Submitted:
17 May 2023
Posted:
18 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
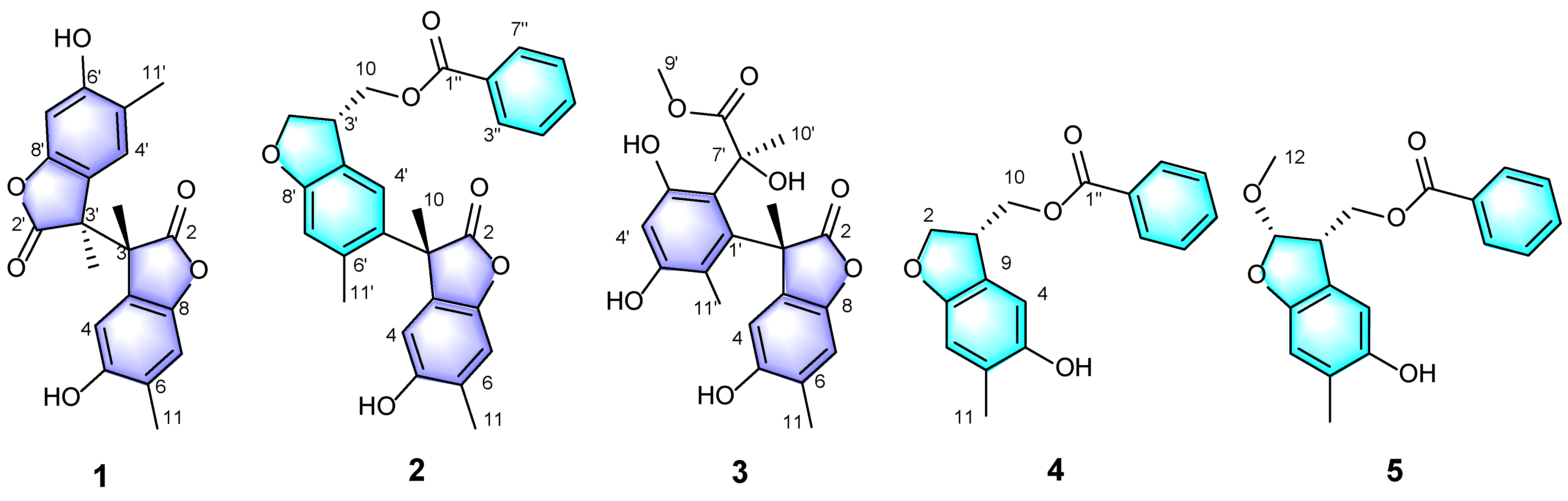
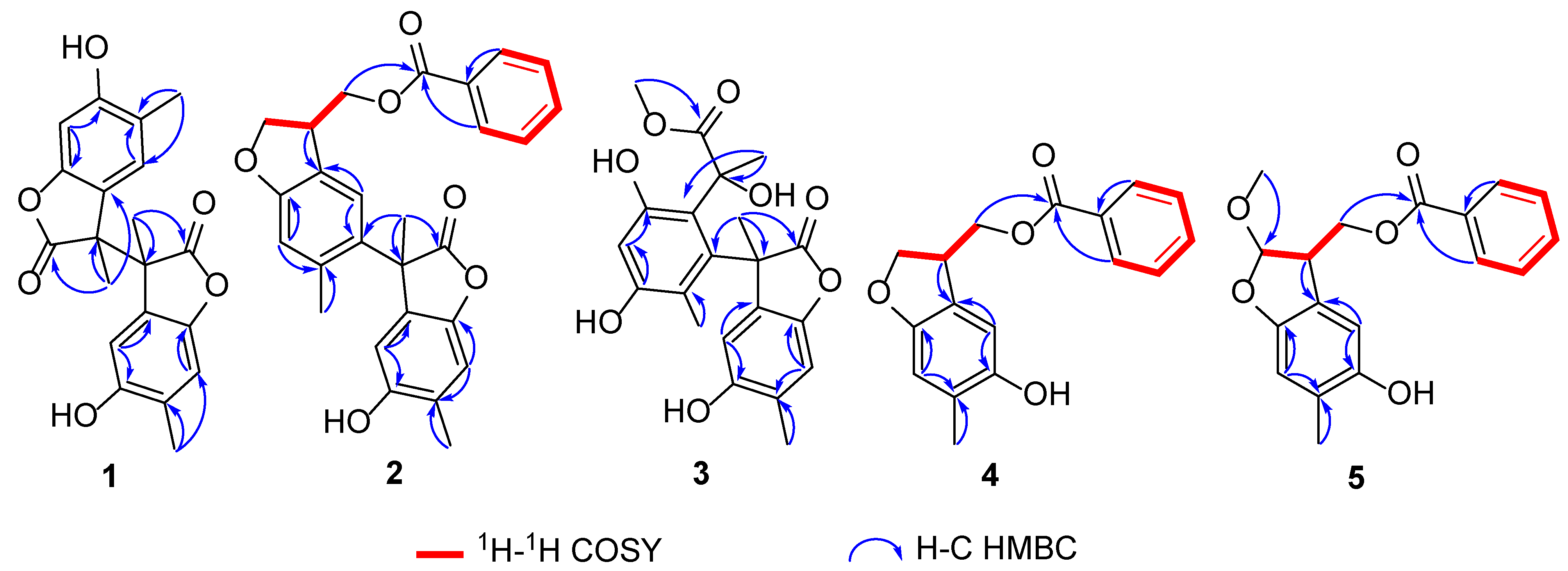
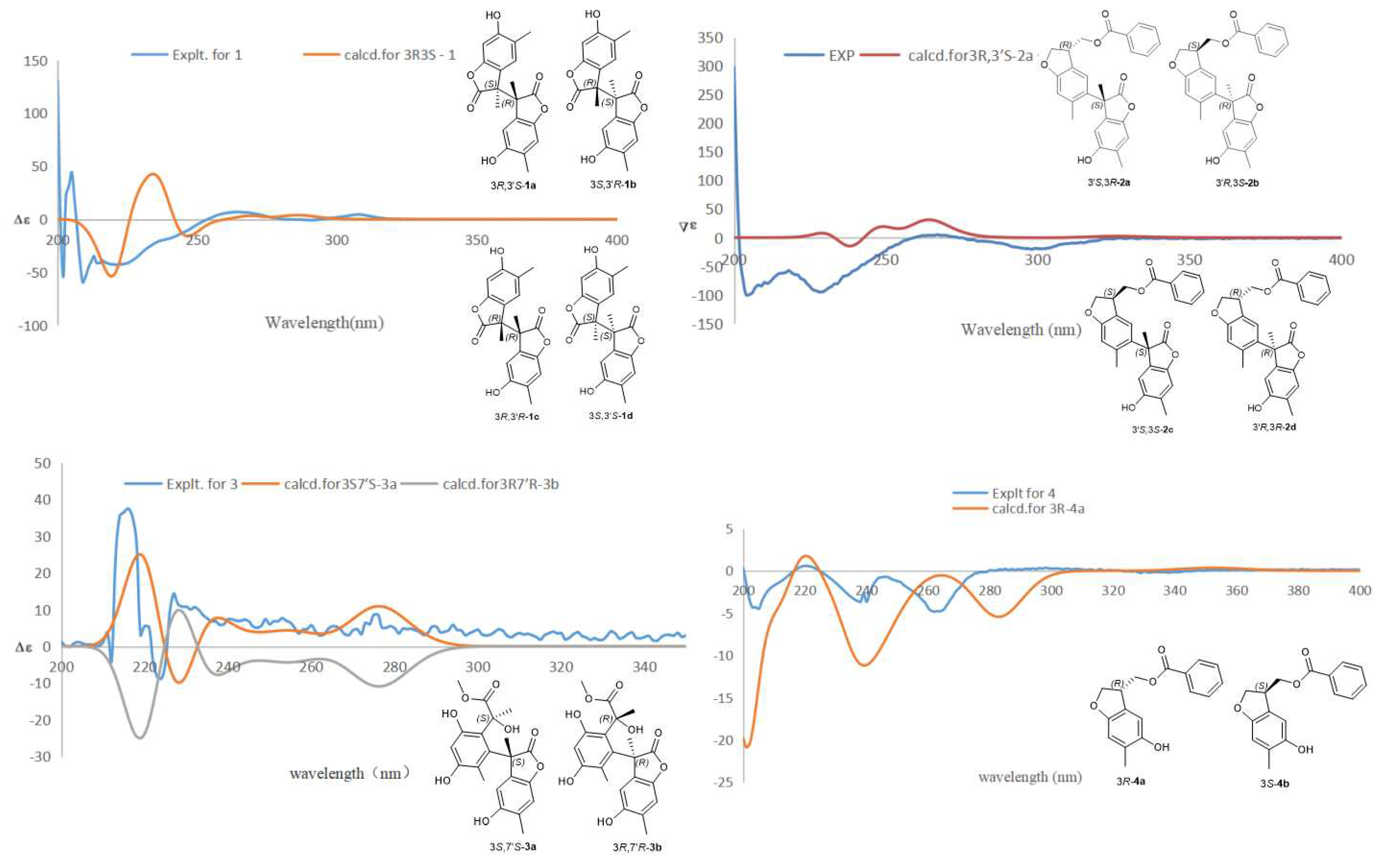
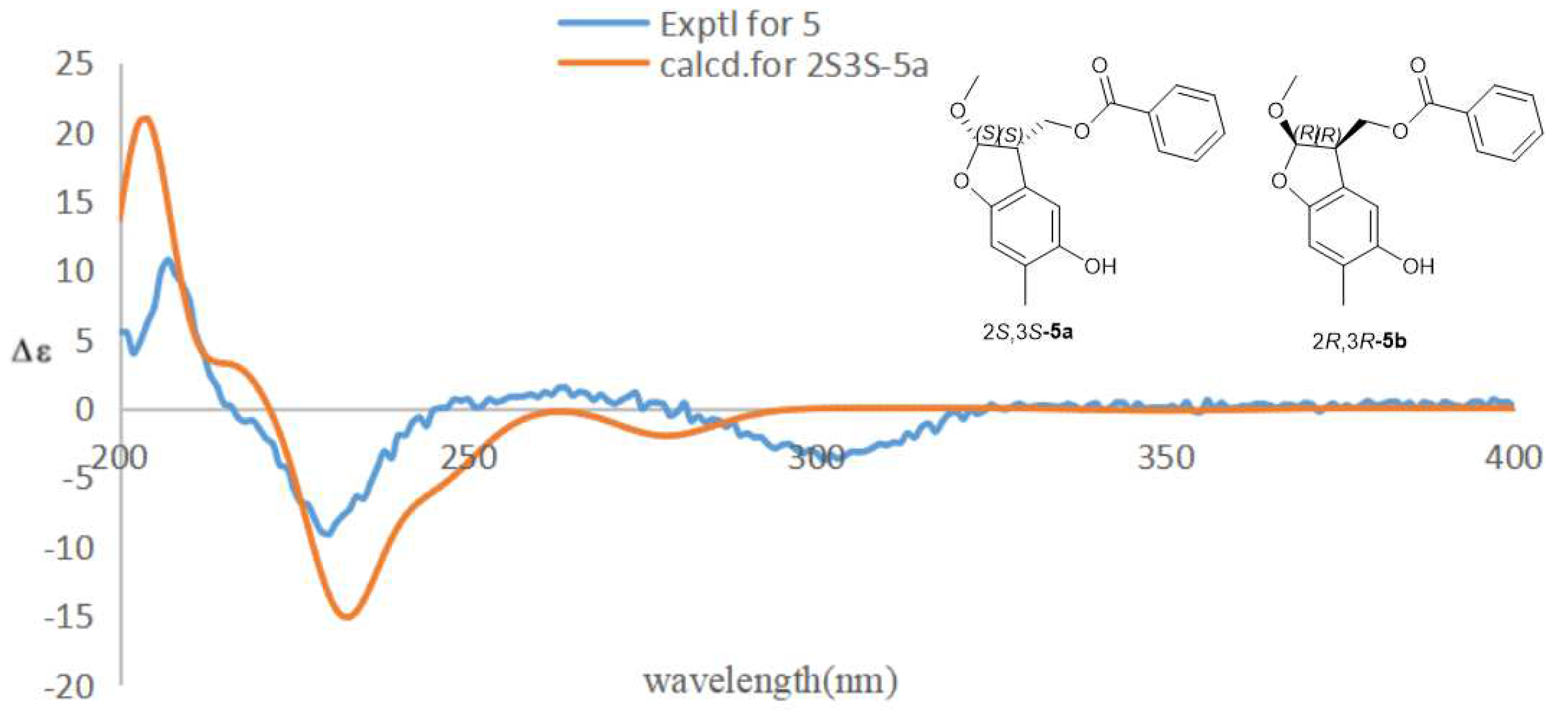
2.1. Structural Elucidation of Compounds 1–5
2.2. Bioactivity Analysis
3. Experimental
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.3.1. Paeobenzofuranone A (1)
3.3.2. Paeobenzofuranone B (2)
3.3.3. Paeobenzofuranone C (3)
3.3.4. Paeobenzofuranone D (4)
3.3.5. Paeobenzofuranone E (5)
3.4. Cytotoxicity Assay
3.5. Anti-Inflammatory Activity Assays
3.6. ECD Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ding, L.; Zhao, F.; Chen, L.; Jiang, Z.; Liu, Y.; Li, Z.; Qiu, F.; Yao, X. New monoterpene glycosides from Paeonia suffruticosa Andrews and their inhibition on NO production in LPS-induced RAW 264.7 cells. 7 cells. Bioorg. Med. Chem. Lett. 2012, 22(23), 7243–7247. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Tang, S.W.; Lam, S.H.; Wang, C.C.; Liu, Y.H.; Lin, H.Y.; Lee, S.S.; Lin, J.Y. Aqueous extract of Paeonia suffruticosa inhibits migration and metastasis of renal cell carcinoma cells via suppressing VEGFR-3 pathway. Evid. Based Compl. Alt. 2012, 409823. [Google Scholar] [CrossRef]
