1. Introduction
Transcription factors (TFs) in the homeotype domain-leucine zipper (HD-ZIP) protein family are unique to plants in which they regulate essential biological processes including morphological development and photosynthetic activity. These proteins contain an HD domain composed of three α helices involved in the recognition of specific DNA sequences [
1], as well as a stable hydrophobic LZ domain composed of multiple leucine-specific polypeptide chains. HD-ZIPs undergo dimerization prior to DNA binding, and the LZ domain is essential for appropriate dimerization and DNA recognition. Members of the HD-ZIP TF family are expressed across plant species and tissue types, governing growth, development, and stress responses. These proteins are broadly classified into four subfamilies (I-IV) based on their conserved domains [
2]. In
Arabidopsis thaliana, the HD-ZIP I subfamily consists of 17 proteins including ATHB1/HAT5, ATHB3/HAT7, ATHB5-7, ATHB12, ATHB13, ATHB16, ATHB20-23, ATHB40, and ATHB-51-54 [
3,
4], while it consists of 21 in
Populus alba [
5,
6]. The intron/exon distributions of HD-ZIP I genes align with their phylogenetic relationships, and they encode ~35 kDa proteins consisting of highly conserved HD domains and less highly conserved LZ domains. A range of environmental factors can influence HD-ZIP expression including high temperatures, light levels, osmotic stress, and drought conditions, with tissue-specific expression patterns being evident for many of these genes in particular plant species [
7].
In general, the numbers of genes in these HD-ZIP subfamilies tend to be relatively similar suggesting that the evolution of these subfamilies is evolutionarily conserved. HD-ZIP I proteins have previously been established as key regulators of signal transduction in response to abiotic stressors and light exposure, functioning as key regulators of processes including leaf development [
8,
9]. HD-ZIP II family members reportedly facilitate plant responses to environmental changes and shape associated developmental processes [
10,
11,
12,
13]. In
A. thaliana, the HD-ZIP III subfamily is composed of 5 genes [
10,
14], which serve as key regulators of leaf polarization, meristem formation, and the development of vascular bundles [
15]. HD-ZIP III genes have been found to play similar roles in other species [
16], while also exhibiting unique species-specific functions in some cases [
17,
18]. In Moss Licorniformis, HD-ZIP Ⅲ family members function as key regulators of leaf development [
19], while in woody plants they govern the growth of secondary vascular tissue and xylem differentiation [
8,
20,
21]. The HD-ZIP IV subfamily in
A. thaliana consists of 16 members including HDG1-HDG6 /FWA, HDG7-HDG12, GLABRA2 (GL2), ANTHOCYANINLESS2 (ANL2), AtML1, and PDF2 [
22], with these genes serving primarily as regulators of epidermal cell differentiation [
23]. HD-ZIP III and IV proteins are similar to one another, harboring START and SAD structural domains, while ZIP IV family members lack MEKHLA structural domains and serve as important regulators of trichome formation and epidermal cell development in many species [
24]. These different HD-ZIP genes have also been linked to valuable agronomic traits in various species, including corn grain development and cotton fiber differentiation.
A limited number of studies to date have demonstrated that HD-ZIP TFs serve as important regulators of the ability of plants to tolerate salt stress. This is noteworthy given that soil salinization represents an increasingly pressing threat to crop growth and agronomic stability. The maize HD-ZIP family member Zmhdz10 is upregulated in response to abscisic acid (ABA) and salt stress, with A. thaliana plants expressing Zmhdz10 exhibiting superior salt stress resistance consistent with a role for Zmhdz10 as an ABA_indicible mediator of salt tolerance. The HD-ZIP family member Oshox22 in O. sativa has similarly been shown to be upregulated readily in response to ABA and saline stress conditions. The knockdown of the S1HB2 HD-ZIP gene in tomato plants resulted in an improved salt tolerance, suggesting that this TF serves as a negative regulator of these stress response signaling pathways. In contrast, the cotton HD-ZIP gene GhHB1 was significantly upregulated in response to salt stress and ABA. ABA signaling thus plays an important role in the regulation of HD-ZIP gene expression in various plants exposed to salt stress and associated signal transduction, with the same being true in P. nana.
