Submitted:
13 May 2023
Posted:
17 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Alzheimer’s disease (AD)
3. Amyloid Precursor Protein
| Mutation | Pathogenicity | Type of Mutation | Biological Effect | Citation |
| c.-488C>A (rs532314089) |
Alzheimer's Disease | Substitution | Predicted to disrupt binding of transcription factor EGR1. PHRED-scaled CADD = 0.26. Negative regulator in multiple cell types including PC12 neuronal-like rat chromaffin cells, SK-N-SH neuroblastoma cells, C6 glial cells and U373 astroctyoma cells among others |
[30] [31] [32] |
| c.24+38G>A (rs373985746) |
Alzheimer's Disease | Substitution | Predicted benign in silico (PHRED-scaled CADD =10). | [32] |
| c.24+288G>A (rs192348494) |
Alzheimer's Disease | Substitution | Predicted benign in silico (PHRED-scaled CADD =12). | [32] |
| c.-23-377A>G (rs150375400) |
Alzheimer's Disease | Substitution | Predicted benign in silico (PHRED-scaled CADD =10). | [32] |
| A18T | Alzheimer's Disease, Cardiovascular Disease | Substitution | Predicted to disrupt signal peptide cleavage and affect ApoE secretion. PHRED-scaled CADD = 22. | [32,33] |
Presenilin1
| Mutation | Pathogenicity | Type of Mutation | Biological Effect | Citation |
| A673T(Icelandic) |
Alzheimer’s Disease - Protective | Substitution | This particular type is linked to limited buildup of amyloid and is believed to guard against amyloid-related issues. It results in a decrease of approximately 40 percent in the production of amyloidogenic Aβ peptides, and the Aβ that is produced has a reduced tendency to form clumps. | [17,18,19,20] |
| A673V | Not Classified | Substitution | According to the CERAD criteria, a clear diagnosis of AD was made, as evidenced by substantial Aβ and tau pathology deposits (Braak stage VI) along with cerebral amyloid angiopathy. The deposits found contained elevated levels of Aβ40 and were notably larger, with fewer preamyloid deposits. Perivascular localization was frequently observed. In laboratory studies, it was discovered that A673V caused a shift in β-secretase processing of APP toward the amyloidogenic pathway and amplified Aβ aggregation. | [21,22] |
| E693Q(Dutch) | Hereditary Cerebral Hemorrhage with Amyloidosis- Pathogenic | Substitution | There is a substantial accumulation of amyloid in the cerebral blood vessels, accompanied by hemorrhages and some diffuse plaques in the brain tissue. In laboratory experiments, it was observed that this condition speeds up Aβ aggregation in vitro, leading to greater fibril formation, and may also modify APP processing. | [24,25] |
| E693del (Osaka, E693∆, E693delta | Alzheimer’s Disease - Pathogenic | Deletion | This variant led to an increased oligomerization and nucleation of Aβ aggregates in vitro. Furthermore, it was found that there was no alteration in the Aβ42/Aβ40 ratio, but there was a decrease in both Aβ42 and Aβ40. This variant was also discovered to be more resistant to degradation by neprilysin and insulin-degrading enzyme. Additionally, this variant had a greater inhibitory effect on long-term potentiation (LTP) compared to wild-type Aβ, which suggests a potential negative impact on synaptic plasticity. | [26,27,28,29] |
| E693K(Italian) | Hereditary Cerebral Hemorrhage with Amyloidosis- Pathogenic | Substitution | The observed symptoms include small to large hematomas, subarachnoid bleeding, scars with hemosiderin deposits, small infarcts, and cortical calcifications. Aβ immunoreactivity was observed in vessel walls and neuropil, but there was an absence of neurofibrillary changes and neuritic plaques. Despite a reduction in the Aβ42/Aβ40 ratio and a decrease in Aβ42 levels, the mutant peptide was found to be toxic in cells and aggregates at a faster rate. | [23] |
| E693G (Arctic, E22G) | Alzheimer’s Disease - Pathogenic | Substitution | Several carriers displayed neuropathology that was indicative of AD. Plaques were observed to have a "targetoid" shape, containing heterogeneous truncated Aβ peptides in the center and surrounded by Aβ42. Cell-based assays revealed a reduction in the production of both Aβ40 and Aβ42. Additionally, there was a decrease in proteolytic degradation of Aβ by neprilysin, a type of enzyme that breaks down proteins. | [23,25] |
Presenilin2
| Mutation | Pathogenicity | Type of Mutation | Biological Effect | Citation |
| A79V | Alzheimer’s Disease - Pathogenic | Substitution | The observed neuropathology was in line with that of AD. It was observed that this variant led to an increase in the Aβ42/Aβ40 ratio and a decrease in the Aβ37/Aβ42 ratios in cells. | [39,40,41] |
| M84V | Alzheimer’s Disease - Pathogenic | Substitution | In two cases, the observed neuropathology was consistent with AD. Additionally, MRI scans revealed cortical and cerebellar atrophy in these two cases. In the third case, frontal and temporal lobe atrophy was observed. Cell studies revealed an increase in both Aβ42 and the Aβ42/Aβ40 ratio. | [42,43] |
| L85P | Alzheimer’s Disease - Pathogenic | Substitution | SPECT and PET scans showed bilateral hypoperfusion and hypometabolism in the occipital and temporal lobes. Cell studies revealed an increase in the Aβ42/Aβ40 ratio as well as increased Aβ42 levels in transfected cells. In vitro studies indicated a decrease in Aβ42 production and the complete absence of Aβ40 production. | [44,45] |
| L113_I114insT (int4del) |
Alzheimer’s Disease - Pathogenic | Substitution | The observed neuropathology was consistent with AD, and included neuron loss in the hippocampus and entorhinal cortex, the presence of neuritic plaques and neurofibrillary tangles in the hippocampus, and amyloid angiopathy which was particularly evident in the cerebellum. The identified mutation involved a deletion of a G in the splice donor site of intron 4, resulting in the production of three aberrant transcripts. Further investigations indicated an increase in both Aβ42 and the Aβ42/Aβ40 ratio, as well as a reduction in Aβ40 and Aβ38 production in patient brain membranes. | [46,47,48] |
| M139V | Alzheimer’s Disease - Pathogenic | Substitution | Decrease in the levels of Aβ40, Aβ38, and Aβ37, and an increase in the levels of Aβ42 and Aβ43. In iPSC-derived neurons, the levels of mutant protein were found to be variable, suggesting protein instability. | [49,50,51] |
Apolipoprotein E
ApoE and tau
ApoE and neuroinflammation
4. Important ApoE Mutations involved in AD onset
c.-488C>A
c.-24+38G>A
c.-24+288G>A
c.-23-377A>G
A18T
| Mutation | Pathogenicity | Type of Mutation | Biological Effect | Citation |
| K82fs | Tauopathy and Pick’s Disease | Deletion | The neuropathological findings were consistent with Pick's disease. A frameshift was identified to start at K82, and the mutant protein was found to be reduced in the frontal cortex and hippocampus. | [54] |
| c.*71C>A | Alzheimer’s Disease - Pathogenic | Substitution | In one case, an MRI scan revealed widening of the sulcus, fissure, and temporal horn, along with a decrease in hippocampal volume. Additionally, FDG-PET showed hypometabolism in the bilateral frontal, parietal, and temporal lobes. Among the five affected carriers, CSF analysis showed Aβ42, total tau, and phospho-tau levels consistent with AD. The study suggests a possible reduction in the binding of PSEN2 expression suppressor miR-183-5p, which may lead to an increased Aβ42/Aβ40 ratio. | [55,56] |
| M239V | Alzheimer’s Disease - Pathogenic | Substitution | The brain pathology showed diffuse cerebral atrophy, senile plaques, neurofibrillary tangles (Braak and Braak stage VI), ectopic neurons in subcortical white matter, and extracellular "ghost" neurofibrillary tangles. In cell-based assays, there was an increase in the Aβ42/Aβ40 ratio and an increase in Aβ42 levels. However, there was no change in the proteolytic products PSEN2-CTF and PSEN2-NTF. | [57,58] |
Microtubule-associated protein tau
5. Important MAPT Mutations involved in AD onset
MAPT IVS10+12 C>T
MAPT A152T
MAPT K257T
MAPT L266V
| Mutation | Pathogenicity | Type of Mutation | Biological Effect | Citation |
| IVS10+12 C>T | Familial Danish Dementia - Pathogenic | Substitution | The mutant protein leads to the formation of tau aggregates in both neurons and glia, and isolated tau filaments exhibit a twisted, ribbon-like morphology and consist of hyperphosphorylated 4-repeat (4R) tau isoforms. The mutation also causes a destabilization of a stem-loop structure that regulates the alternative splicing of exon 10, resulting in a higher frequency of inclusion of exon 10 and an increased proportion of 4R tau isoforms. | [83,84] |
| A152T | Alzheimer’s Disease - Risk | Substitution | The presence of tau pathology is a common feature, often accompanied by Lewy bodies, amyloid plaques, or TDP-43 pathology. The mutant tau has a decreased ability to bind to microtubules, leading to less efficient microtubule assembly and impaired microtubule stability. Additionally, it has an increased propensity to form tau oligomers and is more susceptible to proteolysis by caspases. | [85,86,87] |
| K257T | Tauopathy and Frontotemporal - Pathogenic |
Substitution | The patient exhibited frontotemporal atrophy with significant temporal lobe involvement. Tau-positive Pick bodies were found in the neocortex, hippocampus, and subcortical regions similar to those seen in sporadic Pick's disease. Some cell bodies showed diffuse hyperphosphorylated tau. In vitro analysis showed that recombinant tau protein with the K257T mutation had a decreased ability to promote microtubule assembly. | [88] |
| L266V | Frontotemporal - Pathogenic |
Substitution | The patient had severe atrophy of the frontal and temporal lobes, with extensive neuronal loss and gliosis. Tau-positive inclusions, including Pick bodies, and tau-positive argyrophilic astrocytes with stout filaments and round or irregular argyrophilic inclusions were also observed. In molecular studies, there were increased levels of exon 10+ tau mRNA and soluble four-repeat (4R) tau. The patient showed a decreased rate and extent of tau-induced microtubule assembly, as well as a specific increase in tau self-assembly for the 3R isoform. | [89,90] |
6. The evolving landscape of Alzheimer's disease treatments: exploring current and future perspectives
7. Parkinson’s disease (PD)
8. Perspectives of Treatment
9. Huntington’s disease (HD)
10. Treatment
11. Amyotrophic lateral sclerosis (ALS)
| Mutation | Pathogenicity | Type of Mutation | Biological Effect | Citation |
| A4V | ALS Pathogenic | Substitution | This mutation is responsible for a rapidly progressive dominant form of amyotrophic lateral sclerosis (ALS) that exclusively affects lower motor neurons, and it accounts for 50% of SOD1 mutations associated with familial ALS in North America. However, it is a rare mutation in Europe. | [141,145] |
| G93A | ALS Pathogenic | Substitution | Patients showing different oxidative markers such as glutamate excitotoxicity and dysfunctions at several levels such as mitochondria, due to calcium influx, and axon as well as protein oxidation, modifications observed at SOD1-G93A mice. This mutation is relatively rare in general population but it is very common in familial ALS and also multiple studies on animal models have shown that having the SOD1-G93A mutation is enough to cause motor-neuron degeneration | [146,147,148] |
| L84F | ALS Pathogenic | Substitution | Protein instability and misfolding that can lead to forming protein accumulations. | [143,144] |
| Mutation | Pathogenicity | Type of Mutation | Biological Effect | Citation |
| M337V | ALS Pathogenic | Substitution | production of an abnormal TDP-43 protein that aggregates abnormally and accumulates in motor neurons, ultimately leading to their degeneration and death and ultimately contributing to ALS development | [149,153,154] |