- Qiu, H.; Zhang, L.; Zhu, M.; Zhang, M.; Chen, J.; Feng, L.; Jia, X.; Jacob, J.A. Capture of anti-coagulant active ingredients from Moutan Cortex by platelet immobilized chromatography and evaluation of anticoagulant activity in rats. Biomed. Pharmacother. 2017, 95, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Pei, L.; Jie, S.; Chunnian, H.; Pei, X. Genus Paeonia: A comprehensive review on traditional uses, phytochemistry, pharmacological activities, clinical application, and toxicology. J. Ethnopharmacol. 2021, 269:113708. [CrossRef]
- Mencherini, T.; Picerno, P.; Festa, M.; Russo, P.; Capasso, A.; Aquino, R. Triterpenoid constituents from the roots of Paeonia rockii ssp. rockii. J. Nat. Prod., 2011, 74, 2116–2121. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ohno, O.; Suenaga, K.; Miyamoto, K. Apoptosis-inducing activity and antiproliferative effect of Paeoniflorigenone from moutan cortex. Biosci. Biotechnol. Biochem. 2017, 81, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, Y.; Gao, J.; Lu, Z.; Yin, W.; Deng, R. Resveratrol trimers from seed cake of Paeonia rockii. Molecules. 2014, 19, 19549–19556. [Google Scholar] [CrossRef] [PubMed]
- Furuya, R.; Hu, H.; Zhang, Z.; Shigemori, H. Suffruyabiosides A and B, two new monoterpene diglycosides from Moutan Cortex. Molecules. 2012, 17, 4915–4923. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.K.; Ma, Y.B.; Wu, D.G.; Lu, Y.; Shen, Z.Q.; Zheng, Q.T.; Chen, Z.H. Paeonilide, a novel anti-PAF active monoterpenoid derived metabolite from Paeonia delavayi. Biosci. Biotech. Biochem. 2000, 64, 1511–1514. [Google Scholar] [CrossRef]
