Submitted:
22 May 2023
Posted:
23 May 2023
You are already at the latest version
Abstract
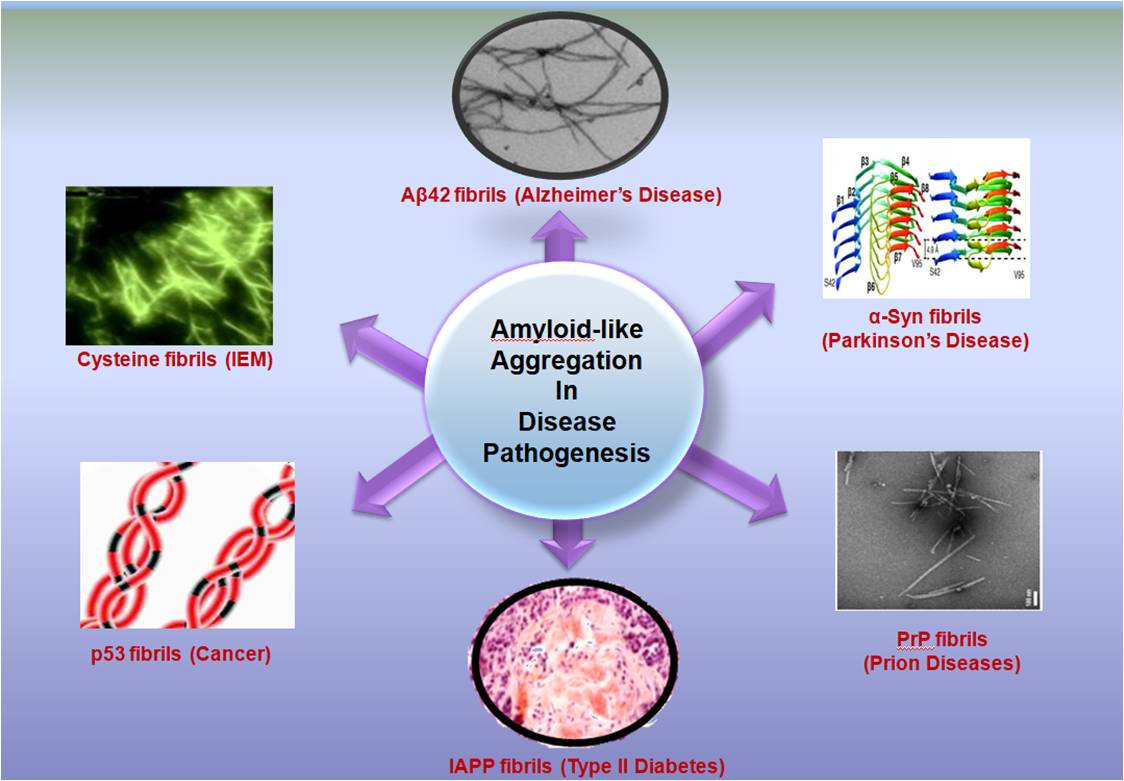
Keywords:
1. Introduction
2. General characteristics of Amyloid structures
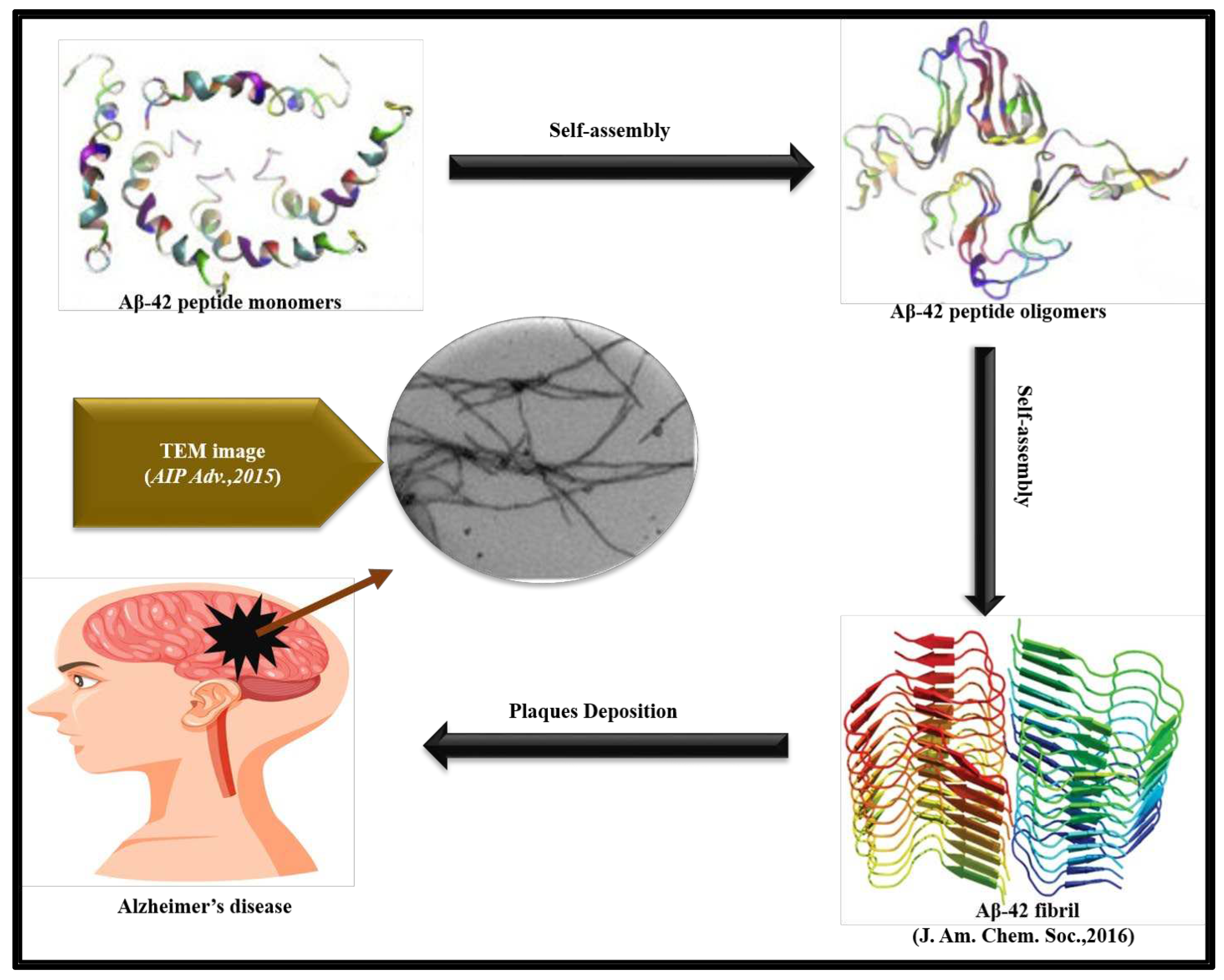
3. Aβ42 and Aβ40 aggregation in Alzheimer’s disease (AD)
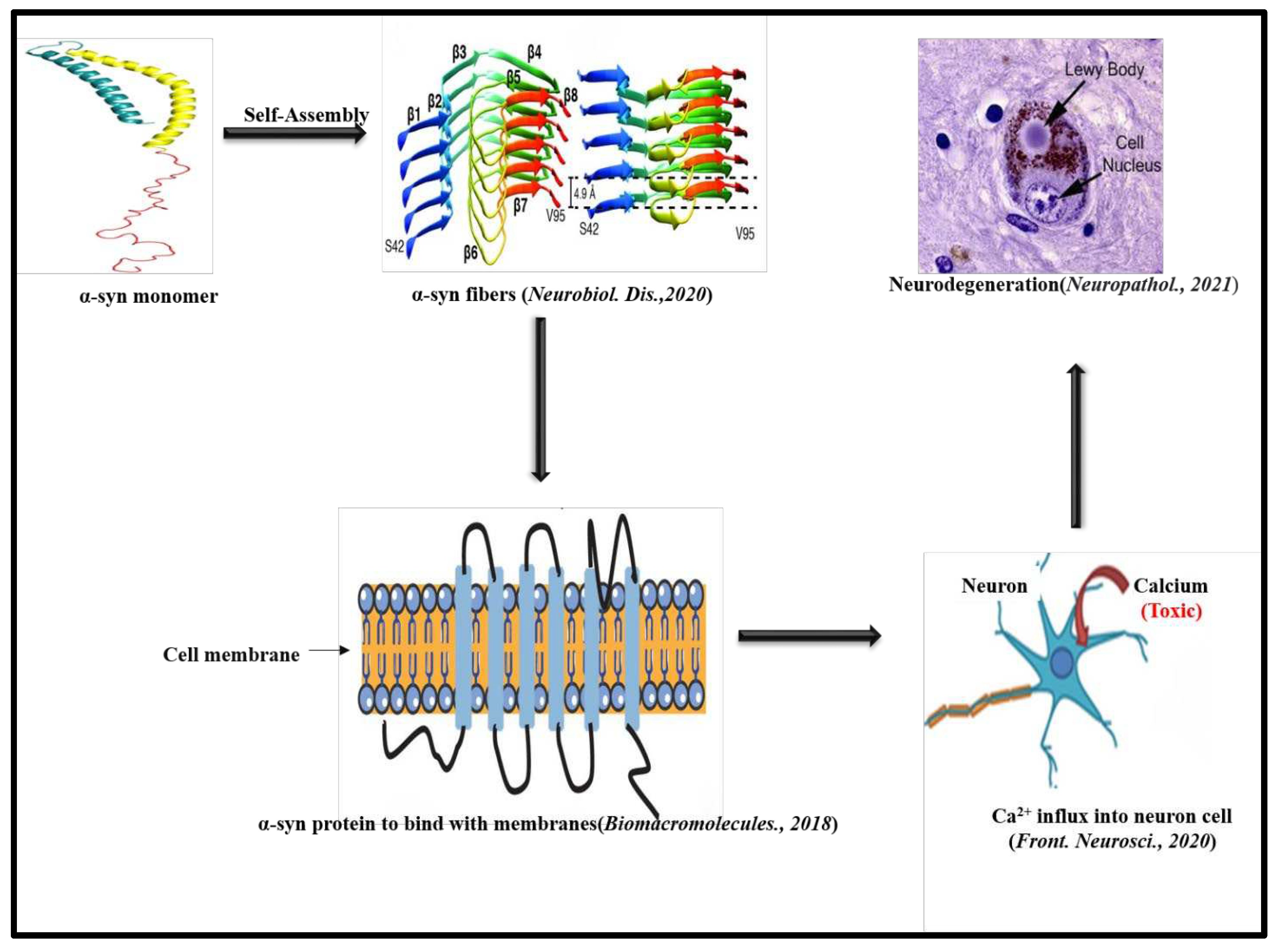
4. α-Synuclein Aggregation and Parkinson’s disease
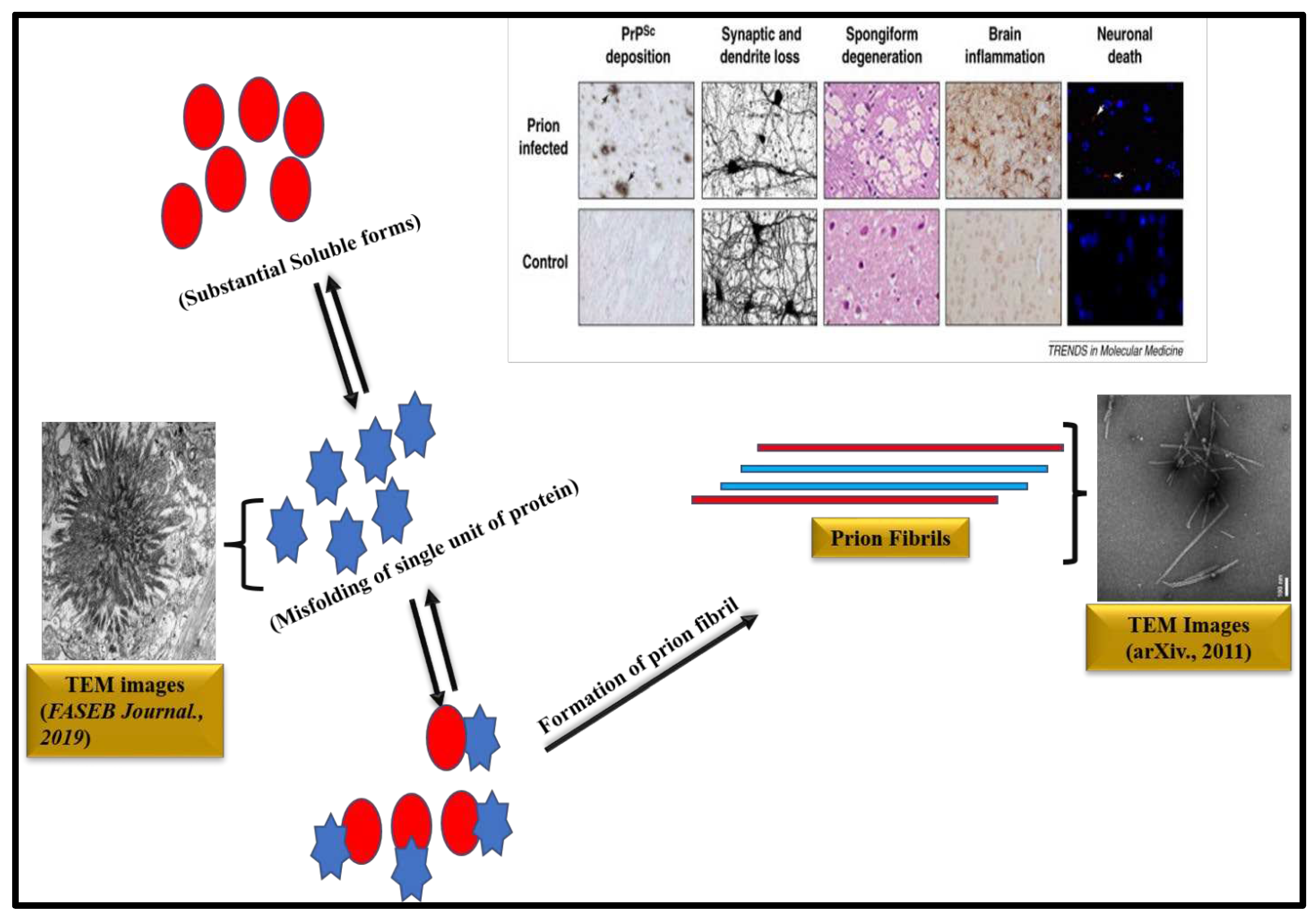
5. Aggregation of Prion protein (PrP) and the associated diseases
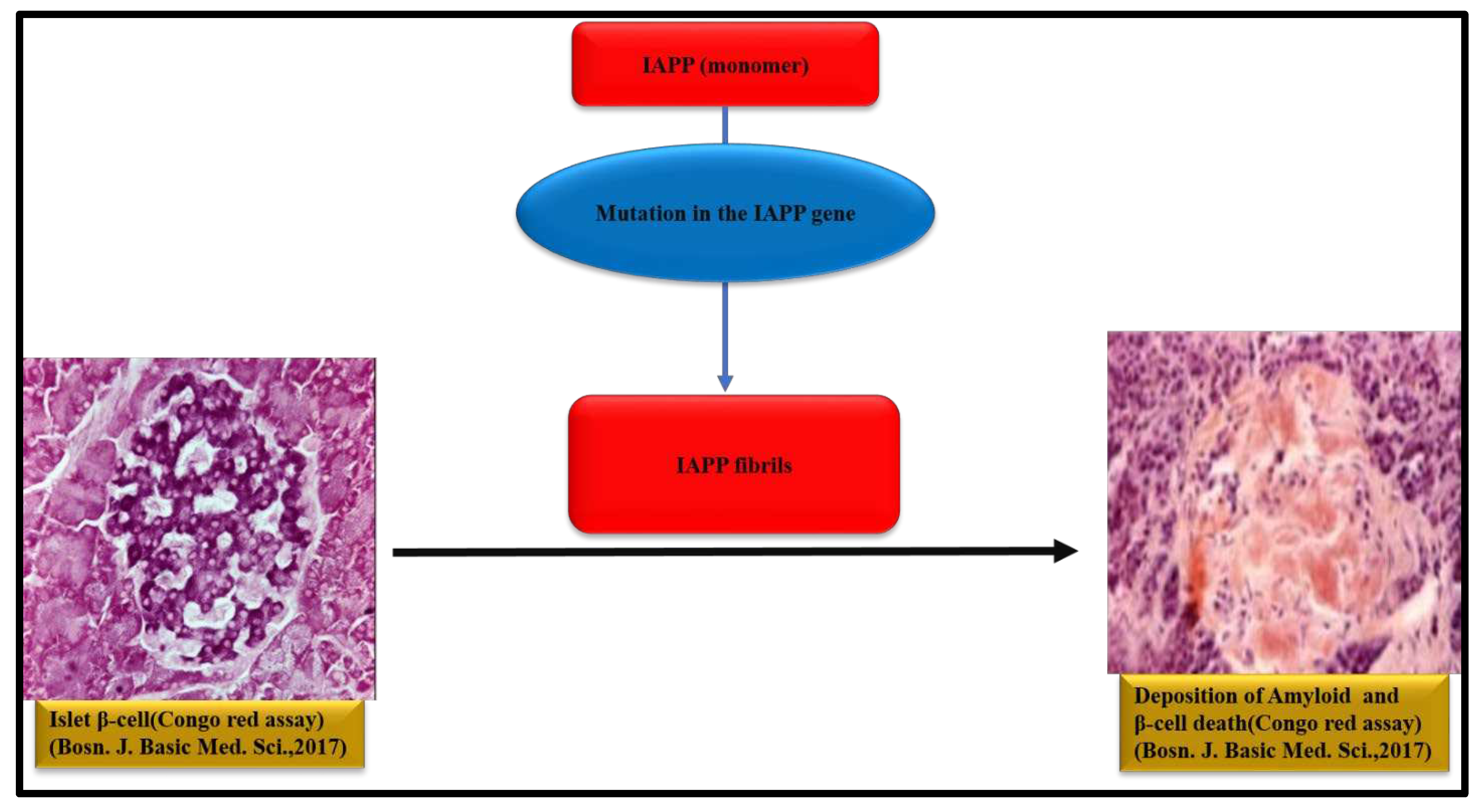
6. IAPP aggregation and Type 2 diabetes
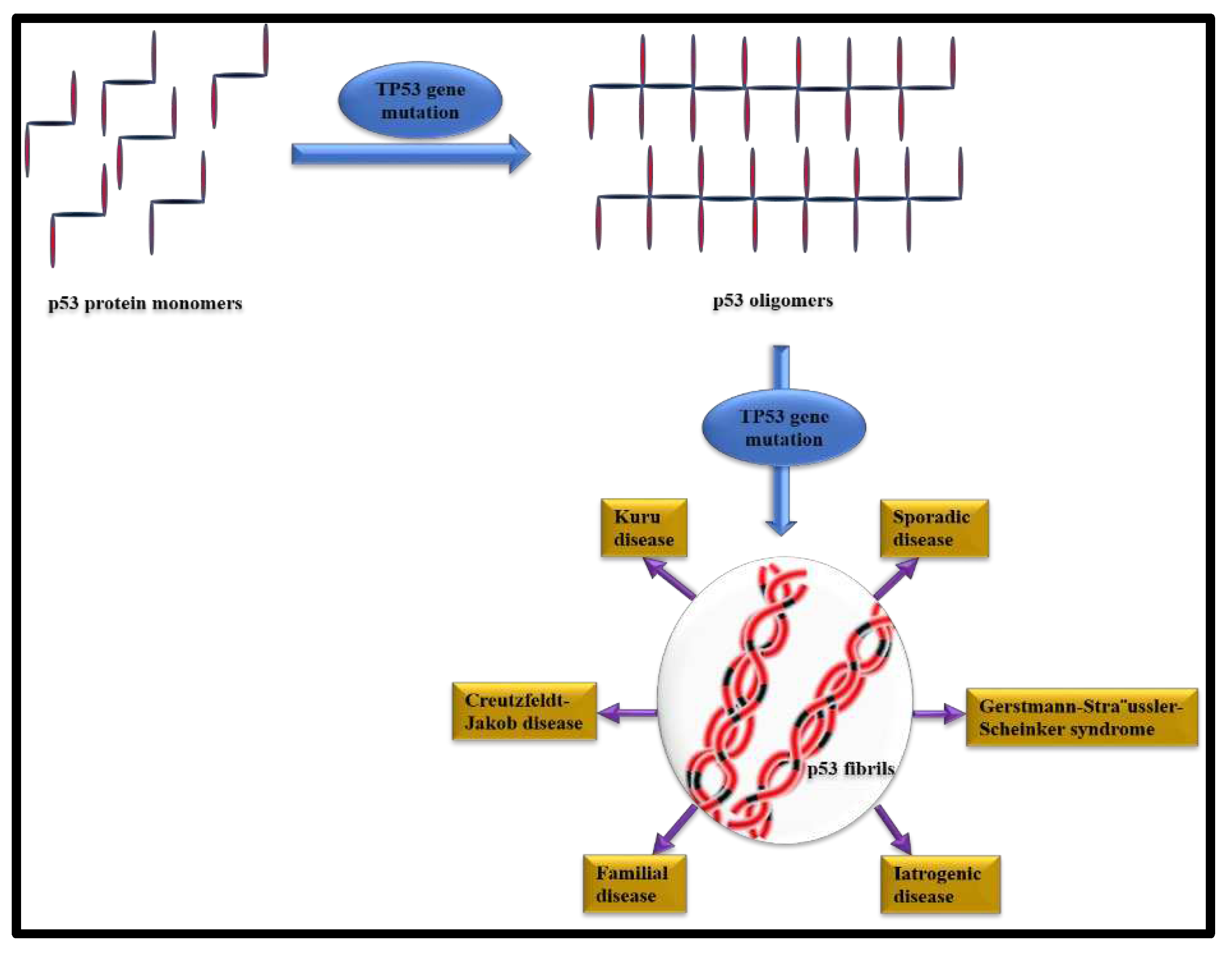
7. Amyloid like aggregation in the patho-physiology of Cancer.
8. Metabolites assemblies as a surprising extension to Generic amyloid Hypothesis.
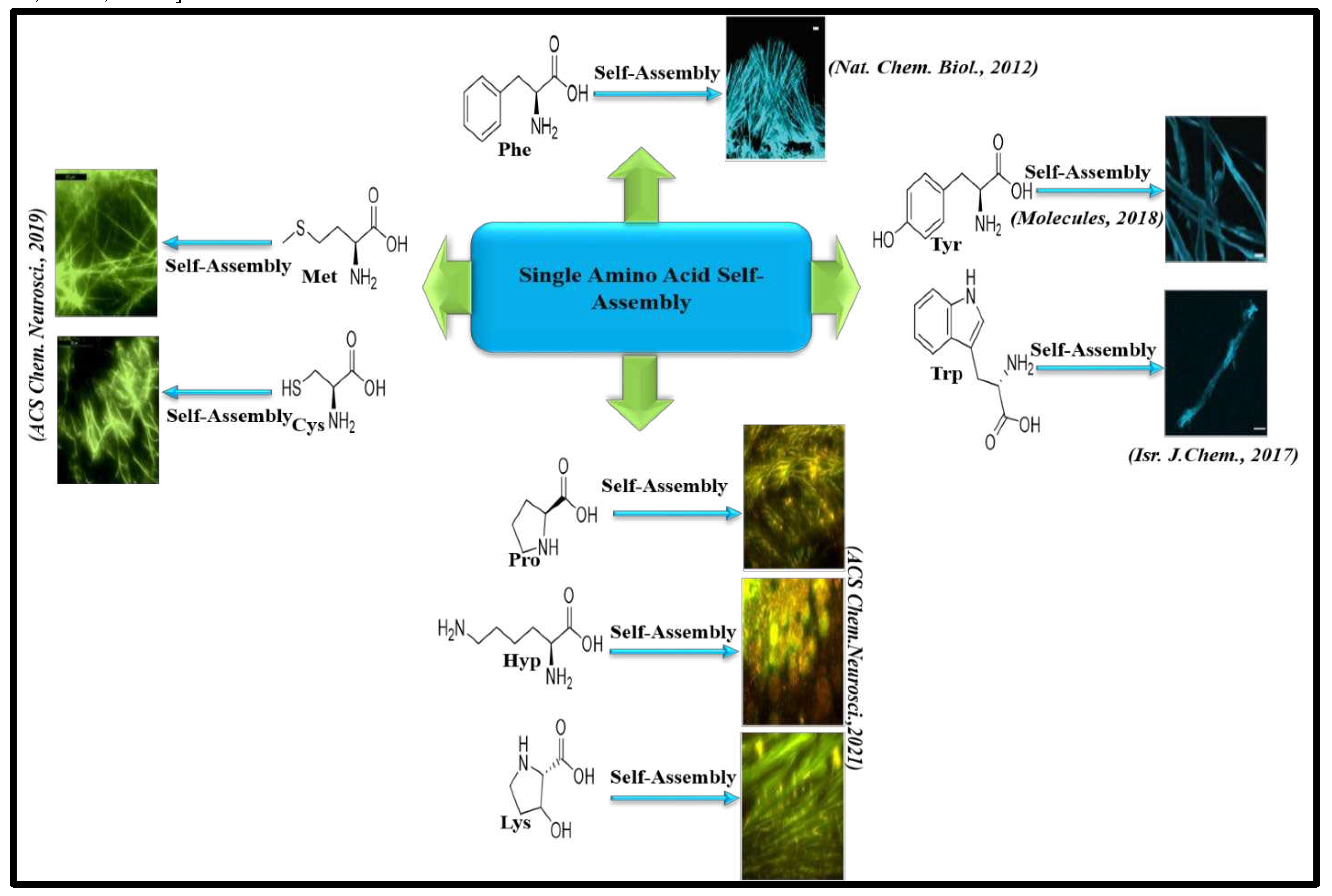
8.1. Amyloid like aggregation of Single amino acids and its implications in IEMs
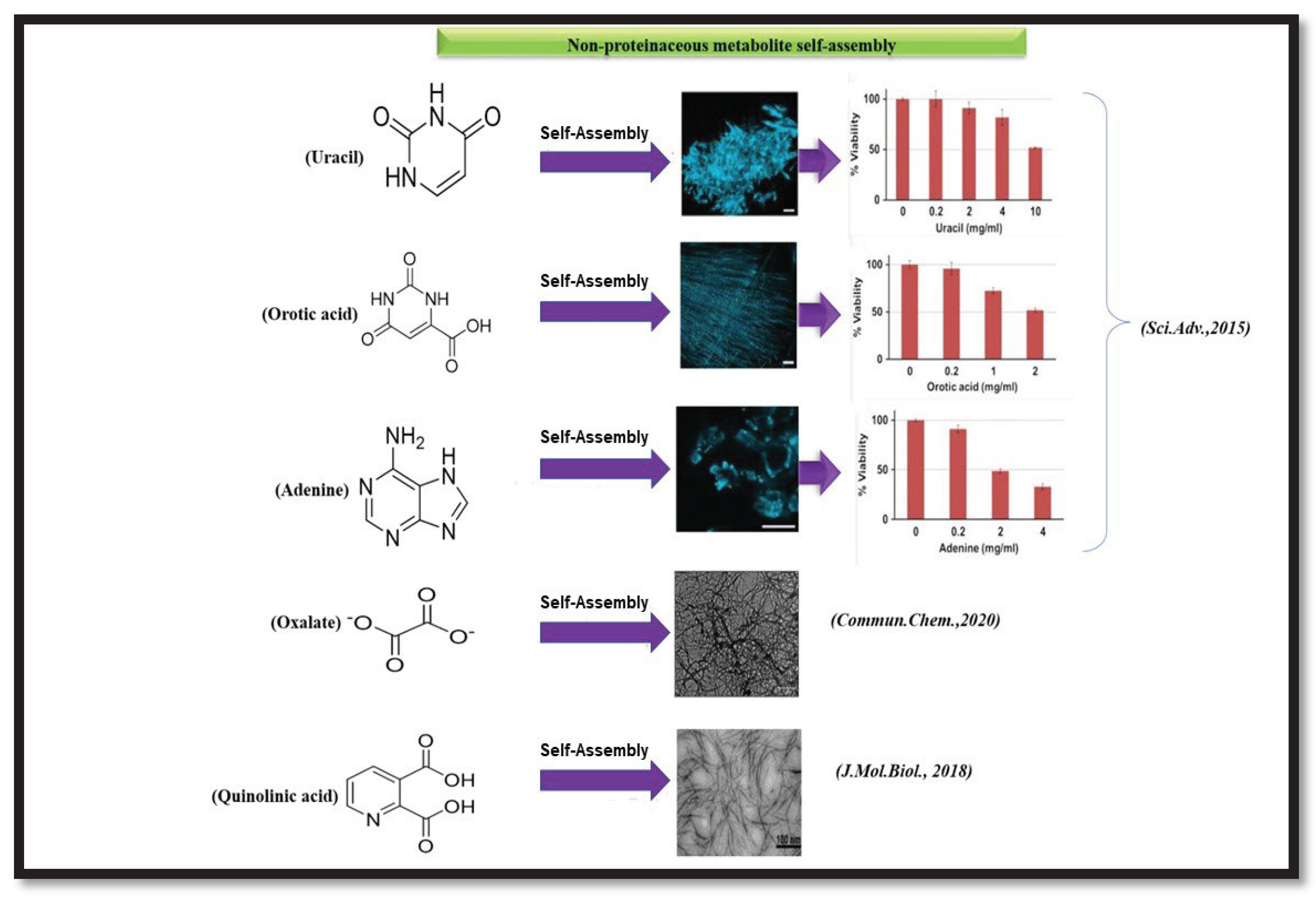
8.2. Non-proteinaceous metabolite assemblies and its implications in IEMs
9. Critical analysis and future outlook
| Type of Amyloids (Disease caused) | Aβ42/40 (AD) | α-Syn (PD) | PrP (prion diseases) | IAPP (Type 2 Diabetes) | p53 (Cancer) | Metabolite Amyloid (IEMs) |
|---|---|---|---|---|---|---|
| Characteristics | 1. Aβ42 – 42 amino acids based-peptide. Aβ40 – 40 amino acids based-peptide. 2.βAPP, PSN1 and PSN2 genes mutations lead to an accumulation of Aβ42 and Aβ40 peptides. 3. Aβ42 accumulation causes AD. 4. Aβ42/40 disrupts cell membrane. |
1. α-Syn is a 140 amino acid based long protein. 2. SNCA gene encodes for α-syn. Mutation in this gene leads to accumulation of α-syn. 3. Accretion of α-Syn causes PD. 4.α-synuclein amyloid fibrils are the major component of Lewy bodies. 5. α-Syn amyloid disrupts cellular membrane. |
1. PrP is 208 amino acids-based protein. 2.Two isomeric forms of PrP are as follows: Cellular form: PrPC Scrapie form: PrPSc. 3. PRNP gene mutation. Mutations in PRNP gene can be missense, insertion or point mutations 4. Prion protein accumulation is known to be associated with the prion diseases. 5. PrP amyloid disrupts cell membrane |
1. It is 37 amino acids-based -polypeptide. 2. It is a regulatory peptide in the islets and it inhibits insulin and glucagon secretion. 3. The human plasma IAPP concentration is only 1-2% of that of insulin. 4.Mutation in the IAPP gene is the main cause. 5. This amyloid causes type 2 diabetes. 6. IAPP amyloids disrupt cell membrane. |
1. It is 393 amino acids based Protein. 2.TP53 gene mutation is responsible for p53 accumulation. 3. Accumulation of p53 is associated with Cancer. 3. It is a tumor suppressor protein. 4.p53 amyloid disrupts cell membrane. |
1. Few amino acids and non- proteinaceous metabolites exhibit amyloid. 2. Mutation in specific genes is the main reason behind accumulation of metabolites. 3.These amyloids can induce cross-seeding. 4.These amyloids can act as functional amyloids. 5. It causes rare Inborn Errors of Metabolism. 6.These amyloids interact with cell membrane leading to disruption of membrane structure. |
| Secondary Structure | β-sheet | β-sheet | β-sheet | β-sheet | β-sheet | β-sheet |
| X-ray | 1. 4.8Å reflection on the meridian. 2. 10-11 Å reflection on equator. |
Meridional diffraction pattern was found at 4.8 Å whereas equatorial diffraction was at 10 Å |
1. A strong meridional diffraction signal at 4.6-4.7 Å. 2. A second strong reflection at equator of 8.5-9.7 Å. |
4.7Å meridional and 10 Å equatorial reflections. | 4.7Å meridional and 10 Å equatorial reflections. | Detects crystalline or non-crystalline nature in assembly process. |
| CD | Aqueous solution of Aβ40 indicates majority portions are β-sheet structure whereas Aβ42 contains only β-sheet structure. |
A mix of α-sheet and random coil to β-sheet enriched structure. | Highly ordered β-sheet conformation. | IAPP aggregates contain mostly β-sheet. | CD spectra shows that both fibrillar and ordered p53 aggregates have β-sheet rich profile. | CD spectra shows β-sheet formation. |
| CR | Apple green birefringence color after staining with CR. | Apple green birefringence color after staining with CR. | Apple green birefringence color after staining with CR. | Apple green birefringence color after staining with CR. | Apple green birefringence color after staining with CR. | It might display Apple green birefringence color after staining with CR. |
| ThT | ThT fluorescence displays an enhanced intensity at 480 nm. | ThT fluorescence displays an enhanced intensity at 480 nm. | ThT fluorescence displays an enhanced intensity at 485 nm. | ThT fluorescence displays an enhanced intensity at 480 nm. | ThT fluorescence displays an enhanced intensity at 480 nm. | It may display an enhanced intensity at 480 nm due to ThT fluorescence. |
| Location | Brain | Brain | Brain | Pancreatic β-cells | Cancer Tissues | Brain and other organs |