| A315T | ALS Pathogenic | Substitution | Transgenic mice carrying the A315T mutation of TDP-43 may succumb to early death due to digestive complications before fully manifesting neurological signs associated with ALS, suggesting it also influences their digestive systems and may contribute to their early demise. Although the exact mechanisms underlying gastrointestinal complications remain poorly understood, experts speculate that abnormal TDP-43 protein build-up in intestinal cells may lead to dysfunction and damage | [155] |
| A382T | Possible ALS Pathogenic | Substitution | Unknown Mechanism | [156,157] |
12. Perspectives for Treatment
13. Discussions
Disclaimer
Abbreviations
| ND | Neurodegenerative Disease |
| AD | Alzheimer’s disease |
| APP | Amyloid precursor protein |
| Aβ | Amyloid beta peptide |
| HCHWA-D | Hereditary cerebral hemorrhage with amyloidosis |
| PiD | Pick's disease |
| GFAP | Glial fibrillary acidic protein |
| SPECT | Single-photon emission computed tomography |
| PET | Positron emission tomography |
| APOE | Apolipoprotein E |
| BBB | Brain Blood Barrier |
| HDL | High density lipoprotein |
| CNS | Central Nervous System |
| NFTs | Neural Focal Thresholds |
| GWAS | Genome-wide association study |
| PP2A | Protein phosphatase 2A |
| FTD | Frontotemporal Dementia |
| CTD | Chronic traumatic encephalopathy |
| CBD | Corticobasal degeneration |
| DAM | Disease-associated microglia |
| MGnD | Microglial neurodegenerative |
| TREM2 | Triggering receptor expressed on myeloid cells 2 |
| CRP | C-reactive protein |
| MCI | Mild cognitive impairment |
| MAPT | Microtubule-associated protein tau |
| PSP | Progressive supranuclear palsy |
| PHF | Abnormally hyperphosphorylated taus |
| CL | Centiloids |
| SUVR | Standardized Uptake Ration |
| PD | Parkinson's disease |
| SNpc | Substantia nigra pars compacta |
| DA | Dopamine |
| DMV | Dorsal motor nucleus of vagus |
| OB | Olfactory bulb |
| LC | Locus coeruleus |
| IML | Intermediolateral nucleus in spinal cord |
| ENS | Enteric nervous system |
| EOPD | Early-onset Parkinson's disease |
| mitoQC | Mitochondrial quality control |
| DFO | Deferrioxamine |
| DFP | Deferiprone |
| HD | Huntington’s disease |
| MSNs | Medium spiny neurons |
| mHTT | Mutation of the huntingtin gene |
| HTT | Huntingtin gene |
| CBP | cAMP response element-binding protein |
| MSK1 | Mitogen-activated and stress-activated protein kinase 1 |
| PGC-1a | Proliferator-activated receptor gamma coactivator alpha |
| HDAC | Histone deacetylase |
| BDNF | Brain-derived neurotrophic factor |
| ASOs | Antisense oligonucleotides |
| RNAi | RNA interference |
| CoQ10 | Coenzyme Q10 |
| CREST-E | Creatine safety tolerability efficacy in Huntington's disease |
| ALS | Amyotrophic lateral sclerosis |
| FALS | Familial amyotrophic lateral sclerosis |
| DPR | Dipeptide repeat proteins |
| FUS | Fused in Sarcoma |
| miRNA | Micro RNA |
| PTM | Post-translational modification |
| UMN | Upper motor neuron |
| LMN | Lower motor neuron |
| PMA | Progressive muscular atrophy |
| PBP | Pseudobulbar Palsy |
References
- Berry, R. M. The Genetic Revolution and the Physician’s Duty of Confidentiality: The Role of the Old Hippocratic Virtues in the Regulation of the New Genetic Intimacy. J. Leg. Med. 1997, 18 (4), 401–441. [CrossRef]
- De Castro, M. Johann Gregor Mendel: Paragon of Experimental Science. Mol. Genet. Genomic Med. 2016, 4 (1), 3–8. [CrossRef]
- Saceleanu, V. M.; Mohan, A. G.; Covache-Busuioc, R. A.; Costin, H. P.; Ciurea, A. V. Wilhelm von Waldeyer: Important Steps in Neural Theory, Anatomy and Citology. Brain Sci. 2022, 12 (2), 224. [CrossRef]
- Kovacs, G. G. Concepts and Classification of Neurodegenerative Diseases. In Handbook of Clinical Neurology; Elsevier, 2018; Vol. 145, pp 301–307. [CrossRef]
- Brettschneider, J.; Tredici, K. D.; Lee, V. M.-Y.; Trojanowski, J. Q. Spreading of Pathology in Neurodegenerative Diseases: A Focus on Human Studies. Nat. Rev. Neurosci. 2015, 16 (2), 109–120. [CrossRef]
- Guo, J. L.; Lee, V. M. Y. Cell-to-Cell Transmission of Pathogenic Proteins in Neurodegenerative Diseases. Nat. Med. 2014, 20 (2), 130–138. [CrossRef]
- Castellani, R. J.; Rolston, R. K.; Smith, M. A. Alzheimer Disease. Dis. Mon. 2010, 56 (9), 484–546. [CrossRef]
- Ciurea, V. A.; Neurosurgery Department, Carol Davila University of Medicine and Pharmacy, Bucharest, Romania; Neurosurgery Department, Sanador Clinical Hospital, Bucharest, Romania; Covache-Busuioc, R.-A.; Carol Davila University of Medicine and Pharmacy, Bucharest, Romania; Mohan, A. G.; Department of Neurosurgery, Bihor County Emergency Clinical Hospital, Oradea, Romania; Neurosurgery Department, Faculty of Medicine, Oradea University, Oradea, Romania; Costin, H. P.; Carol Davila University of Medicine and Pharmacy, Bucharest, Romania; Voicu, V.; Pharmacology, Toxicology and Clinical Psychopharmacology Department, Carol Davila University of Medicine and Pharmacy, Bucharest, Romania; Romanian Academy, Bucharest, Romania. Alzheimer’s Disease: 120 Years of Research and Progress. J. Med. Life 2023, 16 (2), 173–177. [CrossRef]
- Gatz, M.; Reynolds, C. A.; Fratiglioni, L.; Johansson, B.; Mortimer, J. A.; Berg, S.; Fiske, A.; Pedersen, N. L. Role of Genes and Environments for Explaining Alzheimer Disease. Arch. Gen. Psychiatry 2006, 63 (2), 168. [CrossRef]
- An, F.; Zhao, R.; Xuan, X.; Xuan, T.; Zhang, G.; Wei, C. Calycosin Ameliorates Advanced Glycation End Product-Induced Neurodegenerative Changes in Cellular and Rat Models of Diabetes-Related Alzheimer’s Disease. Chem. Biol. Interact. 2022, 368, 110206. [CrossRef]
- Van Cauwenberghe, C.; Van Broeckhoven, C.; Sleegers, K. The Genetic Landscape of Alzheimer Disease: Clinical Implications and Perspectives. Genet. Med. 2016, 18 (5), 421–430. [CrossRef]
- Roberts, S. B.; Ripellino, J. A.; Ingalls, K. M.; Robakis, N. K.; Felsenstein, K. M. Non-Amyloidogenic Cleavage of the Beta-Amyloid Precursor Protein by an Integral Membrane Metalloendopeptidase. J. Biol. Chem. 1994, 269 (4), 3111–3116. [CrossRef]
- Wu, Q.; Sun, J.-X.; Song, X.-H.; Wang, J.; Xiong, C.-Q.; Teng, F.-X.; Gao, C.-X. Blocking Beta 2-Adrenergic Receptor Inhibits Dendrite Ramification in a Mouse Model of Alzheimer’s Disease. Neural Regen. Res. 2017, 12 (9), 1499–1506. [CrossRef]
- Okochi, M.; Tagami, S.; Yanagida, K.; Takami, M.; Kodama, T. S.; Mori, K.; Nakayama, T.; Ihara, Y.; Takeda, M. γ-Secretase Modulators and Presenilin 1 Mutants Act Differently on Presenilin/γ-Secretase Function to Cleave Aβ42 and Aβ43. Cell Rep. 2013, 3 (1), 42–51. [CrossRef]
- Hampel, H.; Hu, Y.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S. H.; Villemagne, V. L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; Masters, C. L.; Cho, M.; Lannfelt, L.; Cummings, J. L.; Vergallo, A. The Amyloid-β Pathway in Alzheimer’s Disease: A Plain Language Summary. Neurodegener. Dis. Manag. 2023, nmt-2022-0037. [CrossRef]
- Tosh, J. L.; Rhymes, E. R.; Mumford, P.; Whittaker, H. T.; Pulford, L. J.; Noy, S. J.; Cleverley, K.; LonDownS Consortium; Strydom, A.; Fisher, E.; Wiseman, F.; Nizetic, D.; Hardy, J.; Tybulewicz, V.; Karmiloff-Smith, A.; Walker, M. C.; Tybulewicz, V. L. J.; Wykes, R. C.; Fisher, E. M. C.; Wiseman, F. K. Genetic Dissection of down Syndrome-Associated Alterations in APP/Amyloid-β Biology Using Mouse Models. Sci. Rep. 2021, 11 (1), 5736. [CrossRef]
- Jonsson, T.; Atwal, J. K.; Steinberg, S.; Snaedal, J.; Jonsson, P. V.; Bjornsson, S.; Stefansson, H.; Sulem, P.; Gudbjartsson, D.; Maloney, J.; Hoyte, K.; Gustafson, A.; Liu, Y.; Lu, Y.; Bhangale, T.; Graham, R. R.; Huttenlocher, J.; Bjornsdottir, G.; Andreassen, O. A.; Jönsson, E. G.; Palotie, A.; Behrens, T. W.; Magnusson, O. T.; Kong, A.; Thorsteinsdottir, U.; Watts, R. J.; Stefansson, K. A Mutation in APP Protects against Alzheimer’s Disease and Age-Related Cognitive Decline. Nature 2012, 488 (7409), 96–99. [CrossRef]
- Benilova, I.; Gallardo, R.; Ungureanu, A.-A.; Castillo Cano, V.; Snellinx, A.; Ramakers, M.; Bartic, C.; Rousseau, F.; Schymkowitz, J.; De Strooper, B. The Alzheimer Disease Protective Mutation A2T Modulates Kinetic and Thermodynamic Properties of Amyloid-β (Aβ) Aggregation. J. Biol. Chem. 2014, 289 (45), 30977–30989. [CrossRef]
- Liu, Y.-W.; He, Y.-H.; Zhang, Y.-X.; Cai, W.-W.; Yang, L.-Q.; Xu, L.-Y.; Kong, Q.-P. Absence of A673T Variant in APP Gene Indicates an Alternative Protective Mechanism Contributing to Longevity in Chinese Individuals. Neurobiol. Aging 2014, 35 (4), 935.e11-935.e12. [CrossRef]
- Mengel-From, J.; Jeune, B.; Pentti, T.; McGue, M.; Christensen, K.; Christiansen, L. The APP A673T Frequency Differs between Nordic Countries. Neurobiol. Aging 2015, 36 (10), 2909.e1-2909.e4. [CrossRef]
- Giaccone, G.; Morbin, M.; Moda, F.; Botta, M.; Mazzoleni, G.; Uggetti, A.; Catania, M.; Moro, M. L.; Redaelli, V.; Spagnoli, A.; Rossi, R. S.; Salmona, M.; Di Fede, G.; Tagliavini, F. Neuropathology of the Recessive A673V APP Mutation: Alzheimer Disease with Distinctive Features. Acta Neuropathol. (Berl.) 2010, 120 (6), 803–812. [CrossRef]
- Villemagne, V. L.; Ataka, S.; Mizuno, T.; Brooks, W. S.; Wada, Y.; Kondo, M.; Jones, G.; Watanabe, Y.; Mulligan, R.; Nakagawa, M.; Miki, T.; Shimada, H.; O’Keefe, G. J.; Masters, C. L.; Mori, H.; Rowe, C. C. High Striatal Amyloid β-Peptide Deposition Across Different Autosomal Alzheimer Disease Mutation Types. Arch. Neurol. 2009, 66 (12). [CrossRef]
- Readhead, B.; Haure-Mirande, J.-V.; Zhang, B.; Haroutunian, V.; Gandy, S.; Schadt, E. E.; Dudley, J. T.; Ehrlich, M. E. Molecular Systems Evaluation of Oligomerogenic APPE693Q and Fibrillogenic APPKM670/671NL/PSEN1Δexon9 Mouse Models Identifies Shared Features with Human Alzheimer’s Brain Molecular Pathology. Mol. Psychiatry 2016, 21 (8), 1099–1111. [CrossRef]
- Natté, R.; Maat-Schieman, M. L. C.; Haan, J.; Bornebroek, M.; Roos, R. A. C.; Van Duinen, S. G. Dementia in Hereditary Cerebral Hemorrhage with Amyloidosis-Dutch Type Is Associated with Cerebral Amyloid Angiopathy but Is Independent of Plaques and Neurofibrillary Tangles: Dementia in HCHWA-D Is Associated with CAA. Ann. Neurol. 2001, 50 (6), 765–772. [CrossRef]
- Nishitsuji, K.; Tomiyama, T.; Ishibashi, K.; Kametani, F.; Ozawa, K.; Okada, R.; Maat-Schieman, M. L.; Roos, R. A. C.; Iwai, K.; Mori, H. Cerebral Vascular Accumulation of Dutch-Type Aβ42, but Not Wild-Type Aβ42, in Hereditary Cerebral Hemorrhage with Amyloidosis, Dutch Type. J. Neurosci. Res. 2007, 85 (13), 2917–2923. [CrossRef]
- Uddin, Md. S.; Tewari, D.; Sharma, G.; Kabir, Md. T.; Barreto, G. E.; Bin-Jumah, M. N.; Perveen, A.; Abdel-Daim, M. M.; Ashraf, G. M. Molecular Mechanisms of ER Stress and UPR in the Pathogenesis of Alzheimer’s Disease. Mol. Neurobiol. 2020, 57 (7), 2902–2919. [CrossRef]
- Alberdi, E.; Wyssenbach, A.; Alberdi, M.; Sánchez-Gómez, M. V.; Cavaliere, F.; Rodríguez, J. J.; Verkhratsky, A.; Matute, C. Ca 2+ -Dependent Endoplasmic Reticulum Stress Correlates with Astrogliosis in Oligomeric Amyloid β-Treated Astrocytes and in a Model of Alzheimer’s Disease. Aging Cell 2013, 12 (2), 292–302. [CrossRef]
- Tomiyama, T.; Nagata, T.; Shimada, H.; Teraoka, R.; Fukushima, A.; Kanemitsu, H.; Takuma, H.; Kuwano, R.; Imagawa, M.; Ataka, S.; Wada, Y.; Yoshioka, E.; Nishizaki, T.; Watanabe, Y.; Mori, H. A New Amyloid β Variant Favoring Oligomerization in Alzheimer’s-Type Dementia. Ann. Neurol. 2008, 63 (3), 377–387. [CrossRef]
- McKnelly, K. J.; Kreutzer, A. G.; Howitz, W. J.; Haduong, K.; Yoo, S.; Hart, C.; Nowick, J. S. Effects of Familial Alzheimer’s Disease Mutations on the Assembly of a β-Hairpin Peptide Derived from Aβ 16–36. Biochemistry 2022, 61 (6), 446–454. [CrossRef]
- Paik, Y. K.; Chang, D. J.; Reardon, C. A.; Walker, M. D.; Taxman, E.; Taylor, J. M. Identification and Characterization of Transcriptional Regulatory Regions Associated with Expression of the Human Apolipoprotein E Gene. J. Biol. Chem. 1988, 263 (26), 13340–13349.