- Yang, X.Y.; Feng, T.; Li, Z.H.; Sheng, Y.; Yin, X.; Leng, Y.; Liu, J.K. Conosilane A, an unprecedented sesquiterpene from the Cultures of Basidiomycete Conocybe siliginea. Org. Lett. 2012, 14, 5382–5384. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gao, Y.; Zhang, L.; Liu, J.K. Bi-linderone, a highly modified methyl-linderone dimer from Lindera aggregata with activity toward improvement of insulin sensitivity in vitro. Org. Lett. 2010, 12, 2354–2357. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Hsieh, K.L.; Liu, J.K. Guajadial: an unusual meroterpenoid from guava leaves Psidium guajava. Org. Lett. 2007, 9, 5135–5138. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Su, J.; Ding, Z.H.; Zheng, Y.T.; Li, Y.; Leng, Y.; Liu, J.K. Chemical constituents and their bioactivities of Tongling White Ginger (Zingiber officinale). J. Agric. Food Chem. 2011, 59, 11690–11695. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.G.; Cheng, C.Q.; Liu, J.K. X-Ray Crystal Structure of Angulatusine A, a new sesquiterpene alkaloid from Celastrus Angulatus. J. Nat. Prod. 1992, 55, 982–985. [Google Scholar] [CrossRef]
- Ha, D.T.; Ngoc, T.M.; Lee, I.S.; Lee, Y. M.; Kim, J.S.; Jung, H.J. Inhibitors of aldose reductase and formation of advanced glycation end-products in moutan cortex (Paeonia suffruticosa). J. Nat. Prod. 2009, 72, 1465–1470. [Google Scholar] [CrossRef]