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kyle, R.A. Amyloidosis: A convoluted story. British journal of haematology 2001, 114, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer, A.; Stelzmann, R.A.; Schnitzlein, H.N.; Murtagh, F.R. An English translation of Alzheimer's 1907 paper," Uber eine eigenartige Erkankung der Hirnrinde". Clinical anatomy (New York, NY) 1995, 8, 429–431. [Google Scholar]
- Plascencia-Villa, G.; Perry, G. Neuropathologic Changes Provide Insights into Key Mechanisms of Alzheimer Disease and Related Dementia. Am. J. Pathol. 2022, 192, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Guo, Z. Alzheimer's Aβ42 and Aβ40 peptides form interlaced amyloid fibrils. Journal of neurochemistry 2013, 126, 305–311. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Sengupta, U.; Sarmiento, J.; Troncoso, J.; Jackson, G.R.; Kayed, R. Identification of oligomers at early stages of tau aggregation in Alzheimer's disease. The FASEB Journal 2012, 26, 1946. [Google Scholar] [CrossRef]
- Wong, Y.C.; Krainc, D. α-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nat. Med. 2017, 23, 1–13. [Google Scholar] [CrossRef]
- Akter, R.; Cao, P.; Noor, H.; Ridgway, Z.; Tu, L.-H.; Wang, H.; Wong, A.G.; Zhang, X.; Abedini, A.; Schmidt, A.M.; et al. Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology. J. Diabetes Res. 2015, 2016, 1–18. [Google Scholar] [CrossRef]
- Wille, H.; Requena, J.R. The structure of PrPSc prions. Pathogens 2018, 7, 20. [Google Scholar] [CrossRef]
- Rangel, L.P.; Costa, D.C.; Vieira, T.C.; Silva, J.L. The aggregation of mutant p53 produces prion-like properties in cancer. Prion 2014, 8, 75–84. [Google Scholar] [CrossRef]
- Adler-Abramovich, L.; Vaks, L.; Carny, O.; Trudler, D.; Magno, A.; Caflisch, A.; Frenkel, D.; Gazit, E. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat. Chem. Biol. 2012, 8, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Gour, N.; Kanth P, C.; Koshti, B.; Kshtriya, V.; Shah, D.; Patel, S.; Agrawal-Rajput, R.; Pandey, M.K. Amyloid-like structures formed by single amino acid self-assemblies of cysteine and methionine. ACS chemical neuroscience 2018, 10, 1230–1239. [Google Scholar] [CrossRef] [PubMed]
- Koshti, B.; Kshtriya, V.; Singh, R.; Walia, S.; Bhatia, D.; Joshi, K.B.; Gour, N. Unusual Aggregates Formed by the Self-Assembly of Proline, Hydroxyproline, and Lysine. ACS Chem. Neurosci. 2021, 12, 3237–3249. [Google Scholar] [CrossRef] [PubMed]
- Shaham-Niv, S.; Adler-Abramovich, L.; Schnaider, L.; Gazit, E. Extension of the generic amyloid hypothesis to nonproteinaceous metabolite assemblies. Sci. Adv. 2015, 1, e1500137–e1500137. [Google Scholar] [CrossRef]
- Koshti, B.; Kshtriya, V.; Naskar, S.; Narode, H.; Gour, N. Controlled aggregation properties of single amino acids modified with protecting groups. New J. Chem. 2022, 46, 4746–4755. [Google Scholar] [CrossRef]
- Gour, N.; Gazit, E. Metabolite assemblies: A surprising extension to the amyloid hypothesis. Curr. Opin. Chem. Biol. 2021, 64, 154–164. [Google Scholar] [CrossRef]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef]
- Khurana, R.; Coleman, C.; Ionescu-Zanetti, C.; Carter, S.A.; Krishna, V.; Grover, R.K.; Roy, R.; Singh, S. Mechanism of thioflavin T binding to amyloid fibrils. J. Struct. Biol. 2005, 151, 229–238. [Google Scholar] [CrossRef]
- Zaman, M.; Khan, A.N.; Wahiduzzaman; Zakariya, S. M.; Khan, R.H. Protein misfolding, aggregation and mechanism of amyloid cytotoxicity: An overview and therapeutic strategies to inhibit aggregation. Int. J. Biol. Macromol. 2019, 134, 1022–1037. [Google Scholar] [CrossRef]
- Amdursky, N.; Stevens, M.M. Circular Dichroism of Amino Acids: Following the Structural Formation of Phenylalanine. Chemphyschem 2015, 16, 2768–2774. [Google Scholar] [CrossRef]
- Carulla, N.; Zhou, M.; Arimon, M.; Gairí, M.; Giralt, E.; Robinson, C.V.; Dobson, C.M. Experimental characterization of disordered and ordered aggregates populated during the process of amyloid fibril formation. Proc. Natl. Acad. Sci. 2009, 106, 7828–7833. [Google Scholar] [CrossRef]
- Adamcik, J.; Mezzenga, R. Study of amyloid fibrils via atomic force microscopy. Curr. Opin. Colloid Interface Sci. 2012, 17, 369–376. [Google Scholar] [CrossRef]
- Stefani, M.; Dobson, C.M. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003, 81, 678–699. [Google Scholar] [CrossRef]
- Arispe, N.; Pollard, H.B.; Rojas, E. The Ability of Amyloid β-Protein [AβP (1–40)] to Form Ca2+ Channels Provides a Mechanism for Neuronal Death in Alzheimer's Disease. Annals of the New York Academy of Sciences 1994, 747, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.; Brennan, S.; Keon, M.; Ooi, L. The role of amyloid oligomers in neurodegenerative pathologies. Int. J. Biol. Macromol. 2021, 181, 582–604. [Google Scholar] [CrossRef]