Here, the HD-ZIP gene family in P. nana was initially identified and characterized through bioinformatics analyses of gene structures, phylogenetic relationships, conserved protein domains, and chromosomal distributions. RNA-seq data was further used to explore the patterns of expression for these HD-ZIP genes across different tissues. A qPCR approach was then used to evaluate changes in HD-ZIP gene expression in P. nana seedlings exposed to cold stress in an effort to more fully understand the potential regulatory roles that these genes may play in response to abiotic stressors. Together, this study serves as the first genome-wide assessment of the P. nana HD-ZIP TF family. These findings offer a basis for future research focused on these genes and the mechanisms governing P. nana cold resistance, potentially offering new genomic tools that can be leveraged to improve the agronomic characteristics of this economically important tree species.
2. Materials and Methods
2.1. P. nana HD-ZIP gene identification
The
Arabidopsis HD-ZIP gene family was initially downloaded from the TAIR database [
28]. Hidden Markov models (HMMs) PF00046 and PF02183 corresponding to the HD-ZIP HD and LZ domains were then downloaded from the Pfam database (
http://pfam-legacy.xfam.org/), after which a preliminary search of the
P. nana genome was performed with the HMMER tool. Validation of HD-ZIP family members was performed with the NCBI-CDD tool (
https://www.ncbi.nlm.nih.gov/cdd accessed on 6 February 2023) with any candidates exhibiting mismatches or incomplete domain sequences being removed, while remaining genes were identified as putative
PnaHD-Zip genes [
34]. Protein classification was conducted for HD-ZIPs from other species (
A. thaliana, P. nana cultivar
‘Nonpareil’, P. avium, P. mume, P. persica, and P. armeniaca) through the same approach employed for
P. nana.
2.2. Analyses of Phylogeny and Gene Structures
Phylogenetic relationships among the HD-ZIP proteins encoded by P. nana and those present in other species were evaluated by generating a phylogenetic tree based upon the protein sequences for these P. nana HD-ZIP family members and those from A. thaliana and P. nana cultivar ‘Nonpareil’. Briefly, a maximum likelihood (ML) tree was constructed using MEGA6 based on full-length protein sequences aligned with Clustal x2.1, with 500 bootstrap replicates. The resultant tree was then displayed with iTOL (
https://itol.embl.de accessed on 6 April 2023).
2.3. Conserved Domains and Motif Analysis
PnaHD-Zip structural predictions were made with GSDS (
http://gsds.gao-lab.org accessed on 8 April 2023), while conserved motifs were detected with MEME (
https://meme-suite.org/meme/tools/fimo accessed on 9 April 2023), with a maximum of 10 motifs, a motif width of 6-100, and all other parameters remaining at default values.
2.4. Chromosomal Mapping Analysis
The chromosomal distributions of PnaHD-Zip genes were assessed with the TBtools “Gene Location Visualize from GTF/GFF” function.
2.5. Cis-acting Regulatory Element Analysis
The promoter regions (2000 bp upstream of the 5′ untranslated region) for identified
PnaHD-Zips were analyzed with the Plant CARE tool (
http://bioinformatics.psb.ugent.be/webtools/plantcare/html accessed on 10 April 2023) in order to predict and classify cis-acting regulatory elements present therein, with the results being visualized in TBtools with the “Gene Structure View (Advanced)” function.
2.6. Plant Growth and Cold Stress Exposure
In total, 9 PnaHD-Zips (PnaHD-Zip1/2/7/9/11/12/20/22/27) were selected as targets for cold stress treatment experiments in P. nana seedlings, as they were found to harbor cold-responsive elements in cis-acting element analyses. Briefly, wild (P. nana) seeds were treated for 24 h using a solution containing gibberellin (200 mM) after which they were planted in a culture chamber and grown under controlled conditions (25℃, light 16 h/dark 8 h). When seedlings were 2 months old, they were transferred to an incubator and cooled at 3°C/h to final target temperatures (25℃, 5℃, -5℃, and -10℃), after which they were incubated for 2 h to assess changes in gene expression in response to cold stress. Leaves were harvested, snap-frozen using liquid nitrogen, followed by storage at -80°C.
