Meristem for Developmental Plasticity. J. Plant Biol. 2018, 61, 358–365. [Google Scholar] [CrossRef]
- Dolzblasz, A.; Smakowska, E.; Gola, E.M.; Sokołowska, K.; Kicia, M.; Janska, H. The mitochondrial protease AtFTSH4 safeguards Arabidopsis shoot apical meristem function. Sci. Rep. 2016, 6, 28315. [Google Scholar] [CrossRef]
- Wilson, M.E.; Mixdorf, M.; Berg, R.H.; Haswell, E.S. Plastid osmotic stress influences cell differentiation at the plant shoot apex. Development 2016, 143, 3382–3393. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