- Maloney, B.; Ge, Y.-W.; Alley, G. M.; Lahiri, D. K. Important Differences between Human and Mouse APOE Gene Promoters: Limitation of Mouse APOE Model in Studying Alzheimer’s Disease. J. Neurochem. 2007, 103 (3), 1237–1257. [CrossRef]
- Yee, A.; Tsui, N. B. Y.; Kwan, R. Y. C.; Leung, A. Y. M.; Lai, C. K. Y.; Chung, T.; Lau, J. Y. N.; Fok, M.; Dai, D. L. K.; Lau, L.-T. Apolipoprotein E Gene Revisited: Contribution of Rare Variants to Alzheimer’s Disease Susceptibility in Southern Chinese. Curr. Alzheimer Res. 2021, 18 (1), 67–79. [CrossRef]
- Zhou, Y.; Mägi, R.; Milani, L.; Lauschke, V. M. Global Genetic Diversity of Human Apolipoproteins and Effects on Cardiovascular Disease Risk. J. Lipid Res. 2018, 59 (10), 1987–2000. [CrossRef]
- Saunders, A. M. Gene Identification in Alzheimer’s Disease. Pharmacogenomics 2001, 2 (3), 239–249. [CrossRef]
- Theuns, J. Genetic Variability in the Regulatory Region of Presenilin 1 Associated with Risk for Alzheimer’s Disease and Variable Expression. Hum. Mol. Genet. 2000, 9 (3), 325–331. [CrossRef]
- Cruts, M.; Theuns, J.; Van Broeckhoven, C. Locus-specific Mutation Databases for Neurodegenerative Brain Diseases. Hum. Mutat. 2012, 33 (9), 1340–1344. [CrossRef]
- Sun, L.; Zhou, R.; Yang, G.; Shi, Y. Analysis of 138 Pathogenic Mutations in Presenilin-1 on the in Vitro Production of Aβ42 and Aβ40 Peptides by γ-Secretase. Proc. Natl. Acad. Sci. 2017, 114 (4). [CrossRef]
- Wines-Samuelson, M.; Schulte, E. C.; Smith, M. J.; Aoki, C.; Liu, X.; Kelleher, R. J.; Shen, J. Characterization of Age-Dependent and Progressive Cortical Neuronal Degeneration in Presenilin Conditional Mutant Mice. PLoS ONE 2010, 5 (4), e10195. [CrossRef]
- Kauwe, J. S. K.; Jacquart, S.; Chakraverty, S.; Wang, J.; Mayo, K.; Fagan, A. M.; Holtzman, D. M.; Morris, J. C.; Goate, A. M. Extreme Cerebrospinal Fluid Amyloid β Levels Identify Family with Late-Onset Alzheimer’s Disease Presenilin 1 Mutation. Ann. Neurol. 2007, 61 (5), 446–453. [CrossRef]
- Kumar-Singh, S.; Theuns, J.; Van Broeck, B.; Pirici, D.; Vennekens, K.; Corsmit, E.; Cruts, M.; Dermaut, B.; Wang, R.; Van Broeckhoven, C. Mean Age-of-Onset of Familial Alzheimer Disease Caused by Presenilin Mutations Correlates with Both Increased Aβ42 and Decreased Aβ40. Hum. Mutat. 2006, 27 (7), 686–695. [CrossRef]
- Koriath, C.; Kenny, J.; Adamson, G.; Druyeh, R.; Taylor, W.; Beck, J.; Quinn, L.; Mok, T. H.; Dimitriadis, A.; Norsworthy, P.; Bass, N.; Carter, J.; Walker, Z.; Kipps, C.; Coulthard, E.; Polke, J. M.; Bernal-Quiros, M.; Denning, N.; Thomas, R.; Raybould, R.; Williams, J.; Mummery, C. J.; Wild, E. J.; Houlden, H.; Tabrizi, S. J.; Rossor, M. N.; Hummerich, H.; Warren, J. D.; Rowe, J. B.; Rohrer, J. D.; Schott, J. M.; Fox, N. C.; Collinge, J.; Mead, S. Predictors for a Dementia Gene Mutation Based on Gene-Panel next-Generation Sequencing of a Large Dementia Referral Series. Mol. Psychiatry 2020, 25 (12), 3399–3412. [CrossRef]
- Gallo, M.; Frangipane, F.; Cupidi, C.; De Bartolo, M.; Turone, S.; Ferrari, C.; Nacmias, B.; Grimaldi, G.; Laganà, V.; Colao, R.; Bernardi, L.; Anfossi, M.; Conidi, M. E.; Vasso, F.; Curcio, S. A. M.; Mirabelli, M.; Smirne, N.; Torchia, G.; Muraca, M. G.; Puccio, G.; Di Lorenzo, R.; Piccininni, M.; Tedde, A.; Maletta, R. G.; Sorbi, S.; Bruni, A. C. The Novel PSEN1 M84V Mutation Associated to Frontal Dysexecutive Syndrome, Spastic Paraparesis, and Cerebellar Atrophy in a Dominant Alzheimer’s Disease Family. Neurobiol. Aging 2017, 56, 213.e7-213.e12. [CrossRef]
- Dominantly Inherited Alzheimer Network (DIAN); Hsu, S.; Gordon, B. A.; Hornbeck, R.; Norton, J. B.; Levitch, D.; Louden, A.; Ziegemeier, E.; Laforce, R.; Chhatwal, J.; Day, G. S.; McDade, E.; Morris, J. C.; Fagan, A. M.; Benzinger, T. L. S.; Goate, A. M.; Cruchaga, C.; Bateman, R. J.; Karch, C. M. Discovery and Validation of Autosomal Dominant Alzheimer’s Disease Mutations. Alzheimers Res. Ther. 2018, 10 (1), 67. [CrossRef]
- Kakuda, N.; Takami, M.; Okochi, M.; Kasuga, K.; Ihara, Y.; Ikeuchi, T. Switched Aβ43 Generation in Familial Alzheimer’s Disease with Presenilin 1 Mutation. Transl. Psychiatry 2021, 11 (1), 558. [CrossRef]
- López-García, S.; Jiménez-Bonilla, J.; López Delgado, A.; Orizaola Balaguer, P.; Infante Ceberio, J.; Banzo Marraco, I.; Rodríguez Rodríguez, E.; Sánchez-Juan, P. A Rare PSEN1 (Leu85Pro) Mutation Causing Alzheimer’s Disease in a 29-Year-Old Woman Presenting as Corticobasal Syndrome. J. Alzheimers Dis. JAD 2019, 70 (3), 655–658. [CrossRef]
- Arber, C.; Villegas-Llerena, C.; Toombs, J.; Pocock, J. M.; Ryan, N. S.; Fox, N. C.; Zetterberg, H.; Hardy, J.; Wray, S. Amyloid Precursor Protein Processing in Human Neurons with an Allelic Series of the PSEN1 Intron 4 Deletion Mutation and Total Presenilin-1 Knockout. Brain Commun. 2019, 1 (1), fcz024. [CrossRef]
- De Jonghe, C.; MarcCruts; Rogaeva, E. A.; Tysoe, C.; Singleton, A.; Vanderstichele, H.; Meschino, W.; Dermaut, B.; Vanderhoeven, I.; Backhovens, H.; Vanmechelen, E.; Morris, C. M.; Hardy, J.; Rubinsztein, D. C.; St George-Hyslop, P. H.; Van Broeckhoven, C. Aberrant Splicing in the Presenilin-1 Intron 4 Mutation Causes Presenile Alzheimer’s Disease by Increased A 42 Secretion. Hum. Mol. Genet. 1999, 8 (8), 1529–1540. [CrossRef]
- Szaruga, M.; Veugelen, S.; Benurwar, M.; Lismont, S.; Sepulveda-Falla, D.; Lleo, A.; Ryan, N. S.; Lashley, T.; Fox, N. C.; Murayama, S.; Gijsen, H.; De Strooper, B.; Chávez-Gutiérrez, L. Qualitative Changes in Human γ-Secretase Underlie Familial Alzheimer’s Disease. J. Exp. Med. 2015, 212 (12), 2003–2013. [CrossRef]
- Fox, N. Clinicopathological Features of Familial Alzheimer’s Disease Associated with the M139V Mutation in the Presenilin 1 Gene. Pedigree but Not Mutation Specific Age at Onset Provides Evidence for a Further Genetic Factor. Brain 1997, 120 (3), 491–501. [CrossRef]
- Chávez-Gutiérrez, L.; Bammens, L.; Benilova, I.; Vandersteen, A.; Benurwar, M.; Borgers, M.; Lismont, S.; Zhou, L.; Van Cleynenbreugel, S.; Esselmann, H.; Wiltfang, J.; Serneels, L.; Karran, E.; Gijsen, H.; Schymkowitz, J.; Rousseau, F.; Broersen, K.; De Strooper, B. The Mechanism of γ-Secretase Dysfunction in Familial Alzheimer Disease: Mechanisms of Alzheimer Disease-Causing Mutations. EMBO J. 2012, 31 (10), 2261–2274. [CrossRef]
- Ringman, J. M.; Gylys, K. H.; Medina, L. D.; Fox, M.; Kepe, V.; Flores, D. L.; Apostolova, L. G.; Barrio, J. R.; Small, G.; Silverman, D. H.; Siu, E.; Cederbaum, S.; Hecimovic, S.; Malnar, M.; Chakraverty, S.; Goate, A. M.; Bird, T. D.; Leverenz, J. B. Biochemical, Neuropathological, and Neuroimaging Characteristics of Early-Onset Alzheimer’s Disease Due to a Novel PSEN1 Mutation. Neurosci. Lett. 2011, 487 (3), 287–292. [CrossRef]
- Devi, G.; Fotiou, A.; Jyrinji, D.; Tycko, B.; DeArmand, S.; Rogaeva, E.; Song, Y.-Q.; Medieros, H.; Liang, Y.; Orlacchio, A.; Williamson, J.; St George-Hyslop, P.; Mayeux, R. Novel Presenilin 1 Mutations Associated With Early Onset of Dementia in a Family With Both Early-Onset and Late-Onset Alzheimer Disease. Arch. Neurol. 2000, 57 (10). [CrossRef]
- Levy-Lahad, E.; Wasco, W.; Poorkaj, P.; Romano, D. M.; Oshima, J.; Pettingell, W. H.; Yu, C.; Jondro, P. D.; Schmidt, S. D.; Wang, K.; Crowley, A. C.; Fu, Y.-H.; Guenette, S. Y.; Galas, D.; Nemens, E.; Wijsman, E. M.; Bird, T. D.; Schellenberg, G. D.; Tanzi, R. E. Candidate Gene for the Chromosome 1 Familial Alzheimer’s Disease Locus. Science 1995, 269 (5226), 973–977. [CrossRef]
- Perrone, F.; Cacace, R.; Van Mossevelde, S.; Van Den Bossche, T.; De Deyn, P. P.; Cras, P.; Engelborghs, S.; Van Der Zee, J.; Van Broeckhoven, C. Genetic Screening in Early-Onset Dementia Patients with Unclear Phenotype: Relevance for Clinical Diagnosis. Neurobiol. Aging 2018, 69, 292.e7-292.e14. [CrossRef]
- Jia, L.; Fu, Y.; Shen, L.; Zhang, H.; Zhu, M.; Qiu, Q.; Wang, Q.; Yan, X.; Kong, C.; Hao, J.; Wei, C.; Tang, Y.; Qin, W.; Li, Y.; Wang, F.; Guo, D.; Zhou, A.; Zuo, X.; Yu, Y.; Li, D.; Zhao, L.; Jin, H.; Jia, J. PSEN1, PSEN2, and APP Mutations in 404 Chinese Pedigrees with Familial Alzheimer’s Disease. Alzheimers Dement. J. Alzheimers Assoc. 2020, 16 (1), 178–191. [CrossRef]
- Pang, Y.; Li, T.; Wang, Q.; Qin, W.; Li, Y.; Wei, Y.; Jia, L. A Rare Variation in the 3’ Untranslated Region of the Presenilin 2 Gene Is Linked to Alzheimer’s Disease. Mol. Neurobiol. 2021, 58 (9), 4337–4347. [CrossRef]
- The collaborators of the GMAJ project; Wallon, D.; Rousseau, S.; Rovelet-Lecrux, A.; Quillard-Muraine, M.; Guyant-Maréchal, L.; Martinaud, O.; Pariente, J.; Puel, M.; Rollin-Sillaire, A.; Pasquier, F.; Le Ber, I.; Sarazin, M.; Croisile, B.; Boutoleau-Bretonnière, C.; Thomas-Antérion, C.; Paquet, C.; Moreaud, O.; Gabelle, A.; Sellal, F.; Sauvée, M.; Laquerrière, A.; Duyckaerts, C.; Delisle, M.-B.; Streichenberger, N.; Lannes, B.; Frebourg, T.; Hannequin, D.; Campion, D. The French Series of Autosomal Dominant Early Onset Alzheimer’s Disease Cases: Mutation Spectrum and Cerebrospinal Fluid Biomarkers. J. Alzheimers Dis. 2012, 30 (4), 847–856. [CrossRef]
- Marcon, G.; Giaccone, G.; Cupidi, C.; Balestrieri, M.; Beltrami, C. A.; Finato, N.; Bergonzi, P.; Sorbi, S.; Bugiani, O.; Tagliavini, F. Neuropathological and Clinical Phenotype of an Italian Alzheimer Family with M239V Mutation of Presenilin 2 Gene. J. Neuropathol. Exp. Neurol. 2004, 63 (3), 199–209. [CrossRef]
- Huang, Y.; Weisgraber, K. H.; Mucke, L.; Mahley, R. W. Apolipoprotein E: Diversity of Cellular Origins, Structural and Biophysical Properties, and Effects in Alzheimer’s Disease. J. Mol. Neurosci. 2004, 23 (3), 189–204. [CrossRef]
- Liu, C.-C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer Disease: Risk, Mechanisms and Therapy. Nat. Rev. Neurol. 2013, 9 (2), 106–118. [CrossRef]
- Kara, E.; Marks, J. D.; Fan, Z.; Klickstein, J. A.; Roe, A. D.; Krogh, K. A.; Wegmann, S.; Maesako, M.; Luo, C. C.; Mylvaganam, R.; Berezovska, O.; Hudry, E.; Hyman, B. T. Isoform- and Cell Type-Specific Structure of Apolipoprotein E Lipoparticles as Revealed by a Novel Forster Resonance Energy Transfer Assay. J. Biol. Chem. 2017, 292 (36), 14720–14729. [CrossRef]
- Lin, Y.-T.; Seo, J.; Gao, F.; Feldman, H. M.; Wen, H.-L.; Penney, J.; Cam, H. P.; Gjoneska, E.; Raja, W. K.; Cheng, J.; Rueda, R.; Kritskiy, O.; Abdurrob, F.; Peng, Z.; Milo, B.; Yu, C. J.; Elmsaouri, S.; Dey, D.; Ko, T.; Yankner, B. A.; Tsai, L.-H. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human IPSC-Derived Brain Cell Types. Neuron 2018, 98 (6), 1141-1154.e7. [CrossRef]
- Kang, S. S.; Ebbert, M. T. W.; Baker, K. E.; Cook, C.; Wang, X.; Sens, J. P.; Kocher, J.-P.; Petrucelli, L.; Fryer, J. D. Microglial Translational Profiling Reveals a Convergent APOE Pathway from Aging, Amyloid, and Tau. J. Exp. Med. 2018, 215 (9), 2235–2245. [CrossRef]
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J. Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; Martorell, A. J.; Ransohoff, R. M.; Hafler, B. P.; Bennett, D. A.; Kellis, M.; Tsai, L.-H. Single-Cell Transcriptomic Analysis of Alzheimer’s Disease. Nature 2019, 570 (7761), 332–337. [CrossRef]
- Therriault, J.; Benedet, A. L.; Pascoal, T. A.; Mathotaarachchi, S.; Chamoun, M.; Savard, M.; Thomas, E.; Kang, M. S.; Lussier, F.; Tissot, C.; Parsons, M.; Qureshi, M. N. I.; Vitali, P.; Massarweh, G.; Soucy, J.-P.; Rej, S.; Saha-Chaudhuri, P.; Gauthier, S.; Rosa-Neto, P. Association of Apolipoprotein E Ε4 With Medial Temporal Tau Independent of Amyloid-β. JAMA Neurol. 2020, 77 (4), 470. [CrossRef]
- Alzheimer’s Disease Neuroimaging Initiative; Shi, Y.; Yamada, K.; Liddelow, S. A.; Smith, S. T.; Zhao, L.; Luo, W.; Tsai, R. M.; Spina, S.; Grinberg, L. T.; Rojas, J. C.; Gallardo, G.; Wang, K.; Roh, J.; Robinson, G.; Finn, M. B.; Jiang, H.; Sullivan, P. M.; Baufeld, C.; Wood, M. W.; Sutphen, C.; McCue, L.; Xiong, C.; Del-Aguila, J. L.; Morris, J. C.; Cruchaga, C.; Fagan, A. M.; Miller, B. L.; Boxer, A. L.; Seeley, W. W.; Butovsky, O.; Barres, B. A.; Paul, S. M.; Holtzman, D. M. ApoE4 Markedly Exacerbates Tau-Mediated Neurodegeneration in a Mouse Model of Tauopathy. Nature 2017, 549 (7673), 523–527. [CrossRef]
- Harris, F. M.; Brecht, W. J.; Xu, Q.; Mahley, R. W.; Huang, Y. Increased Tau Phosphorylation in Apolipoprotein E4 Transgenic Mice Is Associated with Activation of Extracellular Signal-Regulated Kinase. J. Biol. Chem. 2004, 279 (43), 44795–44801. [CrossRef]
- Wadhwani, A. R.; Affaneh, A.; Van Gulden, S.; Kessler, J. A. Neuronal Apolipoprotein E4 Increases Cell Death and Phosphorylated Tau Release in Alzheimer Disease. Ann. Neurol. 2019, 85 (5), 726–739. [CrossRef]
- Gibb, G. M.; Pearce, J.; Betts, J. C.; Lovestone, S.; Hoffmann, M. M.; Maerz, W.; Blackstock, W. P.; Anderton, B. H. Differential Effects of Apolipoprotein E Isoforms on Phosphorylation at Specific Sites on Tau by Glycogen Synthase Kinase-3β Identified by Nano-Electrospray Mass Spectrometry. FEBS Lett. 2000, 485 (2–3), 99–103. [CrossRef]
- Park, J.-S.; Ji, I. J.; An, H. J.; Kang, M.-J.; Kang, S.-W.; Kim, D.-H.; Yoon, S.-Y. Disease-Associated Mutations of TREM2 Alter the Processing of N-Linked Oligosaccharides in the Golgi Apparatus: Impaired Trafficking of TREM2. Traffic 2015, 16 (5), 510–518. [CrossRef]
- Saceleanu, V. M.; Covache-Busuioc, R.-A.; Costin, H.-P.; Glavan, L.-A.; Ciurea, A. V. An Important Step in Neuroscience: Camillo Golgi and His Discoveries. Cells 2022, 11 (24), 4112. [CrossRef]
- Ulrich, J. D.; Ulland, T. K.; Mahan, T. E.; Nyström, S.; Nilsson, K. P.; Song, W. M.; Zhou, Y.; Reinartz, M.; Choi, S.; Jiang, H.; Stewart, F. R.; Anderson, E.; Wang, Y.; Colonna, M.; Holtzman, D. M. ApoE Facilitates the Microglial Response to Amyloid Plaque Pathology. J. Exp. Med. 2018, 215 (4), 1047–1058. [CrossRef]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T. K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; Itzkovitz, S.; Colonna, M.; Schwartz, M.; Amit, I. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169 (7), 1276-1290.e17. [CrossRef]
- Hutton, M.; Lendon, C. L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A.; Hackett, J.; Adamson, J.; Lincoln, S.; Dickson, D.; Davies, P.; Petersen, R. C.; Stevens, M.; De Graaff, E.; Wauters, E.; Van Baren, J.; Hillebrand, M.; Joosse, M.; Kwon, J. M.; Nowotny, P.; Che, L. K.; Norton, J.; Morris, J. C.; Reed, L. A.; Trojanowski, J.; Basun, H.; Lannfelt, L.; Neystat, M.; Fahn, S.; Dark, F.; Tannenberg, T.; Dodd, P. R.; Hayward, N.; Kwok, J. B. J.; Schofield, P. R.; Andreadis, A.; Snowden, J.; Craufurd, D.; Neary, D.; Owen, F.; Oostra, B. A.; Hardy, J.; Goate, A.; Van Swieten, J.; Mann, D.; Lynch, T.; Heutink, P. Association of Missense and 5′-Splice-Site Mutations in Tau with the Inherited Dementia FTDP-17. Nature 1998, 393 (6686), 702–705. [CrossRef]
- Spillantini, M. G.; Bird, T. D.; Ghetti, B. Frontotemporal Dementia and Parkinsonism Linked to Chromosome 17: A New Group of Tauopathies. Brain Pathol. 2006, 8 (2), 387–402. [CrossRef]
- Poorkaj, P.; Bird, T. D.; Wijsman, E.; Nemens, E.; Garruto, R. M.; Anderson, L.; Andreadis, A.; Wiederholt, W. C.; Raskind, M.; Schellenberg, G. D. Tau Is a Candidate Gene for Chromosome 17 Frontotemporal Dementia. Ann. Neurol. 1998, 43 (6), 815–825. [CrossRef]
- Goedert, M.; Spillantini, M. G.; Jakes, R.; Rutherford, D.; Crowther, R. A. Multiple Isoforms of Human Microtubule-Associated Protein Tau: Sequences and Localization in Neurofibrillary Tangles of Alzheimer’s Disease. Neuron 1989, 3 (4), 519–526. [CrossRef]
- Baudier, J.; Cole, R. D. Interactions between the Microtubule-Associated Tau Proteins and S100b Regulate Tau Phosphorylation by the Ca2+/Calmodulin-Dependent Protein Kinase II. J. Biol. Chem. 1988, 263 (12), 5876–5883.
