Submitted:
19 May 2023
Posted:
23 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
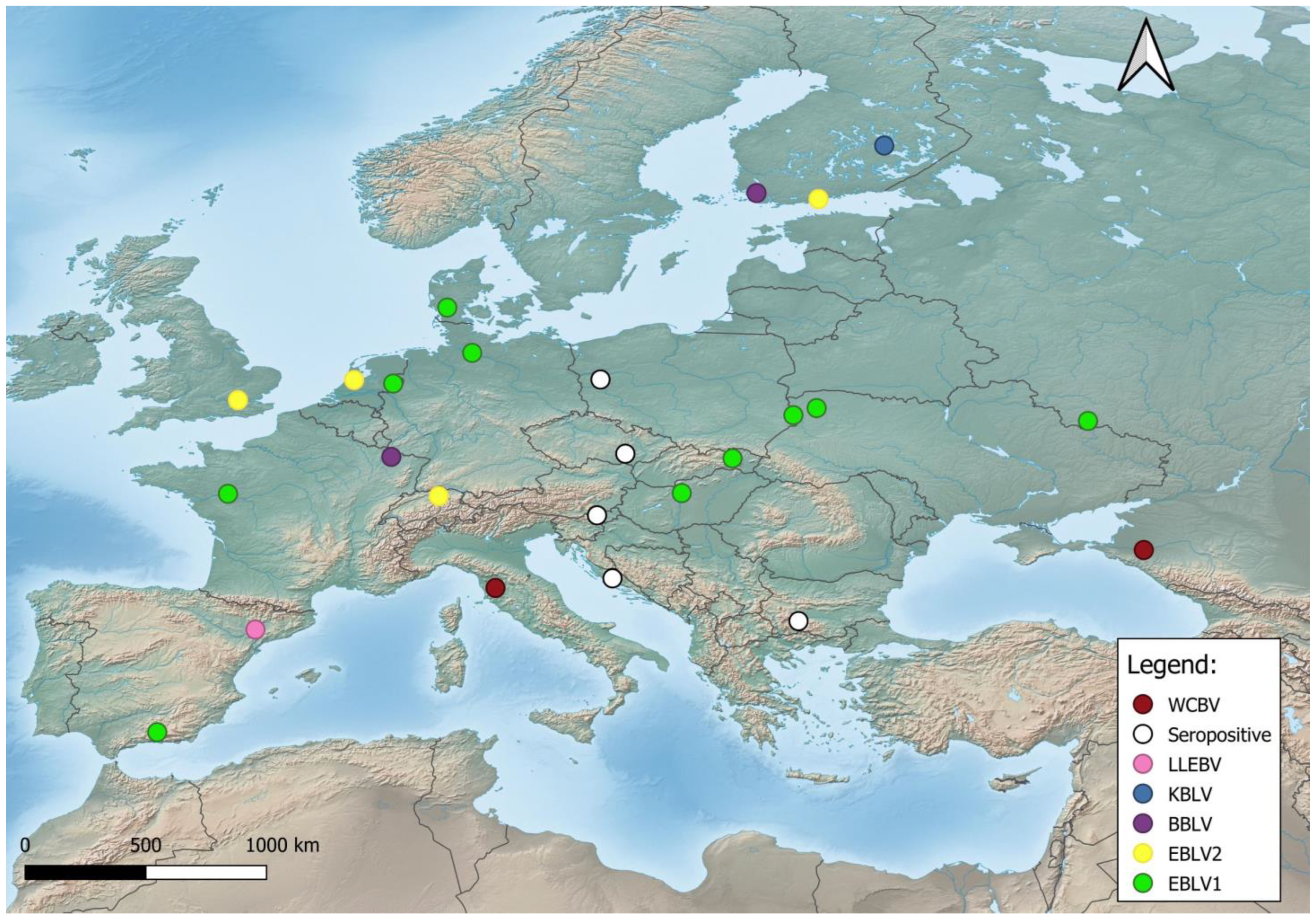
2. Origin, Evolution and Geographic Distribution of Bat Lyssaviruses
3. Virion structure and genome
4. Phylogeny of bat lyssaviruses
5. Transmission routes of bat lyssaviruses
5.1. Bat intra- and cross-species transmission
5.2. Other vertebrates cross-species transmission
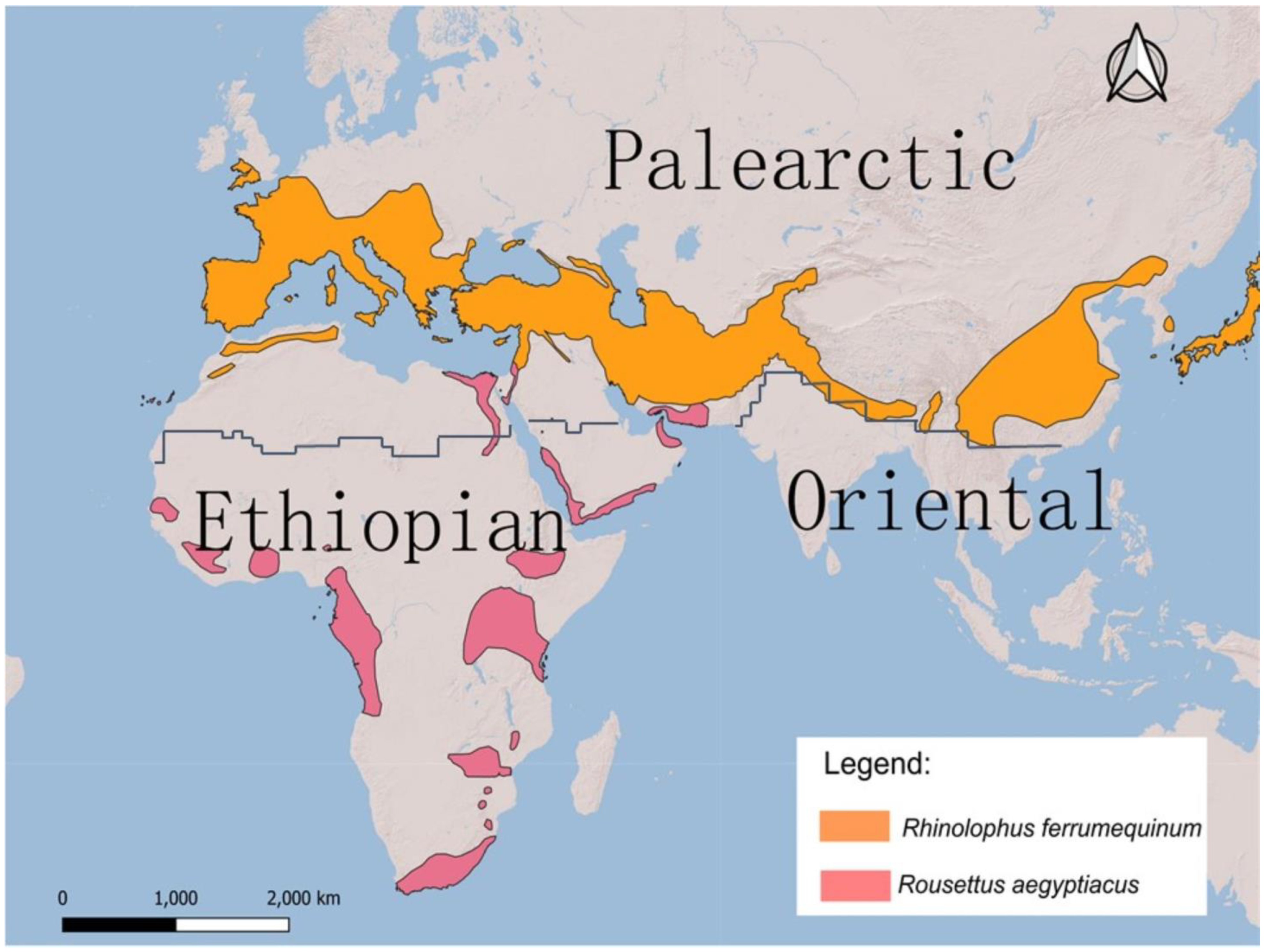
5.3. Within the contact zone of the Palearctic and Ethiopian realms—potential scenarios
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruedi, M.; Stadelmann, B.; Gager, Y.; Douzery, E.J.P.; Francis, C.M.; Lin, L.-K.; Guillén-Servent, A.; Cibois, A. Molecular phylogenetic reconstructions identify East Asia as the cradle for the evolution of the cosmopolitan genus Myotis (Mammalia, Chiroptera). Mol. Phylogenetics Evol. 2013, 69, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Vernes, S.C.; Davalos, L.M.; Ray, D.A.; Gilbert, M.T.P.; Myers, E. ; K Consortium Bat Biology, Genomes, and the Bat1K Project: To Generate Chromosome-Level Genomes for All Living Bat Species. Annu. Rev. Anim. Biosci. 2018, 6, 23–46. [Google Scholar] [CrossRef]
- Teeling, E. C.; Jones, G.; Rossiter, S. J. Phylogeny, genes, and hearing: implications for the evolution of echolocation in bats. In Bat bioacoustics Springer, New York, 2016; pp. 25-54.
- Beltz, L. A. Bats and human health: Ebola, SARS, rabies and beyond. Jonh Wiley & Sons, Ltd. 2017; p. 371.
- Wackermannová, M.; Pinc, L.; Jebavý, L. Olfactory Sensitivity in Mammalian Species. Physiol. Res. 2016, 65, 369–390. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.S.; Adams, D.M.; Haghani, A.; Lu, A.T.; Zoller, J.; Breeze, C.E.; Arnold, B.D.; Ball, H.C.; Carter, G.G.; Cooper, L.N.; et al. DNA methylation predicts age and provides insight into exceptional longevity of bats. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-Y.; Liang, L.; Zhu, Z.-H.; Zhou, W.-P.; Irwin, D.M.; Zhang, Y.-P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. 2010, 107, 8666–8671. [Google Scholar] [CrossRef] [PubMed]
- Subudhi, S.; Rapin, N.; Misra, V. Immune System Modulation and Viral Persistence in Bats: Understanding Viral Spillover. Viruses 2019, 11, 192. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Baker, M.L.; Kulcsar, K.; Misra, V.; Plowright, R.; Mossman, K. Novel Insights Into Immune Systems of Bats. Front. Immunol. 2020, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Begeman, L.; Suu-Ire, R.; Banyard, A.C.; Drosten, C.; Eggerbauer, E.; Freuling, C.M.; Gibson, L.; Goharriz, H.; Horton, D.L.; Jennings, D.; et al. Experimental Lagos bat virus infection in straw-colored fruit bats: A suitable model for bat rabies in a natural reservoir species. PLOS Neglected Trop. Dis. 2020, 14, e0008898. [Google Scholar] [CrossRef]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.-F. Lessons from the host defences of bats, a unique viral reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Brook, C.E.; Dobson, A.P. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef]
- Hayman, D.T. Bats as Viral Reservoirs. Annu. Rev. Virol. 2016, 3, 77–99. [Google Scholar] [CrossRef] [PubMed]
- Latinne, A.; Hu, B.; Olival, K.J.; Zhu, G.; Zhang, L.; Li, H.; Chmura, A.A.; Field, H.E.; Zambrana-Torrelio, C.; Epstein, J.H.; et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- EUROBATS. Action Plan for the Conservation of Bat Species in the European Union 2016–2021; Inf.EUROBATS.AC21. B: EUROBATS, 2006.