- Villegas, S.; Roda, A.; Serra-Mir, G.; Montoliu-Gaya, L.; Tiessler, L. Amyloid-beta peptide and tau protein crosstalk in Alzheimer’s disease. Neural Regen. Res. 2022, 17, 1666–1674. [Google Scholar] [CrossRef]
- Ghiso, J.; Frangione, B. Amyloidosis and Alzheimer’s disease. Advanced drug delivery reviews 2002, 54, 1539–1551. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, W.; Sun, Y.; Dong, X. Insights into the cross-amyloid aggregation of Aβ40 and its N-terminal truncated peptide Aβ11-40 affected by epigallocatechin gallate. Chin. J. Chem. Eng. 2021, 45, 284–293. [Google Scholar] [CrossRef]
- Baazaoui, N.; Iqbal, K. Alzheimer’s Disease: Challenges and a Therapeutic Opportunity to Treat It with a Neurotrophic Compound. Biomolecules 2022, 12, 1409. [Google Scholar] [CrossRef]
- Parkin, E.T.; Sultan, F. The Amyloid Precursor Protein Plays Differential Roles in the UVA Resistance and Proliferation of Human Retinal Pigment Epithelial Cells. Protein Pept. Lett. 2022, 29, 313–327. [Google Scholar] [CrossRef]
- Zhao, J.; O'Connor, T.; Vassar, R. The contribution of activated astrocytes to Aβ production: Implications for Alzheimer's disease pathogenesis. Journal of neuroinflammation 2011, 8, 1–17. [Google Scholar] [CrossRef]
- Calhoun, M.E.; Burgermeister, P.; Phinney, A.L.; Stalder, M.; Tolnay, M.; Wiederhold, K.-H.; Abramowski, D.; Sturchler-Pierrat, C.; Sommer, B.; Staufenbiel, M.; et al. Neuronal overexpression of mutant amyloid precursor protein results in prominent deposition of cerebrovascular amyloid. Proc. Natl. Acad. Sci. 1999, 96, 14088–14093. [Google Scholar] [CrossRef]
- Brothers, H.M.; Gosztyla, M.L.; Robinson, S.R. The physiological roles of amyloid-β peptide hint at new ways to treat Alzheimer's disease. Frontiers in aging neuroscience 2018, 118. [Google Scholar] [CrossRef]
- Sehar, U.; Rawat, P.; Reddy, A.P.; Kopel, J.; Reddy, P.H. Amyloid beta in aging and Alzheimer’s disease. International journal of molecular sciences 2022, 23, 12924. [Google Scholar] [CrossRef]
- Yankner, B.A.; Duffy, L.K.; Kirschner, D.A. Neurotrophic and Neurotoxic Effects of Amyloid β Protein: Reversal by Tachykinin Neuropeptides. Science 1990, 250, 279–282. [Google Scholar] [CrossRef]
- Antonino, M.; Marmo, P.; Freites, C.L.; Quassollo, G.E.; Sánchez, M.F.; Lorenzo, A.; Bignante, E.A. Aβ Assemblies Promote Amyloidogenic Processing of APP and Intracellular Accumulation of Aβ42 Through Go/Gβγ Signaling. Frontiers in Cell and Developmental Biology 2022, 638. [Google Scholar] [CrossRef]
- Kidd, M. Paired helical filaments in electron microscopy of Alzheimer's disease. Nature 1963, 197, 192–193. [Google Scholar] [CrossRef]
- Serpell, L.C. Alzheimer’s amyloid fibrils: Structure and assembly. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2000, 1502, 16–30. [Google Scholar] [CrossRef]
- Harper, J.D.; Wong, S.S.; Lieber, C.M.; Lansbury Jr, P.T. Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chemistry & biology 1997, 4, 119–125. [Google Scholar]
- Kollmer, M.; Close, W.; Funk, L.; Rasmussen, J.; Bsoul, A.; Schierhorn, A.; Schmidt, M.; Sigurdson, C.J.; Jucker, M.; Fändrich, M. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Morris, K.L.; Serpell, L.C. X-ray fibre diffraction studies of amyloid fibrils. Amyloid proteins: Methods and protocols 2012, 121–135.
- Colvin, M.T.; Silvers, R.; Ni, Q.Z.; Can, T.V.; Sergeyev, I.; Rosay, M.; Donovan, K.J.; Michael, B.; Wall, J.; Linse, S. Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. Journal of the American Chemical Society 2016, 138, 9663–9674. [Google Scholar] [CrossRef]
- Wälti, M.A.; Ravotti, F.; Arai, H.; Glabe, C.G.; Wall, J.S.; Böckmann, A.; Güntert, P.; Meier, B.H.; Riek, R. Atomic-resolution structure of a disease-relevant Aβ (1–42) amyloid fibril. Proceedings of the National Academy of Sciences 2016, 113, E4976–E4984. [Google Scholar] [CrossRef]
- Soto, C.; Castaño, E.M.; Frangione, B.; Inestrosa, N.C. The α-Helical to β-Strand Transition in the Amino-terminal Fragment of the Amyloid β-Peptide Modulates Amyloid Formation∗. Journal of Biological Chemistry 1995, 270, 3063–3067. [Google Scholar] [CrossRef]
- Habiba, U.; Merlin, S.; Lim, J.K.; Wong, V.H.; Nguyen, C.T.; Morley, J.W.; Bui, B.V.; Tayebi, M. Age-Specific Retinal and Cerebral Immunodetection of Amyloid-β Plaques and Oligomers in a Rodent Model of Alzheimer’s Disease. J. Alzheimer's Dis. 2020, 76, 1135–1150. [Google Scholar] [CrossRef]
- Zhang, T.; Xia, Y.; Hu, L.; Chen, D.; Gan, C.-L.; Wang, L.; Mei, Y.; Lan, G.; Shui, X.; Tian, Y. Death-associated protein kinase 1 mediates Aβ42 aggregation-induced neuronal apoptosis and tau dysregulation in Alzheimer's disease. International Journal of Biological Sciences 2022, 18, 693. [Google Scholar] [CrossRef]