- Drewes, G.; Ebneth, A.; Preuss, U.; Mandelkow, E.-M.; Mandelkow, E. MARK, a Novel Family of Protein Kinases That Phosphorylate Microtubule-Associated Proteins and Trigger Microtubule Disruption. Cell 1997, 89 (2), 297–308. [CrossRef]
- Drewes, G.; Lichtenberg-Kraag, B.; Döring, F.; Mandelkow, E. M.; Biernat, J.; Goris, J.; Dorée, M.; Mandelkow, E. Mitogen Activated Protein (MAP) Kinase Transforms Tau Protein into an Alzheimer-like State. EMBO J. 1992, 11 (6), 2131–2138. [CrossRef]
- Bancher, C.; Brunner, C.; Lassmann, H.; Budka, H.; Jellinger, K.; Wiche, G.; Seitelberger, F.; Grundke-Iqbal, I.; Iqbal, K.; Wisniewski, H. M. Accumulation of Abnormally Phosphorylated τ Precedes the Formation of Neurofibrillary Tangles in Alzheimer’s Disease. Brain Res. 1989, 477 (1–2), 90–99. [CrossRef]
- Wang, J.; Wu, Q.; Smith, A.; Grundke-Iqbal, I.; Iqbal, K. τ Is Phosphorylated by GSK-3 at Several Sites Found in Alzheimer Disease and Its Biological Activity Markedly Inhibited Only after It Is Prephosphorylated by A-Kinase. FEBS Lett. 1998, 436 (1), 28–34. [CrossRef]
- Yasuda, M.; Takamatsu, J.; D’Souza, I.; Crowther, R. A.; Kawamata, T.; Hasegawa, M.; Hasegawa, H.; Spillantini, M. G.; Tanimukai, S.; Poorkaj, P.; Varani, L.; Varani, G.; Iwatsubo, T.; Goedert, M.; Schellenberg, D. G.; Tanaka, C. A Novel Mutation at Position +12 in the Intron Following Exon 10 of the Tau Gene in Familial Frontotemporal Dementia (FTD-Kumamoto). Ann. Neurol. 2000, 47 (4), 422–429.
- Takamatsu, J.; Kondo, A.; Ikegami, K.; Kimura, T.; Fujii, H.; Mitsuyama, Y.; Hashizume, Y. Selective Expression of Ser 199/202 Phosphorylated Tau in a Case of Frontotemporal Dementia. Dement. Geriatr. Cogn. Disord. 1998, 9 (2), 82–89. [CrossRef]
- Labbé, C.; Ogaki, K.; Lorenzo-Betancor, O.; Soto-Ortolaza, A. I.; Walton, R. L.; Rayaprolu, S.; Fujioka, S.; Murray, M. E.; Heckman, M. G.; Puschmann, A.; McCarthy, A.; Lynch, T.; Siuda, J.; Opala, G.; Rudzinska, M.; Krygowska-Wajs, A.; Barcikowska, M.; Czyzewski, K.; Sanotsky, Y.; Rektorová, I.; McLean, P. J.; Rademakers, R.; Ertekin-Taner, N.; Hassan, A.; Ahlskog, J. E.; Boeve, B. F.; Petersen, R. C.; Maraganore, D. M.; Adler, C. H.; Ferman, T. J.; Parisi, J. E.; Graff-Radford, N. R.; Uitti, R. J.; Wszolek, Z. K.; Dickson, D. W.; Ross, O. A. Role for the Microtubule-Associated Protein Tau Variant p.A152T in Risk of α-Synucleinopathies. Neurology 2015, 85 (19), 1680–1686. [CrossRef]
- Jin, S.; Pastor, P.; Cooper, B.; Cervantes, S.; Benitez, B. A.; Razquin, C.; Goate, A.; Ibero-American Alzheimer Disease Genetics Group Researchers; Cruchaga, C. Pooled-DNA Sequencing Identifies Novel Causative Variants in PSEN1, GRN and MAPT in a Clinical Early-Onset and Familial Alzheimer’s Disease Ibero-American Cohort. Alzheimers Res. Ther. 2012, 4 (4), 34. [CrossRef]
- Coppola, G.; Chinnathambi, S.; Lee, J. J.; Dombroski, B. A.; Baker, M. C.; Soto-Ortolaza, A. I.; Lee, S. E.; Klein, E.; Huang, A. Y.; Sears, R.; Lane, J. R.; Karydas, A. M.; Kenet, R. O.; Biernat, J.; Wang, L.-S.; Cotman, C. W.; DeCarli, C. S.; Levey, A. I.; Ringman, J. M.; Mendez, M. F.; Chui, H. C.; Le Ber, I.; Brice, A.; Lupton, M. K.; Preza, E.; Lovestone, S.; Powell, J.; Graff-Radford, N.; Petersen, R. C.; Boeve, B. F.; Lippa, C. F.; Bigio, E. H.; Mackenzie, I.; Finger, E.; Kertesz, A.; Caselli, R. J.; Gearing, M.; Juncos, J. L.; Ghetti, B.; Spina, S.; Bordelon, Y. M.; Tourtellotte, W. W.; Frosch, M. P.; Vonsattel, J. P. G.; Zarow, C.; Beach, T. G.; Albin, R. L.; Lieberman, A. P.; Lee, V. M.; Trojanowski, J. Q.; Van Deerlin, V. M.; Bird, T. D.; Galasko, D. R.; Masliah, E.; White, C. L.; Troncoso, J. C.; Hannequin, D.; Boxer, A. L.; Geschwind, M. D.; Kumar, S.; Mandelkow, E.-M.; Wszolek, Z. K.; Uitti, R. J.; Dickson, D. W.; Haines, J. L.; Mayeux, R.; Pericak-Vance, M. A.; Farrer, L. A.; Ross, O. A.; Rademakers, R.; Schellenberg, G. D.; Miller, B. L.; Mandelkow, E.; Geschwind, D. H. Evidence for a Role of the Rare p.A152T Variant in MAPT in Increasing the Risk for FTD-Spectrum and Alzheimer’s Diseases. Hum. Mol. Genet. 2012, 21 (15), 3500–3512. [CrossRef]
- Rizzini, C.; Goedert, M.; Hodges, J. R.; Smith, M. J.; Jakes, R.; Hills, R.; Xuereb, J. H.; Crowther, R. A.; Spillantini, M. G. Tau Gene Mutation K257T Causes a Tauopathy Similar to Pick’s Disease. J. Neuropathol. Exp. Neurol. 2000, 59 (11), 990–1001. [CrossRef]
- Kobayashi, T.; Ota, S.; Tanaka, K.; Ito, Y.; Hasegawa, M.; Umeda, Y.; Motoi, Y.; Takanashi, M.; Yasuhara, M.; Anno, M.; Mizuno, Y.; Mori, H. A Novel L266V Mutation of the Tau Gene Causes Frontotemporal Dementia with a Unique Tau Pathology. Ann. Neurol. 2003, 53 (1), 133–137. [CrossRef]
- Hogg, M.; Grujic, Z. M.; Baker, M.; Demirci, S.; Guillozet, A. L.; Sweet, A. P.; Herzog, L. L.; Weintraub, S.; Mesulam, M.-M.; LaPointe, N. E.; Gamblin, T. C.; Berry, R. W.; Binder, L. I.; De Silva, R.; Lees, A.; Espinoza, M.; Davies, P.; Grover, A.; Sahara, N.; Ishizawa, T.; Dickson, D.; Yen, S.-H.; Hutton, M.; Bigio, E. H. The L266V Tau Mutation Is Associated with Frontotemporal Dementia and Pick-like 3R and 4R Tauopathy. Acta Neuropathol. (Berl.) 2003, 106 (4), 323–336. [CrossRef]
- Schilling, S.; Zeitschel, U.; Hoffmann, T.; Heiser, U.; Francke, M.; Kehlen, A.; Holzer, M.; Hutter-Paier, B.; Prokesch, M.; Windisch, M.; Jagla, W.; Schlenzig, D.; Lindner, C.; Rudolph, T.; Reuter, G.; Cynis, H.; Montag, D.; Demuth, H.-U.; Rossner, S. Glutaminyl Cyclase Inhibition Attenuates Pyroglutamate Aβ and Alzheimer’s Disease–like Pathology. Nat. Med. 2008, 14 (10), 1106–1111. [CrossRef]
- Frost, J. L.; Liu, B.; Kleinschmidt, M.; Schilling, S.; Demuth, H.-U.; Lemere, C. A. Passive Immunization against Pyroglutamate-3 Amyloid-β Reduces Plaque Burden in Alzheimer-Like Transgenic Mice: A Pilot Study. Neurodegener. Dis. 2012, 10 (1–4), 265–270. [CrossRef]
- Frost, J. L.; Liu, B.; Rahfeld, J.-U.; Kleinschmidt, M.; O’Nuallain, B.; Le, K. X.; Lues, I.; Caldarone, B. J.; Schilling, S.; Demuth, H.-U.; Lemere, C. A. An Anti-Pyroglutamate-3 Aβ Vaccine Reduces Plaques and Improves Cognition in APPswe/PS1ΔE9 Mice. Neurobiol. Aging 2015, 36 (12), 3187–3199. [CrossRef]
- Hoffmann, T.; Meyer, A.; Heiser, U.; Kurat, S.; Böhme, L.; Kleinschmidt, M.; Bühring, K.-U.; Hutter-Paier, B.; Farcher, M.; Demuth, H.-U.; Lues, I.; Schilling, S. Glutaminyl Cyclase Inhibitor PQ912 Improves Cognition in Mouse Models of Alzheimer’s Disease—Studies on Relation to Effective Target Occupancy. J. Pharmacol. Exp. Ther. 2017, 362 (1), 119–130. [CrossRef]
- DeMattos, R. B.; Lu, J.; Tang, Y.; Racke, M. M.; DeLong, C. A.; Tzaferis, J. A.; Hole, J. T.; Forster, B. M.; McDonnell, P. C.; Liu, F.; Kinley, R. D.; Jordan, W. H.; Hutton, M. L. A Plaque-Specific Antibody Clears Existing β-Amyloid Plaques in Alzheimer’s Disease Mice. Neuron 2012, 76 (5), 908–920. [CrossRef]
- Lowe, S. L.; Willis, B. A.; Hawdon, A.; Natanegara, F.; Chua, L.; Foster, J.; Shcherbinin, S.; Ardayfio, P.; Sims, J. R. Donanemab (LY3002813) Dose-escalation Study in Alzheimer’s Disease. Alzheimers Dement. Transl. Res. Clin. Interv. 2021, 7 (1). [CrossRef]
- Lowe, S. L.; Duggan Evans, C.; Shcherbinin, S.; Cheng, Y.-J.; Willis, B. A.; Gueorguieva, I.; Lo, A. C.; Fleisher, A. S.; Dage, J. L.; Ardayfio, P.; Aguiar, G.; Ishibai, M.; Takaichi, G.; Chua, L.; Mullins, G.; Sims, J. R. Donanemab (LY3002813) Phase 1b Study in Alzheimer’s Disease: Rapid and Sustained Reduction of Brain Amyloid Measured by Florbetapir F18 Imaging. J. Prev. Alzheimers Dis. 2021, 1–11. [CrossRef]
- Shcherbinin, S.; Evans, C. D.; Lu, M.; Andersen, S. W.; Pontecorvo, M. J.; Willis, B. A.; Gueorguieva, I.; Hauck, P. M.; Brooks, D. A.; Mintun, M. A.; Sims, J. R. Association of Amyloid Reduction After Donanemab Treatment With Tau Pathology and Clinical Outcomes: The TRAILBLAZER-ALZ Randomized Clinical Trial. JAMA Neurol. 2022, 79 (10), 1015. [CrossRef]
- Raza, C.; Anjum, R.; Shakeel, N. U. A. Parkinson’s Disease: Mechanisms, Translational Models and Management Strategies. Life Sci. 2019, 226, 77–90. [CrossRef]
- Beitz, J. M. Parkinson s Disease a Review. Front. Biosci. 2014, S6 (1), 65–74. [CrossRef]
- Day, J. O.; Mullin, S. The Genetics of Parkinson’s Disease and Implications for Clinical Practice. Genes 2021, 12 (7), 1006. [CrossRef]
- Ye, H.; Robak, L. A.; Yu, M.; Cykowski, M.; Shulman, J. M. Genetics and Pathogenesis of Parkinson’s Syndrome. Annu. Rev. Pathol. Mech. Dis. 2023, 18 (1), 95–121. [CrossRef]
- Siddiqui, I. J.; Pervaiz, N.; Abbasi, A. A. The Parkinson Disease Gene SNCA: Evolutionary and Structural Insights with Pathological Implication. Sci. Rep. 2016, 6 (1), 24475. [CrossRef]
- Rui, Q.; Ni, H.; Li, D.; Gao, R.; Chen, G. The Role of LRRK2 in Neurodegeneration of Parkinson Disease. Curr. Neuropharmacol. 2018, 16 (9), 1348–1357. [CrossRef]
- Vizziello, M.; Borellini, L.; Franco, G.; Ardolino, G. Disruption of Mitochondrial Homeostasis: The Role of PINK1 in Parkinson’s Disease. Cells 2021, 10 (11), 3022. [CrossRef]
- Crichton, R. R.; Dexter, D. T.; Ward, R. J. Brain Iron Metabolism and Its Perturbation in Neurological Diseases. In Metal Ions in Neurological Systems; Linert, W., Kozlowski, H., Eds.; Springer Vienna: Vienna, 2012; pp 1–15. [CrossRef]
- Chandra, G.; Shenoi, R. A.; Anand, R.; Rajamma, U.; Mohanakumar, K. P. Reinforcing Mitochondrial Functions in Aging Brain: An Insight into Parkinson’s Disease Therapeutics. J. Chem. Neuroanat. 2019, 95, 29–42. [CrossRef]
- C Borgna-Pignatti; S Rugolotto; P De Stefano; H Zhao; MD Cappellini; GC Del Vecchio; MA Romeo; GL Forni; MR Gamberini; R Ghilardi; A Piga; A Cnaan. Survival and Complications in Patients with Thalassemia Major Treated with Transfusion and Deferoxamine. Haematologica 2004, 89 (10), 1187–1193.