- Yu, H. L.; Long, Q.; Yi, W.F.; Yang, B. J.; Ding, X.; Hao, X. J. Two new C21 steroidal glycosides from the roots of Cynanchum paniculatum. Natural Products and Bioprospecting. 2019, 04, 26. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Anh, H. L. T.; Cuc, N. T.; Tai, B. H.; Yen, P. H.; Nhiem, N. X.; ThaoD. T.; Nam, N. H.; Minh, C. V.; Kiem, P. V.; Kim, Y. H. Synthesis of chromonylthiazolidines and their cytotoxicity tohuman cancer cell Lines. Molecules. 2015, 20, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Snene, A.; Mokni, R.E.; Jmii, H.; Jlassi, I.; Jadane, H.; Falconieri, D.; Piras, A.; Dhaouadi, H.; Porcedda, S; Hammami, S. In vitro antimicrobial, antioxidant and antiviral activities of the essential oil and various extracts of wild (Daucus virgatus (Poir.) Maire) from Tunisia. Ind Crops Prod. 2017, 109, 109–115. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumloeffel, A.; Hemberger, Y.; Bringmann, G.; SpecDis: quantifying the comparison of calculated and experimental electronic circular dichroism spectra, Chirality. 2013, 25, 243-249. [CrossRef]
| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 2 | 4.46, m | 5.48, t (1.8) | |||
| 3 | 3.81, t (7.4) | 3.80, t (7.5) | |||
| 4 | 7.18, s | 6.61, s | 6.96, s | 6.74, s | 6.72, s |
| 7 | 6.61, s | 6.52, s | 6.33, s | 4.37, dd (18.0, 9.1) 4.44, dd (10.7, 5.9) | 4.46, dd (11.1, 5.5) 4.33, dd (11.1, 7.8) |