- Tiiman, A.; Krishtal, J.; Palumaa, P.; Tõugu, V. In vitro fibrillization of Alzheimer’s amyloid-β peptide (1-42). Aip Advances 2015, 5, 092401. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R. Mutation in the α-synuclein gene identified in families with Parkinson's disease. science 1997, 276, 2045–2047. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.-Y.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. α-Synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Jiang, Z.; Huang, Y.; Zhang, P.; Han, C.; Lu, Y.; Mo, Z.; Zhang, Z.; Li, X.; Zhao, S.; Cai, F.; et al. Characterization of a pathogenic variant in GBA for Parkinson’s disease with mild cognitive impairment patients. Mol. Brain 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Choi, M.L.; Gandhi, S. Crucial role of protein oligomerization in the pathogenesis of Alzheimer's and Parkinson's diseases. The FEBS journal 2018, 285, 3631–3644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Y.-Q.; Jia, C.; Lim, Y.-J.; Feng, G.; Xu, E.; Long, H.; Kimura, Y.; Tao, Y.; Zhao, C. Mechanistic basis for receptor-mediated pathological α-synuclein fibril cell-to-cell transmission in Parkinson's disease. Proceedings of the National Academy of Sciences 2021, 118, e2011196118. [Google Scholar] [CrossRef]
- Clayton, D.F.; George, J.M. The synucleins: a family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci. 1998, 21, 249–254. [Google Scholar] [CrossRef]
- Weinreb, P.H.; Zhen, W.; Poon, A.W.; Conway, K.A.; Lansbury, P.T. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 1996, 35, 13709–13715. [Google Scholar] [CrossRef] [PubMed]
- Davidson, W.S.; Jonas, A.; Clayton, D.F.; George, J.M. Stabilization of α-Synuclein Secondary Structure upon Binding to Synthetic Membranes. J. Biol. Chem. 1998, 273, 9443–9449. [Google Scholar] [CrossRef] [PubMed]
- Meena, V.K.; Kumar, V.; Karalia, S.; Dangi, R.S.; Sundd, M. Structural and mechanistic insights into modulation of α-Synuclein fibril formation by aloin and emodin. Biochim. et Biophys. Acta (BBA) - Gen. Subj. 2022, 1866, 130151. [Google Scholar] [CrossRef]
- George, J.M. The synucleins. Genome biology 2001, 3, 1–6. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2012, 14, 38–48. [Google Scholar] [CrossRef]
- Volles, M.J.; Lee, S.-J.; Rochet, J.-C.; Shtilerman, M.D.; Ding, T.T.; Kessler, J.C.; Lansbury, P.T. Vesicle permeabilization by protofibrillar α-synuclein: Implications for the pathogenesis and treatment of Parkinson's disease. Biochemistry 2001, 40, 7812–7819. [Google Scholar] [CrossRef]
- Desplats, P.; Lee, H.-J.; Bae, E.-J.; Patrick, C.; Rockenstein, E.; Crews, L.; Spencer, B.; Masliah, E.; Lee, S.-J. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of α-synuclein. Proc. Natl. Acad. Sci. USA 2009, 106, 13010–13015. [Google Scholar] [CrossRef]
- Araki, K.; Yagi, N.; Aoyama, K.; Choong, C.-J.; Hayakawa, H.; Fujimura, H.; Nagai, Y.; Goto, Y.; Mochizuki, H. Parkinson’s disease is a type of amyloidosis featuring accumulation of amyloid fibrils of α-synuclein. Proc. Natl. Acad. Sci. 2019, 116, 17963–17969. [Google Scholar] [CrossRef]
- Musteikytė, G.; Jayaram, A.K.; Xu, C.K.; Vendruscolo, M.; Krainer, G.; Knowles, T.P.J. Interactions of α-synuclein oligomers with lipid membranes. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183536. [Google Scholar] [CrossRef]
- Dean, D.N.; Lee, J.C. Linking Parkinson's Disease and Melanoma: Interplay Between α-Synuclein and Pmel17 Amyloid Formation. Movement Disorders 2021, 36, 1489–1498. [Google Scholar] [CrossRef]
- Mehra, S.; Ghosh, D.; Kumar, R.; Mondal, M.; Gadhe, L.G.; Das, S.; Anoop, A.; Jha, N.N.; Jacob, R.S.; Chatterjee, D.; et al. Glycosaminoglycans have variable effects on α-synuclein aggregation and differentially affect the activities of the resulting amyloid fibrils. J. Biol. Chem. 2018, 293, 12975–12991. [Google Scholar] [CrossRef]
- Reyes, J.F.; Ekmark-Léwen, S.; Perdiki, M.; Klingstedt, T.; Hoffmann, A.; Wiechec, E.; Nilsson, P.; Nilsson, K.P.R.; Alafuzoff, I.; Ingelsson, M. Accumulation of alpha-synuclein within the liver, potential role in the clearance of brain pathology associated with Parkinson’s disease. Acta neuropathologica communications 2021, 9, 1–20. [Google Scholar] [CrossRef]
- Yamaguchi, K.; So, M.; Aguirre, C.; Ikenaka, K.; Mochizuki, H.; Kawata, Y.; Goto, Y. Polyphosphates induce amyloid fibril formation of α-synuclein in concentration-dependent distinct manners. J. Biol. Chem. 2021, 296, 100510. [Google Scholar] [CrossRef]
- Tuttle, M.D.; Comellas, G.; Nieuwkoop, A.J.; Covell, D.J.; Berthold, D.A.; Kloepper, K.D.; Courtney, J.M.; Kim, J.K.; Barclay, A.M.; Kendall, A.; et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 2016, 23, 409–415. [Google Scholar] [CrossRef]
- Guerrero-Ferreira, R.; Taylor, N.M.; Mona, D.; Ringler, P.; Lauer, M.E.; Riek, R.; Britschgi, M.; Stahlberg, H. Cryo-EM structure of alpha-synuclein fibrils. elife 2018, 7, e36402. [Google Scholar] [CrossRef]