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’Brien, S.J.; Murphy, W.J. A Molecular Phylogeny for Bats Illuminates Biogeography and the Fossil Record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef]
- Kohl, C.; Kurth, A. European Bats as Carriers of Viruses with Zoonotic Potential. Viruses 2014, 6, 3110–3128. [Google Scholar] [CrossRef]
- Smreczak, M.; Orłowska, A.; Marzec, A.; Trębas, P.; Müller, T.; Freuling, C. M.; Żmudziński, J. F. Bokeloh bat lyssavirus isolation in a Natterer’s bat, Poland. Zoonoses Public Health. 2018, 65(8), 1015–1019. [Google Scholar] [CrossRef]
- Vos, A.; Kaipf, I.; Denzinger, A.; Fooks, A.R.; Johnson, N.; Müller, T. European bat lyssaviruses — an ecological enigma. Acta Chiropterologica 2007, 9, 283–296. [Google Scholar] [CrossRef]
- Banyard, A. C.; Hayman, D. ; Johnson, McElhinney, N. L; Fooks, A. R. Bats and lyssaviruses. Adv. Virus Res. 2011, 79, 239–89. [Google Scholar] [CrossRef]
- Rupprecht, C.E.; Freuling, C.M.; Mani, R.S.; Palacios, C.; Sabeta, C.T.; Ward, M. A history of rabies—The foundation for global canine rabies elimination. In Rabies, Scientific Basis of the Disease and Its Management, 4th ed.; Fooks, A.R., Jackson, A.C., Eds.; Academic Press: Cambridge, MA, USA, 2020; ISBN 978-0-12-818705-0. [Google Scholar] [CrossRef]
- Afonso, C.L.; Amarasinghe, G.K.; Bányai, K.; Bào, Y.; Basler, C.F.; Bavari, S.; Bejerman, N.; Blasdell, K.R.; Briand, F.-X.; Briese, T.; et al. Taxonomy of the order Mononegavirales: update 2016. Arch. Virol. 2016, 161, 2351–2360. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Hendrickson, R.C.; et al. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch. Virol. 2022, 167, 2429–2440. [Google Scholar] [CrossRef]
- Nokireki, T.; Tammiranta, N.; Kokkonen, U.-M.; Kantala, T.; Gadd, T. Tentative novel lyssavirus in a bat in Finland. Transbound. Emerg. Dis. 2018, 65, 593–596. [Google Scholar] [CrossRef]
- Coertse, J.; Markotter, W.; le Roux, K.; Stewart, D.; Sabeta, C.T.; Nel, L.H. New isolations of the rabies-related Mokola virus from South Africa. BMC Veter- Res. 2016, 13, 1–8. [Google Scholar] [CrossRef]
- Biswas, M.R.; Alzubaidi, M.S.; Shah, U.; Abd-Alrazaq, A.A.; Shah, Z. A Scoping Review to Find out Worldwide COVID-19 Vaccine Hesitancy and Its Underlying Determinants. Vaccines 2021, 9, 1243. [Google Scholar] [CrossRef]
- Marston, D.A.; Horton, D.L.; Ngeleja, C.; Hampson, K.; McElhinney, L.M.; Banyard, A.C.; Haydon, D.; Cleaveland, S.; Rupprecht, C.E.; Bigambo, M.; et al. Ikoma lyssavirus, highly divergent novel lyssavirus in an African civet. Emerg. Infect. Dis. 2012, 18, 664–667. [Google Scholar] [CrossRef] [PubMed]
- Markotter, W.; Kgaladi, J.; Nel, L.H.; Marston, D.; Wright, N.; Coertse, J.; Müller, T.F.; Sabeta, C.T.; Fooks, A.R.; Freuling, C.M. Diversity and Epidemiology of Mokola Virus. PLoS Negl. Trop. Dis. 2013, 7, e2511. [Google Scholar]
- Sabeta, C.T.; Markotter, W.; Mohale, D.K.; Shumba, W.; Wandeler, A.I.; Nel, L.H. Mokola Virus in Domestic Mammals, South Africa. Emerg. Infect. Dis. 2007, 13, 1371–1373. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, C.; Kuzmin, I.; Meslin, F. Lyssaviruses and rabies: current conundrums, concerns, contradictions and controversies. F1000Research 2017, 6, 184. [Google Scholar] [CrossRef]
- Hu, S.-C.; Hsu, C.-L.; Lee, M.-S.; Tu, Y.-C.; Chang, J.-C.; Wu, C.-H.; Lee, S.-H.; Ting, L.-J.; Tsai, K.-R.; Cheng, M.-C.; et al. Lyssavirus in Japanese Pipistrelle, Taiwan. Emerg. Infect. Dis. 2018, 24, 782–785. [Google Scholar] [CrossRef] [PubMed]
- Calvelage, S.; Tammiranta, N.; Nokireki, T.; Gadd, T.; Eggerbauer, E.; Zaeck, L.M.; Potratz, M.; Wylezich, C.; Höper, D.; Müller, T.; et al. Genetic and Antigenetic Characterization of the Novel Kotalahti Bat Lyssavirus (KBLV). Viruses 2021, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-C.; Hsu, C.-L.; Lee, F.; Tu, Y.-C.; Chen, Y.-W.; Chang, J.-C.; Hsu, W.-C. Novel Bat Lyssaviruses Identified by Nationwide Passive Surveillance in Taiwan, 2018–2021. Viruses 2022, 14, 1562. [Google Scholar] [CrossRef]