| 10 | 1.67, s, 3H | 1.75, s, 3H | 1.72, s, 3H | 4.43, m | 4.48, m |
| 11 | 2.15, s, 3H | 2.13, s, 3H | 2.02, s, 3H | 2.14, s, 3H | 2.15, s, 3H |
| 12 | 3.48, s | ||||
| 2′ | 4.43, m; 4.06, m | 8.00, d (1.5) | 7.97, d (1.5) | ||
| 3′ | 3.81, t (7.5) | 7.44, m | 7.45, m | ||
| 4′ | 7.18, s | 6.63, s | 6.88, s | 7.58, m | 7.61, m |
| 5′ | 6.61, s | 6.62, s | 7.49, m | 7.48, m | |
| 6′ | 1.67, s, 3H | 8.01, d (1.3) | 7.99, d (1.3) | ||
| 7′ | 2.15, s, 3H | 6.85, s | |||
| 9′ | 3.73, s | ||||
| 10′ | 4.60, dd (18.0, 9.1); 4.44, dd (10.8, 5.9) | 1.63, s, 3H | |||
| 11′ | 2.05, s, 3H | 1.77, s, 3H | |||
| 3′′ | 8.01, d (1.2) | ||||
| 4′′ | 7.48, t (7.8) | ||||
| 5′′ | 7.56, m | ||||
| 6′′ | 7.48, t (7.8) | ||||
| 7′′ | 8.00, d (1.4) |
| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 2 | 179.4, C | 179.3, C | 182.7, C | 73.5, CH2 | 111.2, CH |
| 3 | 52.6, C | 52.6, C | 50.2, C | 42.0, CH | 50.5, CH |
| 4 | 128.1, CH | 112.2, CH | 115.6, CH | 110.8, CH | 112.6, CH |
| 5 | 113.3, C | 149.3, C | 149.2, C | 148.9, C | 150.9, C |
| 6 | 153.7, C | 126.3, C | 125.0, C | 128.5, CH | 124.4, CH |
| 7 | 110.3, CH | 112.4, CH | 118.9, CH | 110.6, CH | 112.3, CH |
| 8 | 146.2, C | 125.9, C | 144.3, C | 153.4, C | 153.2, C |
| 9 | 127.4, C | 127.4, C | 126.4, C | 125.6, C | 129.8, C |
| 10 | 18.7, CH3 | 18.6, CH3 | 22.3, CH3 | 66.4, CH2 | 66.3, CH2 |