- Flynn, J.D.; McGlinchey, R.P.; Walker, R.L.; Lee, J.C. Structural features of α-synuclein amyloid fibrils revealed by Raman spectroscopy. J. Biol. Chem. 2018, 293, 767–776. [Google Scholar] [CrossRef]
- Kumari, P.; Ghosh, D.; Vanas, A.; Fleischmann, Y.; Wiegand, T.; Jeschke, G.; Riek, R.; Eichmann, C. Structural insights into α-synuclein monomer–fibril interactions. Proceedings of the National Academy of Sciences 2021, 118, e2012171118. [Google Scholar] [CrossRef]
- Zhou, L.; Kurouski, D. Structural Characterization of Individual α-Synuclein Oligomers Formed at Different Stages of Protein Aggregation by Atomic Force Microscopy-Infrared Spectroscopy. Anal. Chem. 2020, 92, 6806–6810. [Google Scholar] [CrossRef]
- Hong, D.-P.; Fink, A.L.; Uversky, V.N. Structural Characteristics of α-Synuclein Oligomers Stabilized by the Flavonoid Baicalein. J. Mol. Biol. 2008, 383, 214–223. [Google Scholar] [CrossRef]
- Faustini, G.; Longhena, F.; Bruno, A.; Bono, F.; Grigoletto, J.; La Via, L.; Barbon, A.; Casiraghi, A.; Straniero, V.; Valoti, E.; et al. Alpha-synuclein/synapsin III pathological interplay boosts the motor response to methylphenidate. Neurobiol. Dis. 2020, 138, 104789. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumari, R.; Kumar, S.; Jangir, D.K.; Maiti, T.K. Extracellular α-Synuclein Disrupts Membrane Nanostructure and Promotes S-Nitrosylation-Induced Neuronal Cell Death. Biomacromolecules 2018, 19, 1118–1129. [Google Scholar] [CrossRef]
- Reis, R.A.d.M.; Freitas, H.R.; de Mello, F.G. Cell Calcium Imaging as a Reliable Method to Study Neuron–Glial Circuits. Front. Neurosci. 2020, 14, 569361. [Google Scholar] [CrossRef] [PubMed]
- Bit-Ivan, E.N.; Bigio, E.H. Neuropathology of neurodegenerative disorders. Progressive Cognitive Impairment and its Neuropathologic Correlates 2016, 1–16. [Google Scholar]
- Ghetti, B.; Piccardo, P.; Frangione, B.; Bugiani, O.; Giaccone, G.; Young, K.; Prelli, F.; Farlow, M.R.; Dlouhy, S.R.; Tagliavini, F. Prion Protein Amyloidosis. Brain Pathol. 1996, 6, 127–145. [Google Scholar] [CrossRef]
- Prusiner, S.B.; McKinley, M.P.; Bowman, K.A.; Bolton, D.C.; Bendheim, P.E.; Groth, D.F.; Glenner, G.G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 1983, 35, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Kamali-Jamil, R.; Vázquez-Fernández, E.; Tancowny, B.; Rathod, V.; Amidian, S.; Wang, X.; Tang, X.; Fang, A.; Senatore, A.; Hornemann, S.; et al. The ultrastructure of infectious L-type bovine spongiform encephalopathy prions constrains molecular models. PLOS Pathog. 2021, 17, e1009628. [Google Scholar] [CrossRef]
- Owen, F.; Poulter, M.; Shah, T.; Collinge, J.; Lofthouse, R.; Baker, H.; Ridley, R.; McVey, J.; Crow, T.J. An in-frame insertion in the prion protein gene in familial Creutzfeldt-Jakob disease. Mol. Brain Res. 1990, 7, 273–276. [Google Scholar] [CrossRef]
- Mastrianni, J.A. Genetics of Prion Disease. Prions and Diseases 2023, 375–424. [Google Scholar]
- Sanz-Hernández, M.; Barritt, J.D.; Sobek, J.; Hornemann, S.; Aguzzi, A.; De Simone, A. Mechanism of misfolding of the human prion protein revealed by a pathological mutation. Proceedings of the National Academy of Sciences 2021, 118, e2019631118. [Google Scholar] [CrossRef]
- Wang, L.-Q.; Zhao, K.; Yuan, H.-Y.; Li, X.-N.; Dang, H.-B.; Ma, Y.; Wang, Q.; Wang, C.; Sun, Y.; Chen, J.; et al. Genetic prion disease–related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, L.; Brown, P.; Little, B.; Cervenakova, L.; Kenney, K.; Gibbs, C.; Gajdusek, D. A new (two-repeat) octapeptide coding insert mutation in Creutzfeldt-Jakob disease. Neurology 1993, 43, 2392–2392. [Google Scholar] [CrossRef] [PubMed]
- Cochran, E.J.; Bennett, D.A.; Cervenakova, L.; Kenney, K.; Bernard, B.; Foster, N.L.; Benson, D.F.; Goldfarb, L.G.; Brown, P. Familial Creutzfeldt-Jakob disease with a five-repeat octapeptide insert mutation. Neurology 1996, 47, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, L.G.; Brown, P.; McCombie, W.R.; Goldgaber, D.; Swergold, G.D.; Wills, P.R.; Cervenakova, L.; Baron, H.; Gibbs Jr, C.; Gajdusek, D.C. Transmissible familial Creutzfeldt-Jakob disease associated with five, seven, and eight extra octapeptide coding repeats in the PRNP gene. Proceedings of the National Academy of Sciences 1991, 88, 10926–10930. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.-H.; Huang, M.-Y.; Lee, Y.-R.; Lin, Y.-K.; Chen, H.-R.; Lee, C.-I. The Effect of Octapeptide Repeats on Prion Folding and Misfolding. Int. J. Mol. Sci. 2021, 22, 1800. [Google Scholar] [CrossRef] [PubMed]