- Boddaert, N.; Le Quan Sang, K. H.; Rötig, A.; Leroy-Willig, A.; Gallet, S.; Brunelle, F.; Sidi, D.; Thalabard, J.-C.; Munnich, A.; Cabantchik, Z. I. Selective Iron Chelation in Friedreich Ataxia: Biologic and Clinical Implications. Blood 2007, 110 (1), 401–408. [CrossRef]
- Devos, D.; Moreau, C.; Devedjian, J. C.; Kluza, J.; Petrault, M.; Laloux, C.; Jonneaux, A.; Ryckewaert, G.; Garçon, G.; Rouaix, N.; Duhamel, A.; Jissendi, P.; Dujardin, K.; Auger, F.; Ravasi, L.; Hopes, L.; Grolez, G.; Firdaus, W.; Sablonnière, B.; Strubi-Vuillaume, I.; Zahr, N.; Destée, A.; Corvol, J.-C.; Pöltl, D.; Leist, M.; Rose, C.; Defebvre, L.; Marchetti, P.; Cabantchik, Z. I.; Bordet, R. Targeting Chelatable Iron as a Therapeutic Modality in Parkinson’s Disease. Antioxid. Redox Signal. 2014, 21 (2), 195–210. [CrossRef]
- McColgan, P.; Tabrizi, S. J. Huntington’s Disease: A Clinical Review. Eur. J. Neurol. 2018, 25 (1), 24–34. [CrossRef]
- Wexler, A.; Wild, E. J.; Tabrizi, S. J. George Huntington: A Legacy of Inquiry, Empathy and Hope. Brain 2016, 139 (8), 2326–2333. [CrossRef]
- Bates, G. P.; Dorsey, R.; Gusella, J. F.; Hayden, M. R.; Kay, C.; Leavitt, B. R.; Nance, M.; Ross, C. A.; Scahill, R. I.; Wetzel, R.; Wild, E. J.; Tabrizi, S. J. Huntington Disease. Nat. Rev. Dis. Primer 2015, 1 (1), 15005. [CrossRef]
- Macdonald, M. A Novel Gene Containing a Trinucleotide Repeat That Is Expanded and Unstable on Huntington’s Disease Chromosomes. Cell 1993, 72 (6), 971–983. [CrossRef]
- Squitieri, F.; Andrew, S. E.; Goldberg, Y. P.; Kremer, B.; Spence, N.; Zelsler, J.; Nichol, K.; Theilmann, J.; Greenberg, J.; Goto, J.; Kanazawa, I.; Vesa, J.; Peltonen, L.; Almqvist, E.; Anvret, M.; Telenius, H.; Lin, B.; Napolitano, G.; Morgan, K.; Hayden, M. R. DNA Haplotype Analysis of Huntington Disease Reveals Clues to the Origins and Mechanisms of CAG Expansion and Reasons for Geographic Variations of Prevalence. Hum. Mol. Genet. 1994, 3 (12), 2103–2114. [CrossRef]
- Persichetti, F.; Srinidhi, J.; Kanaley, L.; Ge, P.; Myers, R. H.; D’Arrigo, K.; Barnes, G. T.; MacDonald, M. E.; Vonsattel, J.-P.; Gusella, J. F.; Bird, E. D. Huntington’s Disease CAG Trinucleotide Repeats in Pathologically Confirmed Post-Mortem Brains. Neurobiol. Dis. 1994, 1 (3), 159–166. [CrossRef]
- Gusella, J. F.; MacDonald, M. E.; Lee, J.-M. Genetic Modifiers of Huntington’s Disease: GENETIC MODIFIERS OF HD. Mov. Disord. 2014, 29 (11), 1359–1365. [CrossRef]
- Lee, J.-M.; Gillis, T.; Mysore, J. S.; Ramos, E. M.; Myers, R. H.; Hayden, M. R.; Morrison, P. J.; Nance, M.; Ross, C. A.; Margolis, R. L.; Squitieri, F.; Griguoli, A.; Di Donato, S.; Gomez-Tortosa, E.; Ayuso, C.; Suchowersky, O.; Trent, R. J.; McCusker, E.; Novelletto, A.; Frontali, M.; Jones, R.; Ashizawa, T.; Frank, S.; Saint-Hilaire, M.-H.; Hersch, S. M.; Rosas, H. D.; Lucente, D.; Harrison, M. B.; Zanko, A.; Abramson, R. K.; Marder, K.; Sequeiros, J.; MacDonald, M. E.; Gusella, J. F. Common SNP-Based Haplotype Analysis of the 4p16.3 Huntington Disease Gene Region. Am. J. Hum. Genet. 2012, 90 (3), 434–444. [CrossRef]
- Landles, C.; Sathasivam, K.; Weiss, A.; Woodman, B.; Moffitt, H.; Finkbeiner, S.; Sun, B.; Gafni, J.; Ellerby, L. M.; Trottier, Y.; Richards, W. G.; Osmand, A.; Paganetti, P.; Bates, G. P. Proteolysis of Mutant Huntingtin Produces an Exon 1 Fragment That Accumulates as an Aggregated Protein in Neuronal Nuclei in Huntington Disease. J. Biol. Chem. 2010, 285 (12), 8808–8823. [CrossRef]
- Ha, A. D.; Fung, V. S. C. Huntingtonʼs Disease: Curr. Opin. Neurol. 2012, 25 (4), 491–498. [CrossRef]
- Ross, C. A.; Tabrizi, S. J. Huntington’s Disease: From Molecular Pathogenesis to Clinical Treatment. Lancet Neurol. 2011, 10 (1), 83–98. [CrossRef]
- G. Vonsattel, J. P.; DiFiglia, M. Huntington Disease: J. Neuropathol. Exp. Neurol. 1998, 57 (5), 369–384. [CrossRef]
- Moss, D. J. H.; Pardiñas, A. F.; Langbehn, D.; Lo, K.; Leavitt, B. R.; Roos, R.; Durr, A.; Mead, S.; Holmans, P.; Jones, L.; Tabrizi, S. J.; Coleman, A.; Santos, R. D.; Decolongon, J.; Sturrock, A.; Bardinet, E.; Ret, C. J.; Justo, D.; Lehericy, S.; Marelli, C.; Nigaud, K.; Valabrègue, R.; Van Den Bogaard, S.; Dumas, E. M.; Van Der Grond, J.; t’Hart, E.; Jurgens, C.; Witjes-Ane, M.-N.; Arran, N.; Callaghan, J.; Stopford, C.; Frost, C.; Jones, R.; Hobbs, N.; Lahiri, N.; Ordidge, R.; Owen, G.; Pepple, T.; Read, J.; Say, M.; Wild, E.; Patel, A.; Fox, N. C.; Gibbard, C.; Malone, I.; Crawford, H.; Whitehead, D.; Keenan, S.; Cash, D. M.; Berna, C.; Bechtel, N.; Bohlen, S.; Man, A. H.; Kraus, P.; Axelson, E.; Wang, C.; Acharya, T.; Lee, S.; Monaco, W.; Campbell, C.; Queller, S.; Whitlock, K.; Campbell, C.; Campbell, M.; Frajman, E.; Milchman, C.; O’Regan, A.; Labuschagne, I.; Stout, J.; Landwehrmeyer, B.; Craufurd, D.; Scahill, R.; Hicks, S.; Kennard, C.; Johnson, H.; Tobin, A.; Rosas, H.; Reilmann, R.; Borowsky, B.; Pourchot, C.; Andrews, S. C.; Bachoud-Lévi, A.-C.; Bentivoglio, A. R.; Biunno, I.; Bonelli, R.; Burgunder, J.-M.; Dunnett, S.; Ferreira, J.; Handley, O.; Heiberg, A.; Illmann, T.; Landwehrmeyer, G. B.; Levey, J.; Ramos-Arroyo, M. A.; Nielsen, J.; Koivisto, S. P.; Päivärinta, M.; Roos, R. A. C.; Sebastián, A. R.; Tabrizi, S.; Vandenberghe, W.; Verellen-Dumoulin, C.; Uhrova, T.; Wahlström, J.; Zaremba, J.; Baake, V.; Barth, K.; Garde, M. B.; Betz, S.; Bos, R.; Callaghan, J.; Come, A.; Guedes, L. C.; Ecker, D.; Finisterra, A. M.; Fullam, R.; Gilling, M.; Gustafsson, L.; Handley, O. J.; Hvalstedt, C.; Held, C.; Koppers, K.; Lamanna, C.; Laurà, M.; Descals, A. M.; Martinez-Horta, S.; Mestre, T.; Minster, S.; Monza, D.; Mütze, L.; Oehmen, M.; Orth, M.; Padieu, H.; Paterski, L.; Peppa, N.; Koivisto, S. P.; Di Renzo, M.; Rialland, A.; Røren, N.; Šašinková, P.; Timewell, E.; Townhill, J.; Cubillo, P. T.; Da Silva, W. V.; Van Walsem, M. R.; Whalstedt, C.; Witjes-Ané, M.-N.; Witkowski, G.; Wright, A.; Zielonka, D.; Zielonka, E.; Zinzi, P.; Bonelli, R. M.; Lilek, S.; Hecht, K.; Herranhof, B.; Holl, A.; Kapfhammer, H.-P.; Koppitz, M.; Magnet, M.; Müller, N.; Otti, D.; Painold, A.; Reisinger, K.; Scheibl, M.; Schöggl, H.; Ullah, J.; Braunwarth, E.-M.; Brugger, F.; Buratti, L.; Hametner, E.-M.; Hepperger, C.; Holas, C.; Hotter, A.; Hussl, A.; Müller, C.; Poewe, W.; Seppi, K.; Sprenger, F.; Wenning, G.; Boogaerts, A.; Calmeyn, G.; Delvaux, I.; Liessens, D.; Somers, N.; Dupuit, M.; Minet, C.; Van Paemel, D.; Ribaï, P.; Verellen-Dumoulin, C.; Boogaerts, A.; Vandenberghe, W.; Van Reijen, D.; Klempír, J.; Majerová, V.; Roth, J.; Stárková, I.; Hjermind, L. E.; Jacobsen, O.; Nielsen, J. E.; Larsen, I. U.; Vinther-Jensen, T.; Hiivola, H.; Hyppönen, H.; Martikainen, K.; Tuuha, K.; Allain, P.; Bonneau, D.; Bost, M.; Gohier, B.; Guérid, M.-A.; Olivier, A.; Prundean, A.; Scherer-Gagou, C.; Verny, C.; Babiloni, B.; Debruxelles, S.; Duché, C.; Goizet, C.; Jameau, L.; Lafoucrière, D.; Spampinato, U.; Barthélémy, R.; De Bruycker, C.; Carette, M. C. A.-S.; Defebvre, E. D. L.; Delliaux, M.; Delval, A.; Destee, A.; Dujardin, K.; Lemaire, M.-H.; Manouvrier, S.; Peter, M.; Plomhouse, L.; Sablonnière, B.; Simonin, C.; Thibault-Tanchou, S.; Vuillaume, I.; Bellonet, M.; Berrissoul, H.; Blin, S.; Courtin, F.; Duru, C.; Fasquel, V.; Godefroy, O.; Krystkowiak, P.; Mantaux, B.; Roussel, M.; Wannepain, S.; Azulay, J.-P.; Delfini, M.; Eusebio, A.; Fluchere, F.; Mundler, L.; Anheim, M.; Julié, C.; Boukbiza, O. L.; Longato, N.; Rudolf, G.; Tranchant, C.; Zimmermann, M.