2.7. qPCR Analyses
The Plant RNA Extraction Kit (Tiangen, Beijing, China) was used to extract RNA from leaf samples, followed by the use of a First Strand cDNA Synthesis Kit (Tiangen) to prepare cDNA. Then, qPCR analyses were performed with an ACEX96 Real-Time PCR Detection instrument (Bio-Rad, CA, USA) and SYBR Premix ExTap (TaKaRa, Dalian, China) using appropriate primers designed with detection tool (
https://sgidtdna.com/scitoolsApplications/RealTimePCR on 15 April 2023). Relative expression was compared via the 2ΔΔCt method, and results were analyzed using one-way ANOVAs.
3. Results
3.1. PnaHD-Zip Gene Family Identification
An initial analysis of the
P. nana genome with the BLAST tool in the Pfam database identified 30 putative HD-ZIP family genes (E value ≤ 0.05), which were designated as
PnaHD-Zip1 through
PnaHD-Zip30. These genes were cloned and validated via RT-PCR and RACE approaches. The physicochemical properties of these genes were further assessed (
Table 1). The proteins encoded by these
PnaHD-Zips were predicted to range from 193-849 amino acids in length with molecular weight values from 22.387-93.019 kDa. The isoelectric point (pI) values for these proteins were between 4.63 and 9.56, with instability index values from 42.18 (
PnaHD-Zip13) to 67.5 (
PnaHD-Zip25), and aliphatic index values from 50.82 (
PnaHD-Zip23) to 89.18 (
PnaHD-Zip15). All of these proteins exhibited GRAVT values below zero, consistent with the hydrophilic properties of these proteins.
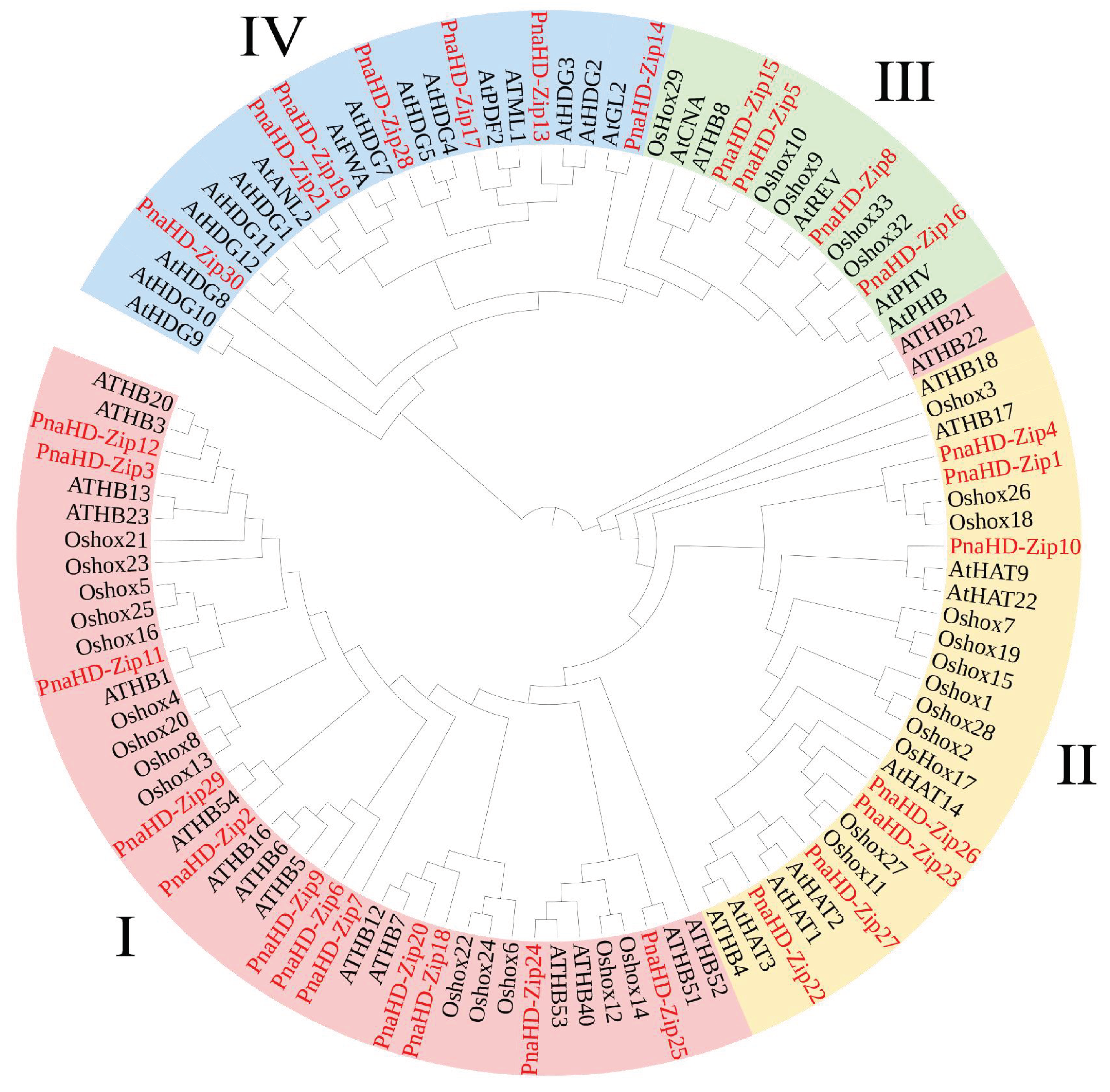
3.2. Phylogenetic Analysis of PnaHD-Zips
The
P. nana HD-ZIP family members were next classified into four subfamilies (I-IV) as reported previously in
A. thaliana [
1,
2,
3,
4], establishing a phylogenetic tree based on these proteins and those from
A. thaliana. The HD-ZIP I gene family was the largest of these subfamilies in
P. nana, consisting of 12 genes (
PnaHD-Zip2/3/6/7/9/11/12/18/20/24/25/29) members. Conversely, 4 HD-ZIP III genes (
PnaHD-Zip5/8/15/16) were identified in
P. nana, and 7 HD-ZIP II and Ⅳ family genes were identified. The different subgroup assignments suggest that these
PnaHD-Zips are likely to exhibit diverse structural and functional properties.
Figure 1.