- Areškevičiūtė, A.; Høgh, P.; Bartoletti-Stella, A.; Melchior, L.C.; Nielsen, P.R.; Parchi, P.; Capellari, S.; Broholm, H.; Scheie, D.; Lund, E.L. A Novel Eight Octapeptide Repeat Insertion in PRNP Causing Prion Disease in a Danish Family. J. Neuropathol. Exp. Neurol. 2019, 78, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Ziaunys, M.; Sneideris, T.; Smirnovas, V. Formation of distinct prion protein amyloid fibrils under identical experimental conditions. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Ziaunys, M.; Sakalauskas, A.; Mikalauskaite, K.; Snieckute, R.; Smirnovas, V. Temperature-Dependent Structural Variability of Prion Protein Amyloid Fibrils. Int. J. Mol. Sci. 2021, 22, 5075. [Google Scholar] [CrossRef]
- Honda, R. Amyloid-β Peptide Induces Prion Protein Amyloid Formation: Evidence for Its Widespread Amyloidogenic Effect. Angew. Chem. Int. Ed. 2018, 57, 6086–6089. [Google Scholar] [CrossRef]
- Artikis, E.; Roy, A.; Verli, H.; Cordeiro, Y.; Caughey, B. Accommodation of In-Register N-Linked Glycans on Prion Protein Amyloid Cores. ACS Chem. Neurosci. 2020, 11, 4092–4097. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.H.; Yokotsuka, M.; Tobiume, M.; Sato, Y.; Hasegawa, H.; Nagao, T.; Gouras, G.K. Accumulation of cellular prion protein within β-amyloid oligomer plaques in aged human brains. Brain Pathol. 2021, 31, e12941. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Sakaguchi, S. N-Terminal Regions of Prion Protein: Functions and Roles in Prion Diseases. Int. J. Mol. Sci. 2020, 21, 6233. [Google Scholar] [CrossRef] [PubMed]
- Torrent, J.; Martin, D.; Noinville, S.; Yin, Y.; Doumic, M.; Moudjou, M.; Béringue, V.; Rezaei, H. Pressure Reveals Unique Conformational Features in Prion Protein Fibril Diversity. Sci. Rep. 2019, 9, 2802. [Google Scholar] [CrossRef] [PubMed]
- Purro, S.A.; Nicoll, A.J.; Collinge, J. Prion Protein as a Toxic Acceptor of Amyloid-β Oligomers. Biol. Psychiatry 2017, 83, 358–368. [Google Scholar] [CrossRef]
- Stöhr, J.; Weinmann, N.; Wille, H.; Kaimann, T.; Nagel-Steger, L.; Birkmann, E.; Panza, G.; Prusiner, S.B.; Eigen, M.; Riesner, D. Mechanisms of prion protein assembly into amyloid. Proceedings of the National Academy of Sciences 2008, 105, 2409–2414. [Google Scholar] [CrossRef]
- Wang, L.Q.; Zhao, K.; Yuan, H.Y.; Wang, Q.; Guan, Z.; Tao, J.; Li, X.N.; Sun, Y.; Yi, C.W.; Chen, J.; et al. Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nat. Struct. Mol. Biol. 2020, 27, 598–602. [Google Scholar] [CrossRef]
- Riek, R.; Eisenberg, D.S. The activities of amyloids from a structural perspective. Nature 2016, 539, 227–235. [Google Scholar] [CrossRef]
- Yamaguchi, K.-I.; Kuwata, K. Formation and properties of amyloid fibrils of prion protein. Biophys. Rev. 2017, 10, 517–525. [Google Scholar] [CrossRef]
- Soto, C.; Satani, N. The intricate mechanisms of neurodegeneration in prion diseases. Trends Mol. Med. 2011, 17, 14–24. [Google Scholar] [CrossRef]
- Matos, C.O.; Passos, Y.M.; do Amaral, M.J.; Macedo, B.; Tempone, M.H.; Bezerra, O.C.L.; Moraes, M.O.; Almeida, M.S.; Weber, G.; Missailidis, S.; et al. Liquid-liquid phase separation and fibrillation of the prion protein modulated by a high-affinity DNA aptamer. FASEB J. 2020, 34, 365–385. [Google Scholar] [CrossRef]
- Ciuperca, I.S.; Hingant, E.; Palade, L.I.; Pujo-Menjouet, L. Fragmentation and monomer lengthening of rod-like polymers, a relevant model for prion proliferation. arXiv preprint arXiv:1112.4342 2011. arXiv:1112.4342 2011.
- Ferreira, S.; Raimundo, A.F.; Menezes, R.; Martins, I.C. Islet amyloid polypeptide & amyloid beta peptide roles in Alzheimer’s disease: Two triggers, one disease. Neural Regeneration Research 2021, 16, 1127. [Google Scholar] [PubMed]
- Novials, A.; Rojas, I.; Casamitjana, R.; Usac, E.; Gomis, R. A novel mutation in islet amyloid polypeptide (IAPP) gene promoter is associated with Type II diabetes mellitus. Diabetologia 2001, 44, 1064–1065. [Google Scholar] [PubMed]
- O'Brien, T.D.; Butler, P.C.; Westermark, P.; Johnson, K.H. Islet Amyloid Polypeptide: A Review of Its Biology and Potential Roles in the Pathogenesis of Diabetes Mellitus. Veter- Pathol. 1993, 30, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, A.; Razzaboni, B.; Weir, G.C.; Yankner, B.A. Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature 1994, 368, 756–760. [Google Scholar] [CrossRef]
- Howard Jr, C.F. Insular amyloidosis and diabetes mellitus in Macaca nigra. Diabetes 1978, 27, 357–364. [Google Scholar] [CrossRef]
- Betsholtz, C.; Christmansson, L.; Engström, U.; Rorsman, F.; Svensson, V.; Johnson, K.H.; Westermark, P.; Mietlicki-Baase, E.G.; Serrano, A.L.; Lomont, J.P.; et al. Sequence divergence in a specific region of islet amyloid polypeptide (IAPP) explains differences in islet amyloid formation between species. FEBS Lett. 1989, 251, 261–264. [Google Scholar] [CrossRef]
- Mirzabekov, T.A.; Lin, M.-C.; Kagan, B.L. Pore Formation by the Cytotoxic Islet Amyloid Peptide Amylin. J. Biol. Chem. 1996, 271, 1988–1992. [Google Scholar] [CrossRef]
- Raimundo, A.F.; Ferreira, S.; Martins, I.C.; Menezes, R. Islet Amyloid Polypeptide: A Partner in Crime With Aβ in the Pathology of Alzheimer's Disease. Frontiers in Molecular Neuroscience 2020, 13, 35. [Google Scholar] [CrossRef]