- Nel, L. H.; Rupprecht, C. E. Emergence of lyssaviruses in the Old World: the case of Africa. Curr Top Microbiol Immunol. 2007, 315, 161–93. [Google Scholar] [CrossRef]
- Hayman, D.T.S.; Fooks, A.R.; Marston, D.A.; Garcia-R, J.C. The Global Phylogeography of Lyssaviruses—Challenging the ‘Out of Africa’ Hypothesis. PLOS Neglected Trop. Dis. 2016, 10, e0005266. [Google Scholar] [CrossRef] [PubMed]
- Longdon, B.; Murray, G.G.R.; Palmer, W.J.; Day, J.P.; Parker, D.J.; Welch, J.J.; Obbard, D.J.; Jiggins, F.M. The evolution, diversity, and host associations of rhabdoviruses. Virus Evol. 2015, 1, vev014. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Firth, C.; Widen, S.G.; Blasdell, K.R.; Guzman, H.; Wood, T.G.; Paradkar, P.N.; Holmes, E.C.; Tesh, R.B.; Vasilakis, N. Evolution of Genome Size and Complexity in the Rhabdoviridae. PLOS Pathog. 2015, 11, e1004664–e1004664. [Google Scholar] [CrossRef]
- Caraballo, D.A.; Lema, C.; Novaro, L.; Gury-Dohmen, F.; Russo, S.; Beltrán, F.J.; Palacios, G.; Cisterna, D.M. A Novel Terrestrial Rabies Virus Lineage Occurring in South America: Origin, Diversification, and Evidence of Contact between Wild and Domestic Cycles. Viruses 2021, 13, 2484. [Google Scholar] [CrossRef]
- Singh, R.; Singh, K.P.; Cherian, S.; Saminathan, M.; Kapoor, S.; Reddy, G.M.; Panda, S.; Dhama, K. Rabies—epidemiology, pathogenesis, public health concerns and advances in diagnosis and control: a comprehensive review. Veter- Q. 2017, 37, 212–251. [Google Scholar] [CrossRef]
- Potratz, M.; Zaeck, L.M.; Weigel, C.; Klein, A.; Freuling, C.M.; Müller, T.; Finke, S. Neuroglia infection by rabies virus after anterograde virus spread in peripheral neurons. Acta Neuropathol. Commun. 2020, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Baghi, H.B.; Rupprecht, C.E. Notes on three periods of rabies focus in the Middle East: From progress during the cradle of civilization to neglected current history. Zoonoses Public Heal. 2020, 68, 697–703. [Google Scholar] [CrossRef]
- E Rupprecht, C.; Turmelle, A.; Kuzmin, I.V. A perspective on lyssavirus emergence and perpetuation. Curr. Opin. Virol. 2011, 1, 662–670. [Google Scholar] [CrossRef]
- Fooks, A.R.; Brookes, S.M.; Johnson, N.; McELHINNEY, L.M.; Hutson, A.M. European bat lyssaviruses: an emerging zoonosis. Epidemiology Infect. 2003, 131, 1029–1039. [Google Scholar] [CrossRef]
- Schatz, J.; Fooks, A.R.; McElhinney, L.; Horton, D.; Echevarria, J.; Vázquez-Moron, S.; Kooi, E.A.; Rasmussen, T.B.; Müller, T.; Freuling, C.M. Bat Rabies Surveillance in Europe. Zoonoses Public Heal. 2012, 60, 22–34. [Google Scholar] [CrossRef]
- McElhinney, L.M.; Marston, D.A.; Wise, E.L.; Freuling, C.M.; Bourhy, H.; Zanoni, R.; Moldal, T.; Kooi, E.A.; Neubauer-Juric, A.; Nokireki, T.; et al. Molecular Epidemiology and Evolution of European Bat Lyssavirus 2. Int. J. Mol. Sci. 2018, 19, 156. [Google Scholar] [CrossRef] [PubMed]
- Šimić, I.; Lojkić, I.; Krešić, N.; Cliquet, F.; Picard-Meyer, E.; Wasniewski, M.; Ćukušić, A.; Zrnčić, V.; Bedeković, T. Molecular and serological survey of lyssaviruses in Croatian bat populations. BMC Veter- Res. 2018, 14, 274. [Google Scholar] [CrossRef]
- Seidlova, V.; Zukal, J.; Brichta, J.; Anisimov, N.; Apoznański, G.; Bandouchova, H.; Bartonička, T.; Berková, H.; Botvinkin, A.D.; Heger, T.; et al. Active surveillance for antibodies confirms circulation of lyssaviruses in Palearctic bats. BMC Veter- Res. 2020, 16, 1–9. [Google Scholar] [CrossRef]
- Coxon, C.; McElhinney, L.; Pacey, A.; Gauntlett, F.; Holland, S. Preliminary Outbreak Assessment: Rabies in a Cat in Italy. Department for Environment, Food and Rural Affairs, Animal and Plant Health Agency, Advice Services—International Disease Monitoring. 2020 Available online at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/897070/rabies-cat-italy-poa.