Phylogenetic analysis of HD-zip genes. Phylogenetic analyses of HD-ZIP genes from A. thaliana and wild P. nana.
Figure 1.
Phylogenetic analysis of HD-zip genes. Phylogenetic analyses of HD-ZIP genes from A. thaliana and wild P. nana.
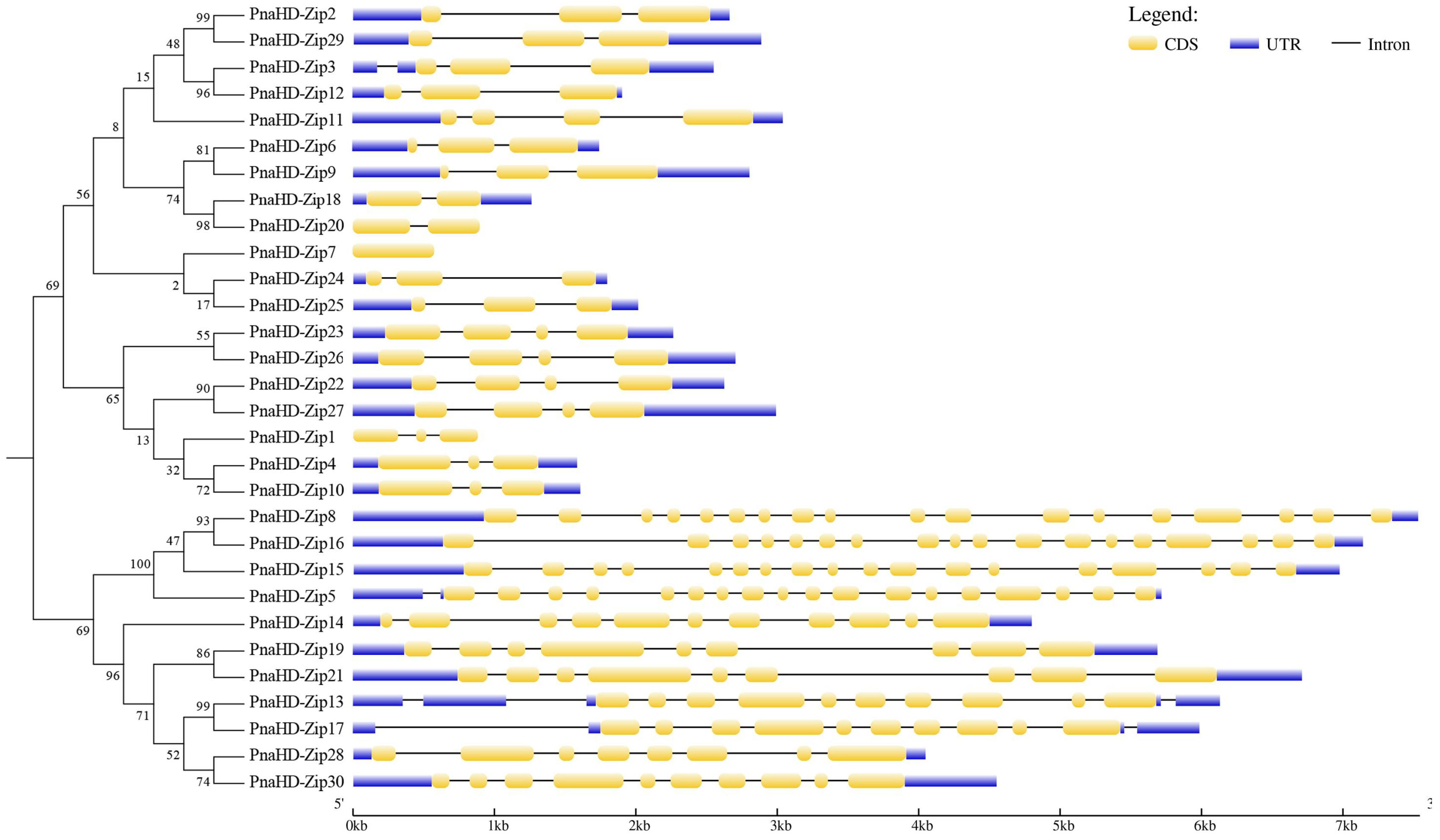
3.3. PnaHD-Zip Gene Structure and Conserved Motif Analyses
The GSDS tool was next employed to examine intron-exon distributions for these 30
PnaHD-Zip genes (
Figure 2), revealing variations in the number of introns present therein. The functions and characteristics of the proteins encoded by these genes were further explored through conserved motif and intron/exon position analyses. The majority of these
PnaHD-Zips had 2-3 introns, although some did not contain any introns. It is worth noting that there are generally 17 introns in group III. The group Ⅳ contains about 8 introns, of which
PnaHD-Zip13 and
PnaHD-Zip17 contain 12 and 11 introns. The high levels of intronic sequence diversity among these
PnaHD-Zips suggest that this gene family may have been subjected to extensive domain shuffling following genomic duplication events.