- Mucibabic, M.; Steneberg, P.; Lidh, E.; Straseviciene, J.; Ziolkowska, A.; Dahl, U.; Lindahl, E.; Edlund, H. α-Synuclein promotes IAPP fibril formation in vitro and β-cell amyloid formation in vivo in mice. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morales-Scheihing, D.; Salvadores, N.; Moreno-Gonzalez, I.; Gonzalez, C.; Taylor-Presse, K.; Mendez, N.; Shahnawaz, M.; Gaber, A.O.; Sabek, O.M.; et al. Induction of IAPP amyloid deposition and associated diabetic abnormalities by a prion-like mechanism. J. Exp. Med. 2017, 214, 2591–2610. [Google Scholar] [CrossRef] [PubMed]
- Kakinen, A.; Xing, Y.; Arachchi, N.H.; Javed, I.; Feng, L.; Faridi, A.; Douek, A.M.; Sun, Y.; Kaslin, J.; Davis, T.P.; et al. Single-Molecular Heteroamyloidosis of Human Islet Amyloid Polypeptide. Nano Lett. 2019, 19, 6535–6546. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P. Quantitative Studies of Amyloid in the Islets of Langerhans. Upsala J. Med. Sci. 1972, 77, 91–94. [Google Scholar] [CrossRef]
- Makin, O.S.; Serpell, L.C. Structures for amyloid fibrils. The FEBS journal 2005, 272, 5950–5961. [Google Scholar] [CrossRef] [PubMed]
- Goldsbury, C.; Kistler, J.; Aebi, U.; Arvinte, T.; Cooper, G.J. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J. Mol. Biol. 1999, 285, 33–39. [Google Scholar] [CrossRef]
- Goldsbury, C.S.; Cooper, G.J.; Goldie, K.N.; Müller, S.A.; Saafi, E.L.; Gruijters, W.; Misur, M.P.; Engel, A.; Aebi, U.; Kistler, J. Polymorphic Fibrillar Assembly of Human Amylin. J. Struct. Biol. 1997, 119, 17–27. [Google Scholar] [CrossRef]
- Röder, C.; Kupreichyk, T.; Gremer, L.; Schäfer, L.U.; Pothula, K.R.; Ravelli, R.B.G.; Willbold, D.; Hoyer, W.; Schröder, G.F. Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-β fibrils. Nat. Struct. Mol. Biol. 2020, 27, 660–667. [Google Scholar] [CrossRef]
- Asthana, S.; Mallick, B.; Alexandrescu, A.T.; Jha, S. IAPP in type II diabetes: Basic research on structure, molecular interactions, and disease mechanisms suggests potential intervention strategies. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1765–1782. [Google Scholar] [CrossRef]
- Tomita, T. Apoptosis in pancreatic β-islet cells in Type 2 diabetes. Bosn. J. Basic Med Sci. 2016, 16, 162–179. [Google Scholar] [CrossRef]
- Gomes, A.S.; Ramos, H.; Inga, A.; Sousa, E.; Saraiva, L. Structural and Drug Targeting Insights on Mutant p53. Cancers 2021, 13, 3344. [Google Scholar] [CrossRef]
- Billant, O.; Friocourt, G.; Roux, P.; Voisset, C. , p53, A Victim of the Prion Fashion. Cancers 2021, 13, 269. 124. Bom, A.P.A.; Rangel, L.P.; Costa, D.C.; de Oliveira, G.A.; Sanches, D.; Braga, C.A.; Gava, L.M.; Ramos, C.H.; Cepeda, A.O.; Stumbo, A.C. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. Journal of Biological Chemistry 2012, 287, 28152–28162. [Google Scholar]
- Ghosh, S.; Salot, S.; Sengupta, S.; Navalkar, A.; Ghosh, D.; Jacob, R.; Das, S.; Kumar, R.; Jha, N.N.; Sahay, S.; et al. p53 amyloid formation leading to its loss of function: implications in cancer pathogenesis. Cell Death Differ. 2017, 24, 1784–1798. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, G.A.P.; Petronilho, E.C.; Pedrote, M.M.; Marques, M.A.; Vieira, T.C.R.G.; Cino, E.A.; Silva, J.L. The Status of p53 Oligomeric and Aggregation States in Cancer. Biomolecules 2020, 10, 548. [Google Scholar] [CrossRef] [PubMed]
- Navalkar, A.; Pandey, S.; Singh, N.; Patel, K.; Datta, D.; Mohanty, B.; Jadhav, S.; Chaudhari, P.; Maji, S.K. Direct evidence of cellular transformation by prion-like p53 amyloid infection. J. Cell Sci. 2021, 134. [Google Scholar] [CrossRef] [PubMed]
- Maritschnegg, E.; Heinzl, N.; Wilson, S.; Deycmar, S.; Niebuhr, M.; Klameth, L.; Holzer, B.M.; Koziel, K.; Concin, N.; Zeillinger, R. Polymer-Ligand-Based ELISA for Robust, High-Throughput, Quantitative Detection of p53 Aggregates. Anal. Chem. 2018, 90, 13273–13279. [Google Scholar] [CrossRef] [PubMed]
- Farmer, K.M.; Ghag, G.; Puangmalai, N.; Montalbano, M.; Bhatt, N.; Kayed, R. P53 aggregation, interactions with tau, and impaired DNA damage response in Alzheimer’s disease. Acta Neuropathologica Communications 2020, 8, 1–21. [Google Scholar] [CrossRef]
- Galea, C.; Bowman, P.; Kriwacki, R.W. Disruption of an intermonomer salt bridge in the p53 tetramerization domain results in an increased propensity to form amyloid fibrils. Protein Sci. 2005, 14, 2993–3003. [Google Scholar] [CrossRef]
- Rajan, R.; Ahmed, S.; Sharma, N.; Kumar, N.; Debas, A.; Matsumura, K. Review of the current state of protein aggregation inhibition from a materials chemistry perspective: special focus on polymeric materials. Mater. Adv. 2021, 2, 1139–1176. [Google Scholar] [CrossRef]