-A.; Kosinski, C. M.; Milkereit, E.; Probst, D.; Reetz, K.; Sass, C.; Schiefer, J.; Schlangen, C.; Werner, C. J.; Gelderblom, H.; Priller, J.; Prüß, H.; Spruth, E. J.; Ellrichmann, G.; Herrmann, L.; Hoffmann, R.; Kaminski, B.; Kotz, P.; Prehn, C.; Saft, C.; Lange, H.; Maiwald, R.; Löhle, M.; Maass, A.; Schmidt, S.; Bosredon, C.; Storch, A.; Wolz, A.; Wolz, M.; Capetian, P.; Lambeck, J.; Zucker, B.; Boelmans, K.; Ganos, C.; Heinicke, W.; Hidding, U.; Lewerenz, J.; Münchau, A.; Orth, M.; Schmalfeld, J.; Stubbe, L.; Zittel, S.; Diercks, G.; Dressler, D.; Gorzolla, H.; Schrader, C.; Tacik, P.; Ribbat, M.; Longinus, B.; Bürk, K.; Möller, J. C.; Rissling, I.; Mühlau, M.; Peinemann, A.; Städtler, M.; Weindl, A.; Winkelmann, J.; Ziegler, C.; Bechtel, N.; Beckmann, H.; Bohlen, S.; Hölzner, E.; Lange, H.; Reilmann, R.; Rohm, S.; Rumpf, S.; Schepers, S.; Weber, N.; Dose, M.; Leythäuser, G.; Marquard, R.; Raab, T.; Wiedemann, A.; Barth, K.; Buck, A.; Connemann, J.; Ecker, D.; Geitner, C.; Held, C.; Kesse, A.; Landwehrmeyer, B.; Lang, C.; Lewerenz, J.; Lezius, F.; Nepper, S.; Niess, A.; Orth, M.; Schneider, A.; Schwenk, D.; Süßmuth, S.; Trautmann, S.; Weydt, P.; Cormio, C.; Sciruicchio, V.; Serpino, C.; De Tommaso, M.; Capellari, S.; Cortelli, P.; Galassi, R.; Rizzo, G.; Poda, R.; Scaglione, C.; Bertini, E.; Ghelli, E.; Ginestroni, A.; Massaro, F.; Mechi, C.; Paganini, M.; Piacentini, S.; Pradella, S.; Romoli, A. M.; Sorbi, S.; Abbruzzese, G.; Di Poggio, M. B.; Ferrandes, G.; Mandich, P.; Marchese, R.; Albanese, A.; Di Bella, D.; Castaldo, A.; Di Donato, S.; Gellera, C.; Genitrini, S.; Mariotti, C.; Monza, D.; Nanetti, L.; Paridi, D.; Soliveri, P.; Tomasello, C.; De Michele, G.; Di Maio, L.; Massarelli, M.; Peluso, S.; Roca, A.; Russo, C. V.; Salvatore, E.; Sorrentino, P.; Amico, E.; Favellato, M.; Griguoli, A.; Mazzante, I.; Petrollini, M.; Squitieri, F.; D’Alessio, B.; Esposito, C.; Bentivoglio, R.; Frontali, M.; Guidubaldi, A.; Ialongo, T.; Jacopini, G.; Piano, C.; Romano, S.; Soleti, F.; Spadaro, M.; Zinzi, P.; Van Hout, M. S. E.; Verhoeven, M. E.; Van Vugt, J. P. P.; De Weert, A. M.; Bolwijn, J. J. W.; Dekker, M.; Kremer, B.; Leenders, K. L.; Van Oostrom, J. C. H.; Van Den Bogaard, S. J. A.; Bos, R.; Dumas, E. M.; ’T Hart, E. P.; Roos, R. A. C.; Kremer, B.; Verstappen, C. C. P.; Aaserud, O.; C, J. F.; Heiberg, A.; Van Walsem, M. R.; Wehus, R.; Bjørgo, K.; Fannemel, M.; Gørvell, P. F.; Lorentzen, E.; Koivisto, S. P.; Retterstøl, L.; Stokke, B.; Bjørnevoll, I.; Sando, S. B.; Dziadkiewicz, A.; Nowak, M.; Robowski, P.; Sitek, E.; Slawek, J.; Soltan, W.; Szinwelski, M.; Blaszcyk, M.; Boczarska-Jedynak, M.; Ciach-Wysocka, E.; Gorzkowska, A.; Jasinska-Myga, B.; Klodowska-Duda, G.; Opala, G.; Stompel, D.; Banaszkiewicz, K.; Bocwinska, D.; Bojakowska-Jaremek, K.; Dec, M.; Krawczyk, M.; Rudzinska, M.; Szczygiel, E.; Szczudlik, A.; Wasielewska, A.; Wójcik, M.; Bryl, A.; Ciesielska, A.; Klimberg, A.; Marcinkowski, J.; Samara, H.; Sempolowicz, J.; Zielonka, D.; Gogol, A.; Janik, P.; Kwiecinski, H.; Jamrozik, Z.; Antczak, J.; Jachinska, K.; Krysa, W.; Rakowicz, M.; Richter, P.; Rola, R.; Ryglewicz, D.; Sienkiewicz-Jarosz, H.; Stepniak, I.; Sulek, A.; Witkowski, G.; Zaremba, J.; Zdzienicka, E.; Zieora-Jakutowicz, K.; Ferreira, J. J.; Coelho, M.; Guedes, L. C.; Mendes, T.; Mestre, T.; Valadas, A.; Andrade, C.; Gago, M.; Garrett, C.; Guerra, M. R.; Herrera, C. D.; Garcia, P. M.; Barbera, M. A.; Guia, D. B.; Hernanz, L. C.; Catena, J. L.; Ferrer, P. Q.; Sebastián, A. R.; Carruesco, G. T.; Bas, J.; Busquets, N.; Calopa, M.; Robert, M. F.; Viladrich, C. M.; Idiago, J. M. R.; Riballo, A. V.; Cubo, E.; Polo, C. G.; Mariscal, N.; Rivadeneyra, P. J.; Barrero, F.; Morales, B.; Fenollar, M.; García, R. G.-R.; Ortega, P.; Villanueva, C.; Alegre, J.; Bascuñana, M.; Caldentey, J. G.; Ventura, M. F.; Ribas, G. G.; De Yébenes, J. G.; Moreno, J. L. L.-S.; Cubillo, P. T.; Alegre, J.; Frech, F. A.; De Yébenes, J. G.; Ruíz, P. J. G.; Martínez-Descals, A.; Guerrero, R.; Artiga, M. J. S.; Sánchez, V.; Perea, M. F. N.; Fortuna, L.; Manzanares, S.; Reinante, G.; Torres, M. M. A.; Moreau, L. V.; González González, S.; Guisasola, L. M.; Salvador, C.; Martín, E. S. S.; Ramirez, I. L.; Gorospe, A.; Lopera, M. R.; Arques, P. N.; Rodríguez, M. J. T.; Pastor, B. V.; Gaston, I.; Martinez-Jaurrieta, M. D.; Ramos-Arroyo, M. A.; Moreno, J. M. G.; Lucena, C. M.; Damas, F.; Cortegana, H. E. P.; Peña, J. C.; Redondo, L.; Carrillo, F.; Teresa Cáceres, M.; Mir, P.; Suarez, M. J. L.; Vargas-González, L.; Bosca, M. E.; Brugada, F. C.; Burguera, J. A.; Campos, A.; Vilaplana, G. C. P.; Berglund, P.; Constantinescu, R.; Fredlund, G.; Høsterey-Ugander, U.; Linnsand, P.; Neleborn-Lingefjärd, L.; Wahlström, J.; Wentzel, M.; Loutfi, G.; Olofsson, C.; Stattin, E.-L.; Westman, L.; Wikström, B.; Burgunder, J.-M.; Stebler, Y.; Kaelin, A.; Romero, I.; Schüpbach, M.; Weber Zaugg, S.; Hauer, M.; Gonzenbach, R.; Jung, H. H.; Mihaylova, V.; Petersen, J.; Jack, R.; Matheson, K.; Miedzybrodzka, Z.; Rae, D.; Simpson, S. A.; Summers, F.; Ure, A.; Vaughan, V.; Akhtar, S.; Crooks, J.; Curtis, A.; De Souza, J.; Piedad, J.; Rickards, H.; Wright, J.; Coulthard, E.; Gethin, L.; Hayward, B.; Sieradzan, K.; Wright, A.; Armstrong, M.; Barker, R. A.; O’Keefe, D.; Di Pietro, A.; Fisher, K.; Goodman, A.; Hill, S.; Kershaw, A.; Mason, S.; Paterson, N.; Raymond, L.; Swain, R.; Guzman, N. V.; Busse, M.; Butcher, C.; Callaghan, J.; Dunnett, S.; Clenaghan, C.; Fullam, R.; Handley, O.; Hunt, S.; Jones, L.; Jones, U.; Khalil, H.; Minster, S.; Owen, M.; Price, K.; Rosser, A.; Townhill, J.; Edwards, M.; Ho, C.; Hughes, T.; McGill, M.; Pearson, P.; Porteous, M.; Smith, P.; Brockie, P.; Foster, J.; Johns, N.; McKenzie, S.; Rothery, J.; Thomas, G.; Yates, S.; Burrows, L.; Chu, C.; Fletcher, A.; Gallantrae, D.; Hamer, S.; Harding, A.; Klöppel, S.; Kraus, A.; Laver, F.; Lewis, M.; Longthorpe, M.; Markova, I.; Raman, A.; Robertson, N.; Silva, M.; Thomson, A.; Wild, S.; Yardumian, P.; Chu, C.; Evans, C.; Gallentrae, D.; Hamer, S.; Kraus, A.; Markova, I.; Raman, A.; Chu, C.; Hamer, S.; Hobson, E.; Jamieson, S.; Kraus, A.; Markova, I.; Raman, A.; Musgrave, H.; Rowett, L.; Toscano, J.; Wild, S.; Yardumian, P.; Bourne, C.; Clapton, J.; Clayton, C.; Dipple, H.; Freire-Patino, D.; Grant, J.; Gross, D.; Hallam, C.; Middleton, J.; Murch, A.; Thompson, C.; Alusi, S.; Davies, R.; Foy, K.; Gerrans, E.; Pate, L.; Andrews, T.; Dougherty, A.; Golding, C.; Kavalier, F.; Laing, H.; Lashwood, A.; Robertson, D.; Ruddy, D.; Santhouse, A.; Whaite, A.; Andrews, T.; Bruno, S.; Doherty, K.; Golding, C.; Haider, S.; Hensman, D.; Lahiri, N.; Lewis, M.; Novak, M.; Patel, A.; Robertson, N.; Rosser, E.; Tabrizi, S.; Taylor, R.; Warner, T.; Wild, E.; Arran, N.; Bek, J.; Callaghan, J.; Craufurd, D.; Fullam, R.; Hare, M.; Howard, L.; Huson, S.; Johnson, L.; Jones, M.; Murphy, H.; Oughton, E.; Partington-Jones, L.; Rogers, D.; Sollom, A.; Snowden, J.; Stopford, C.; Thompson, J.; Trender-Gerhard, I.; Verstraelen, N.; Westmoreland, L.; Armstrong, R.; Dixon, K.; Nemeth, A. H.; Siuda, G.; Valentine, R.; Harrison, D.; Hughes, M.; Parkinson, A.; Soltysiak, B.; Bandmann, O.; Bradbury, A.; Gill, P.; Fairtlough, H.; Fillingham, K.; Foustanos, I.; Kazoka, M.; O’Donovan, K.; Peppa, N.; Taylor, C.; Tidswell, K.; Quarrell, O.; Burgunder, J.-M.; Lau, P. N.; Pica, E.; Tan, L. Identification of Genetic Variants Associated with Huntington’s Disease Progression: A Genome-Wide Association Study. Lancet Neurol. 2017, 16 (9), 701–711. [CrossRef]
- Travessa, A. M.; Rodrigues, F. B.; Mestre, T. A.; Ferreira, J. J. Fifteen Years of Clinical Trials in Huntington’s Disease: A Very Low Clinical Drug Development Success Rate. J. Huntingt. Dis. 2017, 6 (2), 157–163. [CrossRef]
- Kordasiewicz, H. B.; Stanek, L. M.; Wancewicz, E. V.; Mazur, C.; McAlonis, M. M.; Pytel, K. A.; Artates, J. W.; Weiss, A.; Cheng, S. H.; Shihabuddin, L. S.; Hung, G.; Bennett, C. F.; Cleveland, D. W. Sustained Therapeutic Reversal of Huntington’s Disease by Transient Repression of Huntingtin Synthesis. Neuron 2012, 74 (6), 1031–1044. [CrossRef]
- Agustín-Pavón, C.; Mielcarek, M.; Garriga-Canut, M.; Isalan, M. Deimmunization for Gene Therapy: Host Matching of Synthetic Zinc Finger Constructs Enables Long-Term Mutant Huntingtin Repression in Mice. Mol. Neurodegener. 2016, 11 (1), 64. [CrossRef]
- Yang, L.; Calingasan, N. Y.; Wille, E. J.; Cormier, K.; Smith, K.; Ferrante, R. J.; Flint Beal, M. Combination Therapy with Coenzyme Q 10 and Creatine Produces Additive Neuroprotective Effects in Models of Parkinson’s and Huntington’s Diseases. J. Neurochem. 2009, 109 (5), 1427–1439. [CrossRef]
- McGarry, A.; Auinger, P.; Kieburtz, K. D.; Bredlau, A.-L.; Hersch, S. M.; Rosas, H. D. Suicidality Risk Factors Across the CARE-HD, 2CARE, and CREST-E Clinical Trials in Huntington Disease. Neurol. Clin. Pract. 2022, 12 (2), 131–138. [CrossRef]
- Basavarajappa, B. S.; Subbanna, S. Histone Methylation Regulation in Neurodegenerative Disorders. Int. J. Mol. Sci. 2021, 22 (9), 4654. [CrossRef]
- Masrori, P.; Van Damme, P. Amyotrophic Lateral Sclerosis: A Clinical Review. Eur. J. Neurol. 2020, 27 (10), 1918–1929. [CrossRef]
- Renton, A. E.; Majounie, E.; Waite, A.; Simón-Sánchez, J.; Rollinson, S.; Gibbs, J. R.; Schymick, J. C.; Laaksovirta, H.; van Swieten, J. C.; Myllykangas, L.; Kalimo, H.; Paetau, A.; Abramzon, Y.; Remes, A. M.; Kaganovich, A.; Scholz, S. W.; Duckworth, J.; Ding, J.; Harmer, D. W.; Hernandez, D. G.; Johnson, J. O.; Mok, K.; Ryten, M.; Trabzuni, D.; Guerreiro, R. J.; Orrell, R. W.; Neal, J.; Murray, A.; Pearson, J.; Jansen, I. E.; Sondervan, D.; Seelaar, H.; Blake, D.; Young, K.; Halliwell, N.; Callister, J. B.; Toulson, G.; Richardson, A.; Gerhard, A.; Snowden, J.; Mann, D.; Neary, D.; Nalls, M. A.; Peuralinna, T.; Jansson, L.; Isoviita, V.-M.; Kaivorinne, A.-L.; Hölttä-Vuori, M.; Ikonen, E.; Sulkava, R.; Benatar, M.; Wuu, J.; Chiò, A.; Restagno, G.; Borghero, G.; Sabatelli, M.; Heckerman, D.; Rogaeva, E.; Zinman, L.; Rothstein, J. D.; Sendtner, M.; Drepper, C.; Eichler, E. E.; Alkan, C.; Abdullaev, Z.; Pack, S. D.; Dutra, A.; Pak, E.; Hardy, J.; Singleton, A.; Williams, N. M.; Heutink, P.; Pickering-Brown, S.; Morris, H. R.; Tienari, P. J.; Traynor, B. J. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72 (2), 257–268. [CrossRef]