| 11 | 16.6, CH3 | 16.6, CH3 | 15.9, CH3 | 15.6, CH3 | 17.0, CH3 |
| 12 | 56.2, OCH3 | ||||
| 1′ | 133.7, C | ||||
| 2′ | 179.4, C | 73.1, CH2 | 153.2, C | ||
| 3′ | 52.6, C | 43.5, CH | 122.0, C | ||
| 4′ | 128.1, CH | 111.7, CH | 115.9, CH | ||
| 5′ | 113.3, C | 126.4, C | 144.6, C | ||
| 6′ | 153.7, C | 131.4, CH | 125.7, C | ||
| 7′ | 110.3, CH | 110.3, CH | 75.1, C | ||
| 8′ | 146.2, C | 154.9, C | 176.8, C | ||
| 9′ | 127.4, C | 124.4, C | 53.2, OCH3 | ||
| 10′ | 18.7, CH3 | 66.9, CH2 | 26.0, CH3 | ||
| 11′ | 16.6, CH3 | 16.8, CH3 | 10.4, CH3 | ||
| 1′′ | 168.1, C | 166.5, C | 167.9, C | ||
| 2′′ | 130.7, C | 129.7, C | 131.2, C | ||
| 3′′ | 129.7, CH | 129.2, CH | 130.4, CH | ||
| 4′′ | 128.1, CH | 128.2, CH | 129.5, CH | ||
| 5′′ | 134.5, CH | 132.9, CH | 130.7, CH | ||
| 6′′ | 129.7, CH | 128.2, CH | 129.5, CH | ||
| 7′′ | 128.1, CH | 129.1, CH | 130.4, CH |
| Compound | Concentration (μM) | Inhibition Activity (100%) |
|---|---|---|
| L-NMMAa | 50 | 52.0 ± 1.96 |
| 1 | 50 | 43.9 ± 2.07 |
| 2 | 50 | 44.6 ± 0.52 |
| 3 | 50 | 13.0 ± 1.59 |
| 4 | 50 | 33.7 ± 2.24 |
| 5 | 50 | 30.9 ± 1.56 |
| Compound | HL-60 | MDA-MB-231 | SW480 |
|---|---|---|---|
| 2 | 6.8 ± 0.11 | 20.9 ± 0.46 | 12.6 ± 0.73 |
| 4 | 19.1 ± 0.32 | – | 8.9 ± 0.40 |
| 5 | 11.1 ± 1.61 | – | 10.7 ± 0.43 |
| DDPa | 23.5 ± 0.77 | 16.9 ± 1.19 | 25.1 ± 1.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