- Kuzmin, I.V.; Hughes, G.J.; Botvinkin, A.D.; Orciari, L.A.; Rupprecht, C.E. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Res. 2005, 111, 28–43. [Google Scholar] [CrossRef]
- Freuling, C. Novel Lyssavirus in Natterer’s Bat, Germany. Emerg. Infect. Dis. 2011, 17, 1519–22. [Google Scholar] [CrossRef] [PubMed]
- Arechiga Ceballos, N.; Vazquez Moron, S.; Berciano, J.M.; Nicolas, O.; Aznar Lopez, C.; Juste, J.; Rodriguez Nevado, C.; Aguilar Setien, A.; Echevarria, J.E. Novel lyssavirus in bat, Spain. Emerg. Infect. Dis. 2013, 19, 793–795. [Google Scholar] [CrossRef]
- Davis, P.L.; Holmes, E.C.; Larrous, F.; Van der Poel, W.H.M.; Tjørnehøj, K.; Alonso, W.J.; Bourhy, H. Phylogeography, Population Dynamics, and Molecular Evolution of European Bat Lyssaviruses. J. Virol. 2005, 79, 10487–10497. [Google Scholar] [CrossRef]
- Mingo-Casas, P.; Sandonís, V.; Obón, E.; Berciano, J.M.; Vázquez-Morón, S.; Juste, J.; Echevarría, J.E. First cases of European bat lyssavirus type 1 in Iberian serotine bats: Implications for the molecular epidemiology of bat rabies in Europe. PLOS Neglected Trop. Dis. 2018, 12, e0006290. [Google Scholar] [CrossRef]
- Jakava-Viljanen, M.; Nokireki, T.; Sironen, T.; Vapalahti, O.; Sihvonen, L.; Huovilainen, A. Erratum to: Evolutionary trends of European bat lyssavirus type 2 including genetic characterization of Finnish strains of human and bat origin 24 years apart. Arch. Virol. 2015, 160, 1875–1875. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.L.; Aegerter, J.N.; Brookes, S.M.; McElhinney, L.M.; Jones, G.; Smith, G.C.; Fooks, A.R. TARGETED SURVEILLANCE FOR EUROPEAN BAT LYSSAVIRUSES IN ENGLISH BATS (2003–06). J. Wildl. Dis. 2009, 45, 1030–1041. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Kondo, H.; Goodin, M.M.; Kurath, G.; Vasilakis, N. The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 2017, 227, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Lelli, D.; Prosperi, A.; Moreno, A.; Chiapponi, C.; Gibellini, A.M.; De Benedictis, P.; Leopardi, S.; Sozzi, E.; Lavazza, A. Isolation of a novel Rhabdovirus from an insectivorous bat (Pipistrellus kuhlii) in Italy. Virol. J. 2018, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Dietzgen, R.G.; Joubert, D.A.; Blasdell, K.R. Rhabdovirus accessory genes. Virus Res. 2011, 162, 110–125. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Wu, X.; Tordo, N.; Rupprecht, C.E. Complete genomes of Aravan, Khujand, Irkut and West Caucasian bat viruses, with special attention to the polymerase gene and non-coding regions. Virus Res. 2008, 136, 81–90. [Google Scholar] [CrossRef]
- Badrane, H.; Bahloul, C.; Perrin, P.; Tordo, N. Evidence of TwoLyssavirusPhylogroups with Distinct Pathogenicity and Immunogenicity. J. Virol. 2001, 75, 3268–3276. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Orciari, L.A.; Arai, Y.T.; Smith, J.S.; Hanlon, C.A.; Kameoka, Y.; Rupprecht, C.E. Bat lyssaviruses (Aravan and Khujand) from Central Asia: phylogenetic relationships according to N, P and G gene sequences. Virus Res. 2003, 97, 65–79. [Google Scholar] [CrossRef]
- Kuzmin, I.; Novella, I.; Dietzgen, R.; Padhi, A.; Rupprecht, C. The rhabdoviruses: Biodiversity, phylogenetics, and evolution. Infect. Genet. Evol. 2009, 9, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press (OUP): Oxford, Oxfordshire, United Kingdom, 2000. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gunawardena, P.S.; Marston, D.A.; Ellis, R.J.; Wise, E.L.; Karawita, A.C.; Breed, A.C.; McElhinney, L.M.; Johnson, N.; Banyard, A.C.; Fooks, A.R. Lyssavirus in Indian Flying Foxes, Sri Lanka. Emerg. Infect. Dis. 2016, 22, 1456–1459. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I.V.; Mayer, A.E.; Niezgoda, M.; Markotter, W.; Agwanda, B.; Breiman, R.F.; Rupprecht, C.E. Shimoni bat virus, a new representative of the Lyssavirus genus. Virus Res. 2010, 149, 197–210. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Bozick, B.; Guagliardo, S.A.; Kunkel, R.; Shak, J.R.; Tong, S.; Rupprecht, C.E. Bats, emerging infectious diseases, and the rabies paradigm revisited. Emerg. Heal. Threat. J. 2011, 4, 7159. [Google Scholar] [CrossRef]
- Aréchiga -Ceballos, N. A.; Morón, S. V.; Berciano, J. M.; Nicolás, O.; López, C. A.; Juste, J.; Nevado, C. R.; Setién, Á. A.; Echevarría, J. E. Novel lyssavirus in bat Spain. Emerg. Infect. Dis. 2013, 19(5), 793–795. [Google Scholar] [CrossRef]
- Amengual, B.; E Whitby, J.; Cobo, J.S.; King, A.; Bourhy, H. Evolution of European bat lyssaviruses. J. Gen. Virol. 1997, 78, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I. V.; Rupprecht, C.E. Bats and Viruses: A New Frontier of Emerging Infectious Diseases. Bat Lyssaviruses. Published by John Wiley and Sons, Inc., Hoboken, New Jersey Published simultaneously in Canada, 2015 pp. -97. [CrossRef]
- Echevarrı́a, J.E.; Avellón, A.; Juste, J.; Vera, M.; Ibáñez, C. Screening of Active Lyssavirus Infection in Wild Bat Populations by Viral RNA Detection on Oropharyngeal Swabs. J. Clin. Microbiol. 2001, 39, 3678–3683. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cobo, J.; Amengual, B.; Abellan, C.; Bourhy, H. European bat Lyssavirus infection in Spanish bat populations. Emerg. Infect. Dis. 2002, 8, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Picard-Meyer, E.; Servat, A.; Robardet, E.; Moinet, M.; Borel, C.; Cliquet, F. Isolation of Bokeloh bat lyssavirus in Myotis nattereri in France. Arch. Virol. 2013, 158, 2333–2340. [Google Scholar] [CrossRef]