- Woollacott, I. O. C.; Mead, S. The C9ORF72 Expansion Mutation: Gene Structure, Phenotypic and Diagnostic Issues. Acta Neuropathol. (Berl.) 2014, 127 (3), 319–332. [CrossRef]
- Balendra, R.; Isaacs, A. M. C9orf72-Mediated ALS and FTD: Multiple Pathways to Disease. Nat. Rev. Neurol. 2018, 14 (9), 544–558. [CrossRef]
- Herdewyn, S.; Zhao, H.; Moisse, M.; Race, V.; Matthijs, G.; Reumers, J.; Kusters, B.; Schelhaas, H. J.; van den Berg, L. H.; Goris, A.; Robberecht, W.; Lambrechts, D.; Van Damme, P. Whole-Genome Sequencing Reveals a Coding Non-Pathogenic Variant Tagging a Non-Coding Pathogenic Hexanucleotide Repeat Expansion in C9orf72 as Cause of Amyotrophic Lateral Sclerosis. Hum. Mol. Genet. 2012, 21 (11), 2412–2419. [CrossRef]
- Majounie, E.; Renton, A. E.; Mok, K.; Dopper, E. G.; Waite, A.; Rollinson, S.; Chiò, A.; Restagno, G.; Nicolaou, N.; Simon-Sanchez, J.; Van Swieten, J. C.; Abramzon, Y.; Johnson, J. O.; Sendtner, M.; Pamphlett, R.; Orrell, R. W.; Mead, S.; Sidle, K. C.; Houlden, H.; Rohrer, J. D.; Morrison, K. E.; Pall, H.; Talbot, K.; Ansorge, O.; Hernandez, D. G.; Arepalli, S.; Sabatelli, M.; Mora, G.; Corbo, M.; Giannini, F.; Calvo, A.; Englund, E.; Borghero, G.; Floris, G. L.; Remes, A. M.; Laaksovirta, H.; McCluskey, L.; Trojanowski, J. Q.; Van Deerlin, V. M.; Schellenberg, G. D.; Nalls, M. A.; Drory, V. E.; Lu, C.-S.; Yeh, T.-H.; Ishiura, H.; Takahashi, Y.; Tsuji, S.; Le Ber, I.; Brice, A.; Drepper, C.; Williams, N.; Kirby, J.; Shaw, P.; Hardy, J.; Tienari, P. J.; Heutink, P.; Morris, H. R.; Pickering-Brown, S.; Traynor, B. J. Frequency of the C9orf72 Hexanucleotide Repeat Expansion in Patients with Amyotrophic Lateral Sclerosis and Frontotemporal Dementia: A Cross-Sectional Study. Lancet Neurol. 2012, 11 (4), 323–330. [CrossRef]
- Xi, Z.; Zinman, L.; Grinberg, Y.; Moreno, D.; Sato, C.; Bilbao, J. M.; Ghani, M.; Hernández, I.; Ruiz, A.; Boada, M.; Morón, F. J.; Lang, A. E.; Marras, C.; Bruni, A.; Colao, R.; Maletta, R. G.; Puccio, G.; Rainero, I.; Pinessi, L.; Galimberti, D.; Morrison, K. E.; Moorby, C.; Stockton, J. D.; Masellis, M.; Black, S. E.; Hazrati, L.-N.; Liang, Y.; Van Haersma De With, J.; Fornazzari, L.; Villagra, R.; Rojas-Garcia, R.; Clarimón, J.; Mayeux, R.; Robertson, J.; St George-Hyslop, P.; Rogaeva, E. Investigation of C9orf72 in 4 Neurodegenerative Disorders. Arch. Neurol. 2012, 69 (12), 1583. [CrossRef]
- Česnik, A. B.; Darovic, S.; Mihevc, S. P.; Štalekar, M.; Malnar, M.; Motaln, H.; Lee, Y.-B.; Mazej, J.; Pohleven, J.; Grosch, M.; Modic, M.; Fonovič, M.; Turk, B.; Drukker, M.; Shaw, C. E.; Rogelj, B. Nuclear RNA Foci from C9ORF72 Expansion Mutation Form Paraspeckle-like Bodies. J. Cell Sci. 2019, jcs.224303. [CrossRef]
- Saccon, R. A.; Bunton-Stasyshyn, R. K. A.; Fisher, E. M. C.; Fratta, P. Is SOD1 Loss of Function Involved in Amyotrophic Lateral Sclerosis? Brain 2013, 136 (8), 2342–2358. [CrossRef]
- Bosco, D. A.; Morfini, G.; Karabacak, N. M.; Song, Y.; Gros-Louis, F.; Pasinelli, P.; Goolsby, H.; Fontaine, B. A.; Lemay, N.; McKenna-Yasek, D.; Frosch, M. P.; Agar, J. N.; Julien, J.-P.; Brady, S. T.; Brown, R. H. Wild-Type and Mutant SOD1 Share an Aberrant Conformation and a Common Pathogenic Pathway in ALS. Nat. Neurosci. 2010, 13 (11), 1396–1403. [CrossRef]
- Bruijn, L. I.; Houseweart, M. K.; Kato, S.; Anderson, K. L.; Anderson, S. D.; Ohama, E.; Reaume, A. G.; Scott, R. W.; Cleveland, D. W. Aggregation and Motor Neuron Toxicity of an ALS-Linked SOD1 Mutant Independent from Wild-Type SOD1. Science 1998, 281 (5384), 1851–1854. [CrossRef]
- Saeed, M.; Yang, Y.; Deng, H.-X.; Hung, W.-Y.; Siddique, N.; Dellefave, L.; Gellera, C.; Andersen, P. M.; Siddique, T. Age and Founder Effect of SOD1 A4V Mutation Causing ALS. Neurology 2009, 72 (19), 1634–1639. [CrossRef]
- Hensley, K.; Mhatre, M.; Mou, S.; Pye, Q. N.; Stewart, C.; West, M.; Williamson, K. S. On the Relation of Oxidative Stress to Neuroinflammation: Lessons Learned from the G93A-SOD1 Mouse Model of Amyotrophic Lateral Sclerosis. Antioxid. Redox Signal. 2006, 8 (11–12), 2075–2087. [CrossRef]
- Curti, D.; Rognoni, F.; Alimonti, D.; Malaspina, A.; Feletti, F.; Tessera, S.; Finotti, N.; Rehak, L.; Mazzini, L.; Zerbi, F.; Poloni, T.; Ceroni, M. SOD1 Activity and Protective Factors in Familial ALS Patients with L84F SOD1 Mutation. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2002, 3 (3), 115–122. [CrossRef]
- Ceroni, M.; Malaspina, A.; Poloni, T. E.; Alimonti, D.; Rognoni, F.; Habgood, J.; Imbesi, F.; Antonelli, P.; Alfonsi, E.; Curti, D.; deBelleroche, J. Clustering of ALS Patients in Central Italy Due to the Occurrence of the L84F SOD1 Gene Mutation. Neurology 1999, 53 (5), 1064–1064. [CrossRef]
- Juneja, T.; Pericak-Vance, M. A.; Laing, N. G.; Dave, S.; Siddique, T. Prognosis in Familial Amyotrophic Lateral Sclerosis: Progression and Survival in Patients with Glu100gly and Ala4val Mutations in Cu,Zn Superoxide Dismutase. Neurology 1997, 48 (1), 55–57. [CrossRef]
- Zhou, Y.; Tang, J.; Lan, J.; Zhang, Y.; Wang, H.; Chen, Q.; Kang, Y.; Sun, Y.; Feng, X.; Wu, L.; Jin, H.; Chen, S.; Peng, Y. Honokiol Alleviated Neurodegeneration by Reducing Oxidative Stress and Improving Mitochondrial Function in Mutant SOD1 Cellular and Mouse Models of Amyotrophic Lateral Sclerosis. Acta Pharm. Sin. B 2023, 13 (2), 577–597. [CrossRef]
- Cassina, P.; Cassina, A.; Pehar, M.; Castellanos, R.; Gandelman, M.; De León, A.; Robinson, K. M.; Mason, R. P.; Beckman, J. S.; Barbeito, L.; Radi, R. Mitochondrial Dysfunction in SOD1 G93A -Bearing Astrocytes Promotes Motor Neuron Degeneration: Prevention by Mitochondrial-Targeted Antioxidants. J. Neurosci. 2008, 28 (16), 4115–4122. [CrossRef]
- Zhang, X.; Li, L.; Chen, S.; Yang, D.; Wang, Y.; Zhang, X.; Wang, Z.; Le, W. Rapamycin Treatment Augments Motor Neuron Degeneration in SOD1 G93A Mouse Model of Amyotrophic Lateral Sclerosis. Autophagy 2011, 7 (4), 412–425. [CrossRef]
- Daoud, H.; Valdmanis, P. N.; Kabashi, E.; Dion, P.; Dupre, N.; Camu, W.; Meininger, V.; Rouleau, G. A. Contribution of TARDBP Mutations to Sporadic Amyotrophic Lateral Sclerosis. J. Med. Genet. 2008, 46 (2), 112–114. [CrossRef]
- Lattante, S.; Rouleau, G. A.; Kabashi, E. TARDBP and FUS Mutations Associated with Amyotrophic Lateral Sclerosis: Summary and Update. Hum. Mutat. 2013, 34 (6), 812–826. [CrossRef]
- Kabashi, E.; Valdmanis, P. N.; Dion, P.; Spiegelman, D.; McConkey, B. J.; Velde, C. V.; Bouchard, J.-P.; Lacomblez, L.; Pochigaeva, K.; Salachas, F.; Pradat, P.-F.; Camu, W.; Meininger, V.; Dupre, N.; Rouleau, G. A. TARDBP Mutations in Individuals with Sporadic and Familial Amyotrophic Lateral Sclerosis. Nat. Genet. 2008, 40 (5), 572–574. [CrossRef]
- Corcia, P.; Valdmanis, P.; Millecamps, S.; Lionnet, C.; Blasco, H.; Mouzat, K.; Daoud, H.; Belzil, V.; Morales, R.; Pageot, N.; Danel-Brunaud, V.; Vandenberghe, N.; Pradat, P. F.; Couratier, P.; Salachas, F.; Lumbroso, S.; Rouleau, G. A.; Meininger, V.; Camu, W. Phenotype and Genotype Analysis in Amyotrophic Lateral Sclerosis with TARDBP Gene Mutations. Neurology 2012, 78 (19), 1519–1526. [CrossRef]
- Tamaoka, A.; Arai, M.; Itokawa, M.; Arai, T.; Hasegawa, M.; Tsuchiya, K.; Takuma, H.; Tsuji, H.; Ishii, A.; Watanabe, M.; Takahashi, Y.; Goto, J.; Tsuji, S.; Akiyama, H. TDP-43 M337V Mutation in Familial Amyotrophic Lateral Sclerosis in Japan. Intern. Med. 2010, 49 (4), 331–334. [CrossRef]
- Watanabe, S.; Oiwa, K.; Murata, Y.; Komine, O.; Sobue, A.; Endo, F.; Takahashi, E.; Yamanaka, K. ALS-Linked TDP-43M337V Knock-in Mice Exhibit Splicing Deregulation without Neurodegeneration. Mol. Brain 2020, 13 (1), 8. [CrossRef]
- Esmaeili, M. A.; Panahi, M.; Yadav, S.; Hennings, L.; Kiaei, M. Premature Death of TDP-43 (A315T) Transgenic Mice Due to Gastrointestinal Complications Prior to Development of Full Neurological Symptoms of Amyotrophic Lateral Sclerosis. Int. J. Exp. Pathol. 2013, 94 (1), 56–64. [CrossRef]
- Orrù, S.; Coni, P.; Floris, A.; Littera, R.; Carcassi, C.; Sogos, V.; Brancia, C. Reduced Stress Granule Formation and Cell Death in Fibroblasts with the A382T Mutation of TARDBP Gene: Evidence for Loss of TDP-43 Nuclear Function. Hum. Mol. Genet. 2016, ddw276. [CrossRef]