- Zaguri, D.; Shaham-Niv, S.; Chakraborty, P.; Arnon, Z.A.; Makam, P.; Bera, S.; Rencus-Lazar, S.; Stoddart, P.R.; Gazit, E.; Reynolds, N.P. Nanomechanical Properties and Phase Behavior of Phenylalanine Amyloid Ribbon Assemblies and Amorphous Self-Healing Hydrogels. ACS Appl. Mater. Interfaces 2020, 12, 21992–22001. [Google Scholar] [CrossRef]
- Aliu, E.; Kanungo, S.; Arnold, G.L. Amino acid disorders. Annals of translational medicine 2018, 6. [Google Scholar] [CrossRef]
- Ménard-Moyon, C.; Venkatesh, V.; Krishna, K.V.; Bonachera, F.; Verma, S.; Bianco, A. Self-Assembly of Tyrosine into Controlled Supramolecular Nanostructures. Chem. – A Eur. J. 2015, 21, 11681–11686. [Google Scholar] [CrossRef] [PubMed]
- Zaguri, D.; Kreiser, T.; Shaham-Niv, S.; Gazit, E. Antibodies towards Tyrosine Amyloid-Like Fibrils Allow Toxicity Modulation and Cellular Imaging of the Assemblies. Molecules 2018, 23, 1273. [Google Scholar] [CrossRef] [PubMed]
- Anand, B.G.; Prajapati, K.P.; Shekhawat, D.S.; Kar, K. Tyrosine-Generated Nanostructures Initiate Amyloid Cross-Seeding in Proteins Leading to a Lethal Aggregation Trap. Biochemistry 2018, 57, 5202–5209. [Google Scholar] [CrossRef] [PubMed]
- Shaham-Niv, S.; Rehak, P.; Vuković, L.; Adler-Abramovich, L.; Král, P.; Gazit, E. Formation of Apoptosis-Inducing Amyloid Fibrils by Tryptophan. Isr. J. Chem. 2016, 57, 729–737. [Google Scholar] [CrossRef]
- Shaham-Niv, S.; Rehak, P.; Zaguri, D.; Kolusheva, S.; Král, P.; Gazit, E. Metabolite amyloid-like fibrils interact with model membranes. Chem. Commun. 2018, 54, 4561–4564. [Google Scholar] [CrossRef]
- Gour, N.; Jaiswal, A.; Patel, M.; Revi, N.; Rengan, A. Aggregation characteristics of non-aromatic polar amino acids and its association to amyloids. ChemRxiv. Cambridge: Cambridge Open Engage; 2023; This content is a preprint and has not been peer-reviewed.
- Singh, P.; Pandey, S.K.; Grover, A.; Sharma, R.K.; Wangoo, N. Understanding the self-ordering of amino acids into supramolecular architectures: co-assembly-based modulation of phenylalanine nanofibrils. Mater. Chem. Front. 2020, 5, 1971–1981. [Google Scholar] [CrossRef]
- Bera, S.; Mondal, S.; Rencus-Lazar, S.; Gazit, E. Organization of Amino Acids into Layered Supramolecular Secondary Structures. Accounts Chem. Res. 2018, 51, 2187–2197. [Google Scholar] [CrossRef]
- Yazdi, D.S.; Bar-Yosef, D.L.; Adsi, H.; Kreiser, T.; Sigal, S.; Bera, S.; Zaguri, D.; Shaham-Niv, S.; Oluwatoba, D.S.; Levy, D.; et al. Homocysteine fibrillar assemblies display cross-talk with Alzheimer’s disease β-amyloid polypeptide. Proc. Natl. Acad. Sci. 2021, 118. [Google Scholar] [CrossRef]
- Zaguri, D.; Shaham-Niv, S.; Naaman, E.; Mimouni, M.; Magen, D.; Pollack, S.; Kreiser, T.; Leibu, R.; Rencus-Lazar, S.; Adler-Abramovich, L.; et al. Induction of retinopathy by fibrillar oxalate assemblies. Commun. Chem. 2020, 3, 1–9. [Google Scholar] [CrossRef]
- Tavassoly, O.; Sade, D.; Bera, S.; Shaham-Niv, S.; Vocadlo, D.J.; Gazit, E. Quinolinic Acid Amyloid-like Fibrillar Assemblies Seed α-Synuclein Aggregation. J. Mol. Biol. 2018, 430, 3847–3862. [Google Scholar] [CrossRef]
- Laor, D.; Sade, D.; Shaham-Niv, S.; Zaguri, D.; Gartner, M.; Basavalingappa, V.; Raveh, A.; Pichinuk, E.; Engel, H.; Iwasaki, K.; et al. Fibril formation and therapeutic targeting of amyloid-like structures in a yeast model of adenine accumulation. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Jacoby, G.; Bar-Yosef, D.L.; Beck, R.; Gazit, E.; Segal, D. Glucosylceramide Associated with Gaucher Disease Forms Amyloid-like Twisted Ribbon Fibrils That Induce α-Synuclein Aggregation. ACS Nano 2021, 15, 11854–11868. [Google Scholar] [CrossRef] [PubMed]
- Anand, B.G.; Prajapati, K.P.; Dubey, K.; Ahamad, N.; Shekhawat, D.S.; Rath, P.C.; Joseph, G.K.; Kar, K. Self-Assembly of Artificial Sweetener Aspartame Yields Amyloid-like Cytotoxic Nanostructures. ACS Nano 2019, 13, 6033–6049. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, K.P.; Anand, B.G.; Ansari, M.; Temgire, M.; Tiku, A.B.; Kar, K. Amyloid-mimicking toxic nanofibers generated via self-assembly of dopamine. Nanoscale 2022, 14, 8649–8662. [Google Scholar] [CrossRef] [PubMed]
- Gour N, Patel M, Gupta S, Wilson B. Metabolite assemblies of urea cycle and uric acid pathway. ChemRxiv. Cambridge: Cambridge Open Engage; 2022; This content is a preprint and has not been peer-reviewed.
| Amino acids | Plasma concentration 5range (µmol/L) |
|---|---|
| Alanine | 564-644 |
| Arginine | 44-51 |
| Asparagine | 92-98 |
| Aspartate | 64-69 |
| Glutamate | 261-296 |
| Glutamine | 506-547 |
| Glycine | 390-452 |
| Histidine | 114-122 |
| Isoleucine | 96-102 |
| Leucine | 191-210 |
| Lysine | 210-242 |
| Methionine | 27-31 |
| Proline | 244-265 |
| Phenylalanine | 95-101 |
| Serine | 191-201 |
| Tyrosine | 73-83 |
| Threonine | 164-168 |
| Tryptophan | 65-72 |
| Valine | 217-233 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