- Parize, P.; Robledo, I.C.T.; Cervantes-Gonzalez, M.; Kergoat, L.; Larrous, F.; Serra-Cobo, J.; Dacheux, L.; Bourhy, H. Circumstances of Human–Bat interactions and risk of lyssavirus transmission in metropolitan France. Zoonoses Public Heal. 2020, 67, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Salicini, I.; Ibáñez, C.; Juste, J. Multilocus phylogeny and species delimitation within the Natterer’s bat species complex in the Western Palearctic. Mol. Phylogenetics Evol. 2011, 61, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Salicini, I.; Ibáñez, C.; Juste, J. Deep differentiation between and within Mediterranean glacial refugia in a flying mammal, the Myotis nattereri bat complex. J. Biogeogr. 2012, 40, 1182–1193. [Google Scholar] [CrossRef]
- Puechmaille, S.J.; Allegrini, B.; Boston, E.S.; Dubourg-Savage, M.-J.; Evin, A.; Knochel, A.; Le Bris, Y.; Lecoq, V.; Lemaire, M.; Rist, D.; et al. Genetic analyses reveal further cryptic lineages within the Myotis nattereri species complex. Mamm. Biol. 2012, 77, 224–228. [Google Scholar] [CrossRef]
- Eggerbauer, E.; Pfaff, F.; Finke, S.; Höper, D.; Beer, M.; Mettenleiter, T.C.; Nolden, T.; Teifke, J.-P.; Müller, T.; Freuling, C.M. Comparative analysis of European bat lyssavirus 1 pathogenicity in the mouse model. PLOS Neglected Trop. Dis. 2017, 11, e0005668. [Google Scholar] [CrossRef]
- Çoraman, E.; Dietz, C.; Hempel, E.; Ghazaryan, A.; Levin, E.; Presetnik, P.; Zagmajster, M.; Mayer, F. Reticulate evolutionary history of a Western Palaearctic Bat Complex explained by multiple mtDNA introgressions in secondary contacts. J. Biogeogr. 2019, 46, 343–354. [Google Scholar] [CrossRef]
- Biswas, M.R.; Alzubaidi, M.S.; Shah, U.; Abd-Alrazaq, A.A.; Shah, Z. A Scoping Review to Find out Worldwide COVID-19 Vaccine Hesitancy and Its Underlying Determinants. Vaccines 2021, 9, 1243. [Google Scholar] [CrossRef] [PubMed]
- Dietz, C.; von Helversen, O.; Nill, D. Bats of Britain. Europe and Northwest Africa. A and C Black, London. 2009. [Google Scholar]
- Wright, P.G.R.; Newton, J.; Agnelli, P.; Budinski, I.; Di Salvo, I.; Flaquer, C.; Fulco, A.; Georgiakakis, P.; Martinoli, A.; Mas, M.; et al. Hydrogen isotopes reveal evidence of migration of Miniopterus schreibersii in Europe. BMC Ecol. 2020, 20, 1–7. [Google Scholar] [CrossRef]
- Freuling, C.; Vos, A.; Johnson, N.; Kaipf, I.; Denzinger, A.; Neubert, L.; Mansfield, K.; Hicks, D.; Nuñez, A.; Tordo, N.; et al. Experimental infection of serotine bats (Eptesicus serotinus) with European bat lyssavirus type 1a. J. Gen. Virol. 2009, 90, 2493–2502. [Google Scholar] [CrossRef]
- de Thoisy, B.; Bourhy, H.; Delaval, M.; Pontier, D.; Dacheux, L.; Darcissac, E.; Donato, D.; Guidez, A.; Larrous, F.; Lavenir, R.; et al. Bioecological Drivers of Rabies Virus Circulation in a Neotropical Bat Community. PLOS Neglected Trop. Dis. 2016, 10, e0004378–e0004378. [Google Scholar] [CrossRef]
- Bonnaud, E.M.; Troupin, C.; Dacheux, L.; Holmes, E.C.; Monchatre-Leroy, E.; Tanguy, M.; Bouchier, C.; Cliquet, F.; Barrat, J.; Bourhy, H. Comparison of intra- and inter-host genetic diversity in rabies virus during experimental cross-species transmission. PLOS Pathog. 2019, 15, e1007799. [Google Scholar] [CrossRef]
- Colombi, D.; Serra-Cobo, J.; Métras, R.; Apolloni, A.; Poletto, C.; López-Roig, M.; Bourhy, H.; Colizza, V. Mechanisms for lyssavirus persistence in non-synanthropic bats in Europe: insights from a modeling study. Sci. Rep. 2019, 9, 537. [Google Scholar] [CrossRef]
- Horton, D.L.; Breed, A.C.; Arnold, M.E.; Smith, G.C.; Aegerter, J.N.; McElhinney, L.M.; Johnson, N.; Banyard, A.C.; Raynor, R.; Mackie, I.; et al. Between roost contact is essential for maintenance of European bat lyssavirus type-2 in Myotis daubentonii bat reservoir: ‘The Swarming Hypothesis’. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tájek, P.; Tájková, P. A long distance migration in Nyctalus leisleri from the Czech Republic to southern France (Chiroptera: Vespertilionidae). Lynx new Ser. 2021, 51, 223–226. [Google Scholar] [CrossRef]
- Dundarova, H.; Michev, B.; Pandourski, I. Bats over the Western Black Sea open water area. Acta Zool. Bulg. 2021, 73(4), 543–546. [Google Scholar]
- Vega, S.; Lorenzo-Rebenaque, L.; Marin, C.; Domingo, R.; Fariñas, F. Tackling the Threat of Rabies Reintroduction in Europe. Front. Veter- Sci. 2021, 7. [Google Scholar] [CrossRef]
- Bouma, H.R.; Carey, H.V.; Kroese, F.G.M. Hibernation: the immune system at rest? J. Leukoc. Biol. 2010, 88, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Field, K.A.; Sewall, B.J.; Prokkola, J.M.; Turner, G.G.; Gagnon, M.F.; Lilley, T.M.; White, J.P.; Johnson, J.S.; Hauer, C.L.; Reeder, D.M. Effect of torpor on host transcriptomic responses to a fungal pathogen in hibernating bats. Mol. Ecol. 2018, 27, 3727–3743. [Google Scholar] [CrossRef]
- Constantine, D.G. Bat rabies and other lyssavirus infections. Reston, Va., U.S. Geological Survey Circular 1329. 2009, 68 p.