- Zanini, G.; Selleri, V.; Nasi, M.; De Gaetano, A.; Martinelli, I.; Gianferrari, G.; Lofaro, F. D.; Boraldi, F.; Mandrioli, J.; Pinti, M. Mitochondrial and Endoplasmic Reticulum Alterations in a Case of Amyotrophic Lateral Sclerosis Caused by TDP-43 A382T Mutation. Int. J. Mol. Sci. 2022, 23 (19), 11881. [CrossRef]
- Deng, H.; Gao, K.; Jankovic, J. The Role of FUS Gene Variants in Neurodegenerative Diseases. Nat. Rev. Neurol. 2014, 10 (6), 337–348. [CrossRef]
- Corrado, L.; Del Bo, R.; Castellotti, B.; Ratti, A.; Cereda, C.; Penco, S.; Soraru, G.; Carlomagno, Y.; Ghezzi, S.; Pensato, V.; Colombrita, C.; Gagliardi, S.; Cozzi, L.; Orsetti, V.; Mancuso, M.; Siciliano, G.; Mazzini, L.; Comi, G. P.; Gellera, C.; Ceroni, M.; D’Alfonso, S.; Silani, V. Mutations of FUS Gene in Sporadic Amyotrophic Lateral Sclerosis. J. Med. Genet. 2010, 47 (3), 190–194. [CrossRef]
- Rademakers, R.; Stewart, H.; Dejesus-Hernandez, M.; Krieger, C.; Graff-Radford, N.; Fabros, M.; Briemberg, H.; Cashman, N.; Eisen, A.; Mackenzie, I. R. A. Fus Gene Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Muscle Nerve 2010, 42 (2), 170–176. [CrossRef]
- Ticozzi, N.; Silani, V.; LeClerc, A. L.; Keagle, P.; Gellera, C.; Ratti, A.; Taroni, F.; Kwiatkowski, T. J.; McKenna-Yasek, D. M.; Sapp, P. C.; Brown, R. H.; Landers, J. E. Analysis of FUS Gene Mutation in Familial Amyotrophic Lateral Sclerosis within an Italian Cohort. Neurology 2009, 73 (15), 1180–1185. [CrossRef]
- Weinreich, M.; Shepheard, S. R.; Verber, N.; Wyles, M.; Heath, P. R.; Highley, J. R.; Kirby, J.; Shaw, P. J. Neuropathological Characterization of a Novel TANK Binding Kinase ( TBK1 ) Gene Loss of Function Mutation Associated with Amyotrophic Lateral Sclerosis. Neuropathol. Appl. Neurobiol. 2020, 46 (3), 279–291. [CrossRef]
- Oakes, J. A.; Davies, M. C.; Collins, M. O. TBK1: A New Player in ALS Linking Autophagy and Neuroinflammation. Mol. Brain 2017, 10 (1), 5. [CrossRef]
- Gurfinkel, Y.; Polain, N.; Sonar, K.; Nice, P.; Mancera, R. L.; Rea, S. L. Functional and Structural Consequences of TBK1 Missense Variants in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Neurobiol. Dis. 2022, 174, 105859. [CrossRef]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A Comprehensive Review of Amyotrophic Lateral Sclerosis. Surg. Neurol. Int. 2015, 6 (1), 171. [CrossRef]
- Deans, C.; Maggert, K. A. What Do You Mean, “Epigenetic”? Genetics 2015, 199 (4), 887–896. [CrossRef]
- Burggren, W. Epigenetic Inheritance and Its Role in Evolutionary Biology: Re-Evaluation and New Perspectives. Biology 2016, 5 (2), 24. [CrossRef]
- Neumann, M.; Sampathu, D. M.; Kwong, L. K.; Truax, A. C.; Micsenyi, M. C.; Chou, T. T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C. M.; McCluskey, L. F.; Miller, B. L.; Masliah, E.; Mackenzie, I. R.; Feldman, H.; Feiden, W.; Kretzschmar, H. A.; Trojanowski, J. Q.; Lee, V. M.-Y. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 2006, 314 (5796), 130–133. [CrossRef]
- Fecto, F. SQSTM1 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Arch. Neurol. 2011, 68 (11), 1440. [CrossRef]
- Maruyama, H.; Morino, H.; Ito, H.; Izumi, Y.; Kato, H.; Watanabe, Y.; Kinoshita, Y.; Kamada, M.; Nodera, H.; Suzuki, H.; Komure, O.; Matsuura, S.; Kobatake, K.; Morimoto, N.; Abe, K.; Suzuki, N.; Aoki, M.; Kawata, A.; Hirai, T.; Kato, T.; Ogasawara, K.; Hirano, A.; Takumi, T.; Kusaka, H.; Hagiwara, K.; Kaji, R.; Kawakami, H. Mutations of Optineurin in Amyotrophic Lateral Sclerosis. Nature 2010, 465 (7295), 223–226. [CrossRef]
- Johnson, J. O.; Mandrioli, J.; Benatar, M.; Abramzon, Y.; Van Deerlin, V. M.; Trojanowski, J. Q.; Gibbs, J. R.; Brunetti, M.; Gronka, S.; Wuu, J.; Ding, J.; McCluskey, L.; Martinez-Lage, M.; Falcone, D.; Hernandez, D. G.; Arepalli, S.; Chong, S.; Schymick, J. C.; Rothstein, J.; Landi, F.; Wang, Y.-D.; Calvo, A.; Mora, G.; Sabatelli, M.; Monsurrò, M. R.; Battistini, S.; Salvi, F.; Spataro, R.; Sola, P.; Borghero, G.; Galassi, G.; Scholz, S. W.; Taylor, J. P.; Restagno, G.; Chiò, A.; Traynor, B. J. Exome Sequencing Reveals VCP Mutations as a Cause of Familial ALS. Neuron 2010, 68 (5), 857–864. [CrossRef]
- Boeynaems, S.; Bogaert, E.; Van Damme, P.; Van Den Bosch, L. Inside out: The Role of Nucleocytoplasmic Transport in ALS and FTLD. Acta Neuropathol. (Berl.) 2016, 132 (2), 159–173. [CrossRef]
- Vilarino-Guell, C.; Wider, C.; Soto-Ortolaza, A. I.; Cobb, S. A.; Kachergus, J. M.; Keeling, B. H.; Dachsel, J. C.; Hulihan, M. M.; Dickson, D. W.; Wszolek, Z. K.; Uitti, R. J.; Graff-Radford, N. R.; Boeve, B. F.; Josephs, K. A.; Miller, B.; Boylan, K. B.; Gwinn, K.; Adler, C. H.; Aasly, J. O.; Hentati, F.; Destee, A.; Krygowska-Wajs, A.; Chartier-Harlin, M.-C.; Ross, O. A.; Rademakers, R.; Farrer, M. J. Characterization of DCTN1 Genetic Variability in Neurodegeneration. Neurology 2009, 72 (23), 2024–2028. [CrossRef]
- Araki, E.; Tsuboi, Y.; Daechsel, J.; Milnerwood, A.; Vilarino-Guell, C.; Fujii, N.; Mishima, T.; Oka, T.; Hara, H.; Fukae, J.; Farrer, M. J. A Novel DCTN1 Mutation with Late-Onset Parkinsonism and Frontotemporal Atrophy: A Novel DCTN1 Mutation. Mov. Disord. 2014, 29 (9), 1201–1204. [CrossRef]
- Al-Chalabi, A.; Hardiman, O.; Kiernan, M. C.; Chiò, A.; Rix-Brooks, B.; Van Den Berg, L. H. Amyotrophic Lateral Sclerosis: Moving towards a New Classification System. Lancet Neurol. 2016, 15 (11), 1182–1194. [CrossRef]
- Chio, A.; Calvo, A.; Moglia, C.; Mazzini, L.; Mora, G.; PARALS study group. Phenotypic Heterogeneity of Amyotrophic Lateral Sclerosis: A Population Based Study. J. Neurol. Neurosurg. Psychiatry 2011, 82 (7), 740–746. [CrossRef]
- Finegan, E.; Chipika, R. H.; Li Hi Shing, S.; Hardiman, O.; Bede, P. Pathological Crying and Laughing in Motor Neuron Disease: Pathobiology, Screening, Intervention. Front. Neurol. 2019, 10, 260. [CrossRef]
- Wijesekera, L. C.; Mathers, S.; Talman, P.; Galtrey, C.; Parkinson, M. H.; Ganesalingam, J.; Willey, E.; Ampong, M. A.; Ellis, C. M.; Shaw, C. E.; Al-Chalabi, A.; Leigh, P. N. Natural History and Clinical Features of the Flail Arm and Flail Leg ALS Variants. Neurology 2009, 72 (12), 1087–1094. [CrossRef]
- Doble, A. The Pharmacology and Mechanism of Action of Riluzole. Neurology 1996, 47 (Issue 6, Supplement 4), 233S-241S. [CrossRef]
- Amyotrophic Lateral Sclerosis/Riluzole Study Group II; Lacomblez, L.; Bensimon, G.; Meininger, V.; Leigh, P. N.; Guillet, P. Dose-Ranging Study of Riluzole in Amyotrophic Lateral Sclerosis. The Lancet 1996, 347 (9013), 1425–1431. [CrossRef]
- Hinchcliffe, M.; Smith, A. Riluzole: Real-World Evidence Supports Significant Extension of Median Survival Times in Patients with Amyotrophic Lateral Sclerosis. Degener. Neurol. Neuromuscul. Dis. 2017, Volume 7, 61–70. [CrossRef]
- Mora, J. S.; Genge, A.; Chio, A.; Estol, C. J.; Chaverri, D.; Hernández, M.; Marín, S.; Mascias, J.; Rodriguez, G. E.; Povedano, M.; Paipa, A.; Dominguez, R.; Gamez, J.; Salvado, M.; Lunetta, C.; Ballario, C.; Riva, N.; Mandrioli, J.; Moussy, A.; Kinet, J.-P.; Auclair, C.; Dubreuil, P.; Arnold, V.; Mansfield, C. D.; Hermine, O.; on behalf of the AB10015 STUDY GROUP. Masitinib as an Add-on Therapy to Riluzole in Patients with Amyotrophic Lateral Sclerosis: A Randomized Clinical Trial. Amyotroph. Lateral Scler. Front. Degener. 2020, 21 (1–2), 5–14. [CrossRef]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C. E.; Cummings, J.; Van Der Flier, W. M. Alzheimer’s Disease. The Lancet 2021, 397 (10284), 1577–1590. [CrossRef]
- Bloem, B. R.; Henderson, E. J.; Dorsey, E. R.; Okun, M. S.; Okubadejo, N.; Chan, P.; Andrejack, J.; Darweesh, S. K. L.; Munneke, M. Integrated and Patient-Centred Management of Parkinson’s Disease: A Network Model for Reshaping Chronic Neurological Care. Lancet Neurol. 2020, 19 (7), 623–634. [CrossRef]
- Tabrizi, S. J.; Flower, M. D.; Ross, C. A.; Wild, E. J. Huntington Disease: New Insights into Molecular Pathogenesis and Therapeutic Opportunities. Nat. Rev. Neurol. 2020, 16 (10), 529–546. [CrossRef]
- Deda, H.; Inci, M.; Kürekçi, A.; Sav, A.; Kayıhan, K.; Özgün, E.; Üstünsoy, G.; Kocabay, S. Treatment of Amyotrophic Lateral Sclerosis Patients by Autologous Bone Marrow-Derived Hematopoietic Stem Cell Transplantation: A 1-Year Follow-Up. Cytotherapy 2009, 11 (1), 18–25. [CrossRef]
- Moviglia, G. A.; Moviglia-Brandolino, M. T.; Varela, G. S.; Albanese, G.; Piccone, S.; Echegaray, G.; Martinez, G.; Blasseti, N.; Farias, J.; Farina, P.; Perusso, A.; Gaeta, C. A. Feasibility, Safety, and Preliminary Proof of Principles of Autologous Neural Stem Cell Treatment Combined with T-Cell Vaccination for ALS Patients. Cell Transplant. 2012, 21 (1_suppl), 57–63. [CrossRef]
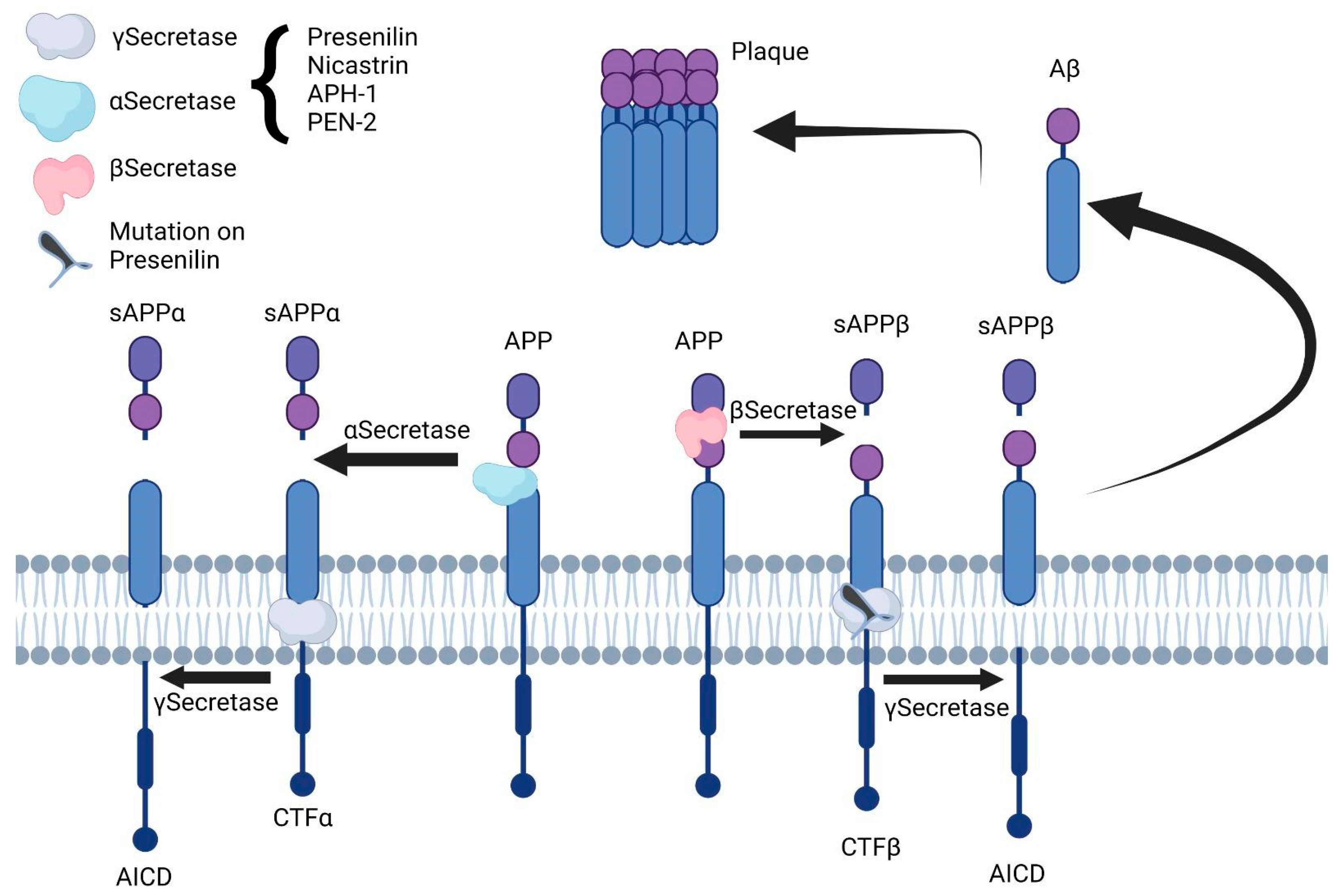
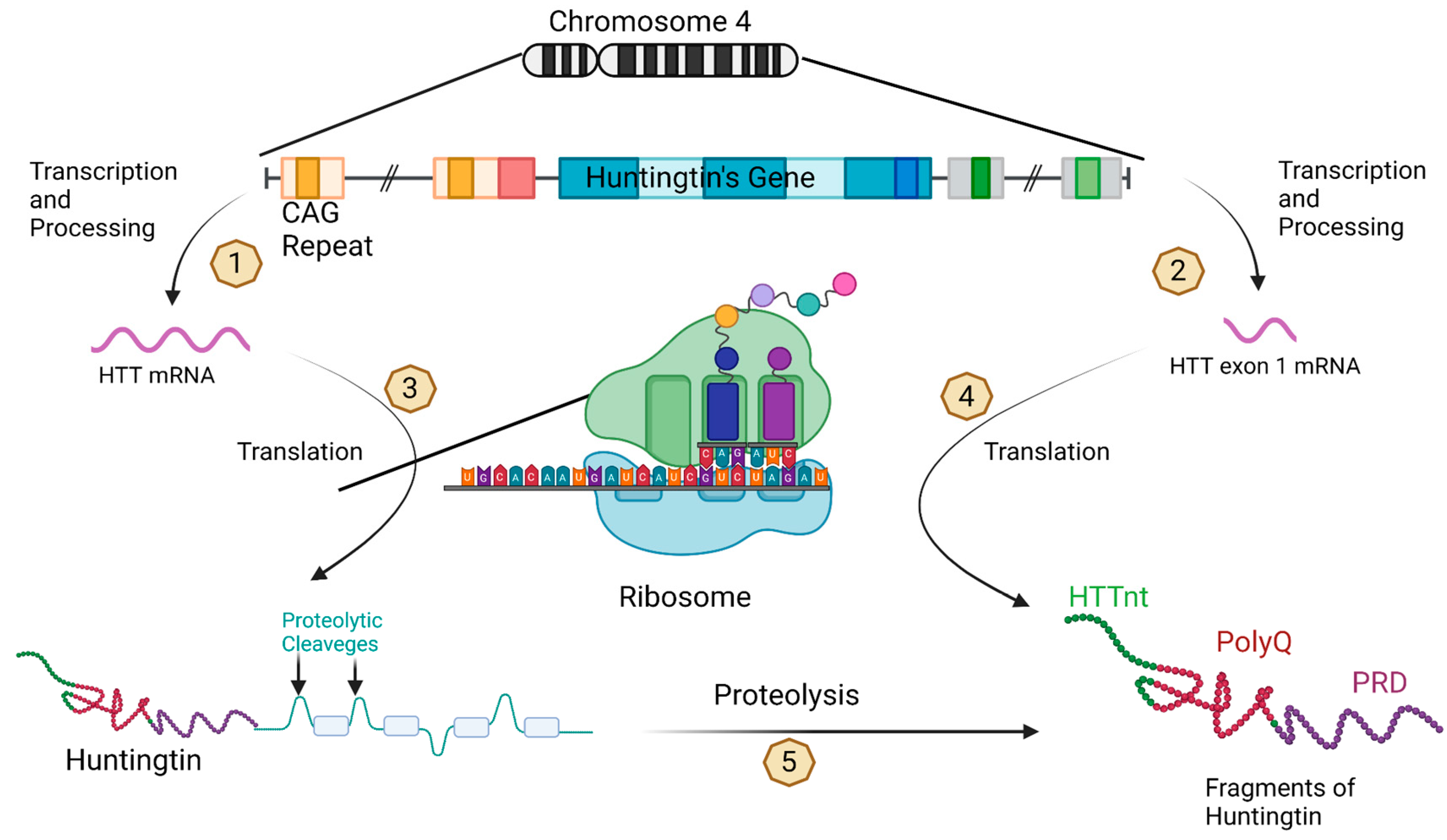
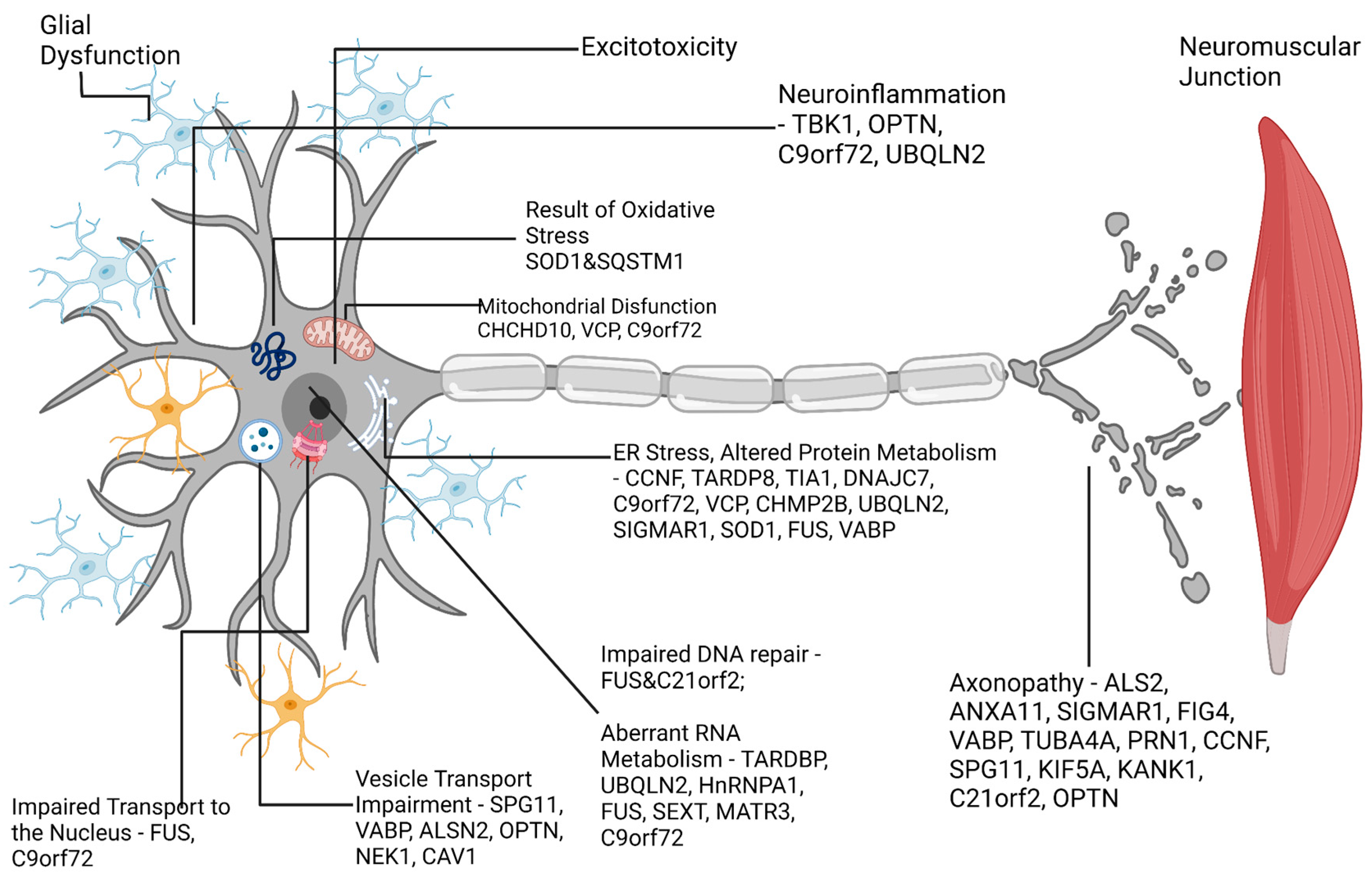
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