- Harazim, M.; Perrot, J.; Varet, H.; Bourhy, H.; Lannoy, J.; Pikula, J.; Seidlová, V.; Dacheux, L.; Martínková, N. Transcriptomic responses of bat cells to European bat lyssavirus 1 infection under conditions simulating euthermia and hibernation. BMC Immunol. 2023, 24, 1–13. [Google Scholar] [CrossRef]
- Gilbert, A. T.; McCracken, G. F.; Sheeler, L. L.; Muller, L. I.; O’Rourke, D.; Kelch, W. J.; New Jr., J. C. Rabies surveillance among bats in Tennessee, USA, 1996–2010. J. Wildl. Dis. 2015, 51(4), 821-832.
- Jin J, Lu Z, Li Y, Cowart LA, Lopes-Virella MF, Huang Y. Docosahexaenoic acid antagonizes the boosting effect of palmitic acid on LPS inflammatory signaling by inhibiting gene transcription and ceramide synthesis. PLoS One. 2018, 13: e0193343. [CrossRef]
- Shipley, R.; Wright, E.; Selden, D.; Wu, G.; Aegerter, J.; Fooks, A.R.; Banyard, A.C. Bats and Viruses: Emergence of Novel Lyssaviruses and Association of Bats with Viral Zoonoses in the EU. Trop. Med. Infect. Dis. 2019, 4, 31. [Google Scholar] [CrossRef]
- Constantine, D.G.; Emmons, R.W.; Woodie, J.D. Rabies Virus in Nasal Mucosa of Naturally Infected Bats. Science 1972, 175, 1255–1256. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, S.D.; Cortez, A.; Heinemann, M.B.; Harary, C.M.A.; Antunes, J.M.A.; Peres, M.G.; Vicente, A.F.; Sodré, M.M.; da Rosa, A.R.; Megid, J. Rabies virus distribution in tissues and molecular characterization of strains from naturally infected non-hematophagous bats. Virus Res. 2012, 165, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.R.; Streicker, D.G.; Schnell, M.J. The spread and evolution of rabies virus: conquering new frontiers. Nat. Rev. Microbiol. 2018, 16, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.D.; Rudd, R.J.; Bowen, R.A. Effects of Aerosolized Rabies Virus Exposure on Bats and Mice. J. Infect. Dis. 2007, 195, 1144–1150. [Google Scholar] [CrossRef]
- Johnson, N.; Phillpotts, R.; Fooks, A.R. Airborne transmission of lyssaviruses. J. Med Microbiol. 2006, 55, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Victoria, J.G.; Wang, C.; Jones, M.; Fellers, G.M.; Kunz, T.H.; Delwart, E. Bat Guano Virome: Predominance of Dietary Viruses from Insects and Plants plus Novel Mammalian Viruses. J. Virol. 2010, 84, 6955–6965. [Google Scholar] [CrossRef] [PubMed]
- Lojkić, I.; Šimić, I.; Bedeković, T.; Krešić, N. Current Status of Rabies and Its Eradication in Eastern and Southeastern Europe. Pathogens 2021, 10, 742. [Google Scholar] [CrossRef] [PubMed]
- Dacheux, L.; Larrous, F.; Mailles, A.; Boisseleau, D.; Delmas, O.; Biron, C.; Bouchier, C.; Capek, I.; Muller, M.; Ilari, F.; et al. European Bat Lyssavirus Transmission among Cats, Europe. Emerg. Infect. Dis. 2009, 15, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Leopardi, S.; Barneschi, E.; Manna, G.; Zecchin, B.; Priori, P.; Drzewnioková, P.; Festa, F.; Lombardo, A.; Parca, F.; Scaravelli, D.; et al. Spillover of West Caucasian Bat Lyssavirus (WCBV) in a Domestic Cat and Westward Expansion in the Palearctic Region. Viruses 2021, 13, 2064. [Google Scholar] [CrossRef]
- Banyard, A.C.; Evans, J.S.; Luo, T.R.; Fooks, A.R. Lyssaviruses and Bats: Emergence and Zoonotic Threat. Viruses 2014, 6, 2974–2990. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Niezgoda, M.; Franka, R.; Agwanda, B.; Markotter, W.; Beagley, J.C.; Urazova, O.Y.; Breiman, R.F.; Rupprecht, C.E. Lagos Bat Virus in Kenya. J. Clin. Microbiol. 2008, 46, 1451–1461. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Turmelle, A.S.; Agwanda, B.; Markotter, W.; Niezgoda, M.; Breiman, R.F.; Rupprecht, C.E.; Woldehanna, S.; Zimicki, S.; Nel, L.; et al. Commerson’s Leaf-Nosed Bat (Hipposideros commersoni) is the Likely Reservoir of Shimoni Bat Virus. Vector-Borne Zoonotic Dis. 2011, 11, 1465–1470. [Google Scholar] [CrossRef]
- Lučan, R.K.; Bartonička, T.; Benda, P.; Bilgin, R.; Jedlička, P.; Nicolaou, H.; Reiter, A.; Shohdi, W.M.; Šálek, M.; Řeřucha. ; et al. Reproductive seasonality of the Egyptian fruit bat (Rousettus aegyptiacus) at the northern limits of its distribution. J. Mammal. 2014, 95, 1036–1042. [Google Scholar] [CrossRef]
- Benda, P.; Abi-Said, M.; Bartonička, T.; Bilgin, R.; Faizolahi, K.; Lučan, R. K.; Nicolaou, H.; Raiter, A.; Shohdi, W. M.; Uhrin, M.; Horáček, I. Rousettus aegyptiacus (Pteropodidae) in the Palaearctic: list of records and revision of the distribution range. Vespertilio. 2011, 15, 3—36. [Google Scholar]
- Benda, P.; Abi Said, M. R.; Bou Jaoude, I.; Karanouh, R.; Lučan, R. K.; Sadek, R. ; Ševčik, Uhrin, M. ; Horáček, I. Bats (Mammalia: Chiroptera) of the Eastern Mediterranean and Middle East. Part 13. Review of distribution and ectoparasites of bats in Lebanon. Acta Soc. Zool. Bohem. 2016, 80, 207–316. [Google Scholar]
- Willoughby, A.R.; Phelps, K.L.; PREDICT Consortium; Olival, K. J. A Comparative Analysis of Viral Richness and Viral Sharing in Cave-Roosting Bats. Diversity 2017, 9, 35. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, G.; Huang, M.; Dai, Q.; Hu, Y.; Zhou, J.; Wei, F. Global patterns of phylogenetic diversity and transmission of bat coronavirus. Sci. China Life Sci. 2022, 66, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Albery, G.F.; Becker, D.J.; Brierley, L.; Brook, C.E.; Christofferson, R.C.; Cohen, L.E.; Dallas, T.A.; Eskew, E.A.; Fagre, A.; Farrell, M.J.; et al. The science of the host–virus network. Nat. Microbiol. 2021, 6, 1483–1492. [Google Scholar] [CrossRef]

| Species | IUCN status |
Bat lyssaviruses in Europe | ||||||
|---|---|---|---|---|---|---|---|---|
| Rhinolophidae | EBLV-1 | EBLV-2 | BBLV | KBLV | LLEBV | WCBV | ||
| Rhinolophus blasii | Blasius’s horseshoe bat | VU | ||||||
| Rhinolophus euryale | Mediterranean horseshoe bat | VU | ||||||
| Rhinolophus ferrumequinum | Greater horseshoe bat | NT | + | |||||
| Rhinolophus hipposideros | Lesser horseshoe bat | NT | ||||||
| Rhinolophus mehelyi | Mehely’s horseshoe bat | VU | ||||||
| Vespertilionidae | ||||||||
| Barbastella barbastellus | Western Barbastelle bat | VU | ||||||
| Eptesicus bottae | Botta’s Serotine | N/A | ||||||
| Eptesicus nilssonii | Northern bat | LC | ||||||
| Eptesicus isabellinus | Isabelline Serotine bat | N/A | + | |||||
| Eptesicus serotinus | Common Serotine | LC | + | |||||
| Hypsugo savii | Savi’s pipistrelle | LC | ||||||
| Myotis alcathoe | Alcathoe whiskered bat | DD | ||||||
| Myotis aurascens | Steppe whiskered bat | LC | ||||||
| Myotis bechsteinii | Myotis bechsteinii | VU | ||||||
| Myotis blythii | Lesser mouse-eared bat | NT | + | |||||
| Myotis brandtii | Brandt’s bat | LC | + | |||||
| Myotis capaccinii | Long-fingered bat | VU | ||||||
| Myotis dasycneme | Pond bat | NT | + | |||||
| Myotis daubentonii | Daubenton’s bat | LC | + | |||||
| Myotis escalerai | Escalerai bat | N/A | ||||||
| Myotis emarginatus | Geoffroy’s bat | LC | ||||||
| Myotis myotis | Greater mouse-eared bat | LC | + | |||||
| Myotis mystacinus | Whiskered bat | LC | ||||||
| Myotis nattereri | Natterer’s bat | LC | + | + | ||||
| Myotis punicus | Maghreb mouse-eared bat | NT | ||||||
| Nyctalus azoreum | Azorean bat | EN | ||||||
| Nyctalus lasiopterus | Greater noctule bat | DD | ||||||
| Nyctalus leisleri | Leisler’s bat | LC | ||||||
| Nyctalus noctula | Common noctule | LC | + | |||||
| Pipistrellus kuhlii | Kuhl’s pipistrelle | LC | ||||||
| Pipistrellus hanaki | Hanaki’s Dwarf Bat | N/A | ||||||
| Pipistrellus maderensis | Madeira pipistrelle | EN | ||||||
| Pipistrellus nathusii | Nathusius’s pipistrelle | LC | + | |||||
| Pipistrellus pipistrellus | Common pipistrelle | LC | + | |||||
| Pipistrellus pygmaeus | Pygmy pipistrelle | LC | ||||||
| Plecotus auritus | Brown long-eared bat | LC | + | |||||
| Plecotus austriacus | Grey long-eared bat | LC | ||||||
| Plecotus kolombatovici | Kolombatovic’s Long-eared bat | NT | ||||||
| Plecotus macrobullaris | Mountain long-eared bat | NT | ||||||
| Plecotus sardus | Sardinian long-eared bat | VU | ||||||
| Plecotus teneriffae | Tenerife long-eared bat | EN | ||||||
| Vespertilio murinus | Parti-coloured bat | LC | + | |||||
| Miniopteridae | ||||||||
| Miniopterus schreibersii | Schreibers’s long-fingered bat | NT | + | + | + | |||
| Molossidae | ||||||||
| Tadarida teniotis | European free-tailed bat | LC | + | |||||
| Pteropodidae | ||||||||
| Rousettus aegyptiacus | Egyptian fruit bat | N/A (EN?) | + | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