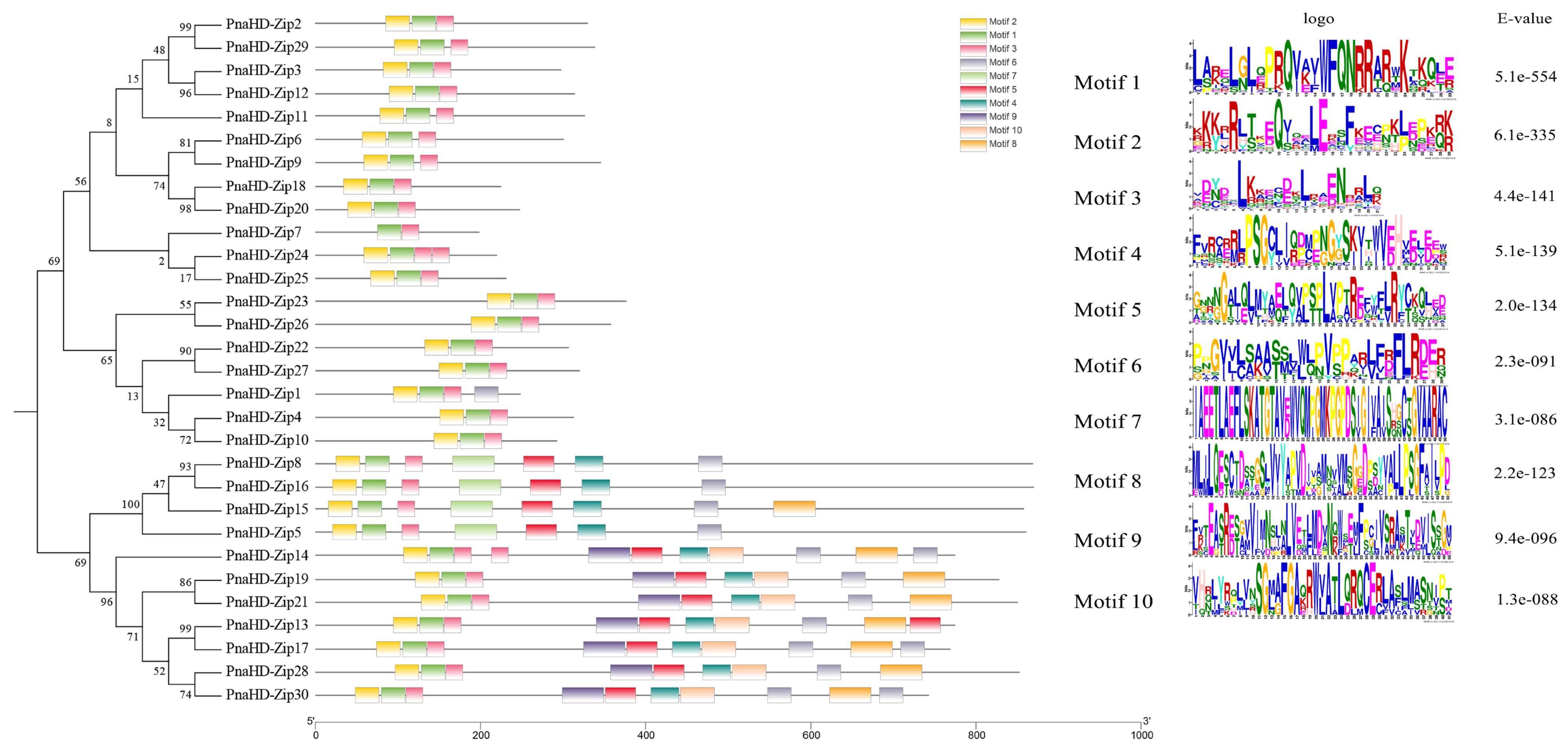
The MEME software enabled the identification of 10 motifs that were conserved in these 30
PnaHD-Zip proteins (
Figure 3). The motif composition of HD-ZIPs in the same subfamilies was largely consistent, suggesting functional similarities within these subfamilies. Motifs 1-3 were present in all 30
PnaHD-Zips
, consistent with the characteristic structural properties of HD-ZIP proteins. Motif 4 and 5 was present in 11
PnaHD-Zips (
PnaHD-Zip4/5/8/13/15/16/17/19/21/28/30), while motif 6 was present in 12 (
PnaHD-Zip1/4/5/8/13/15/16/17/19/21/28/30), and motif 7 was similarly present in 2 (
PnaHD-Zip5 and 7). These findings highlighted the high degree of conservation for motifs 1-3 in the
PnaHD-Zip gene family over the course of evolution (
Figure 3).
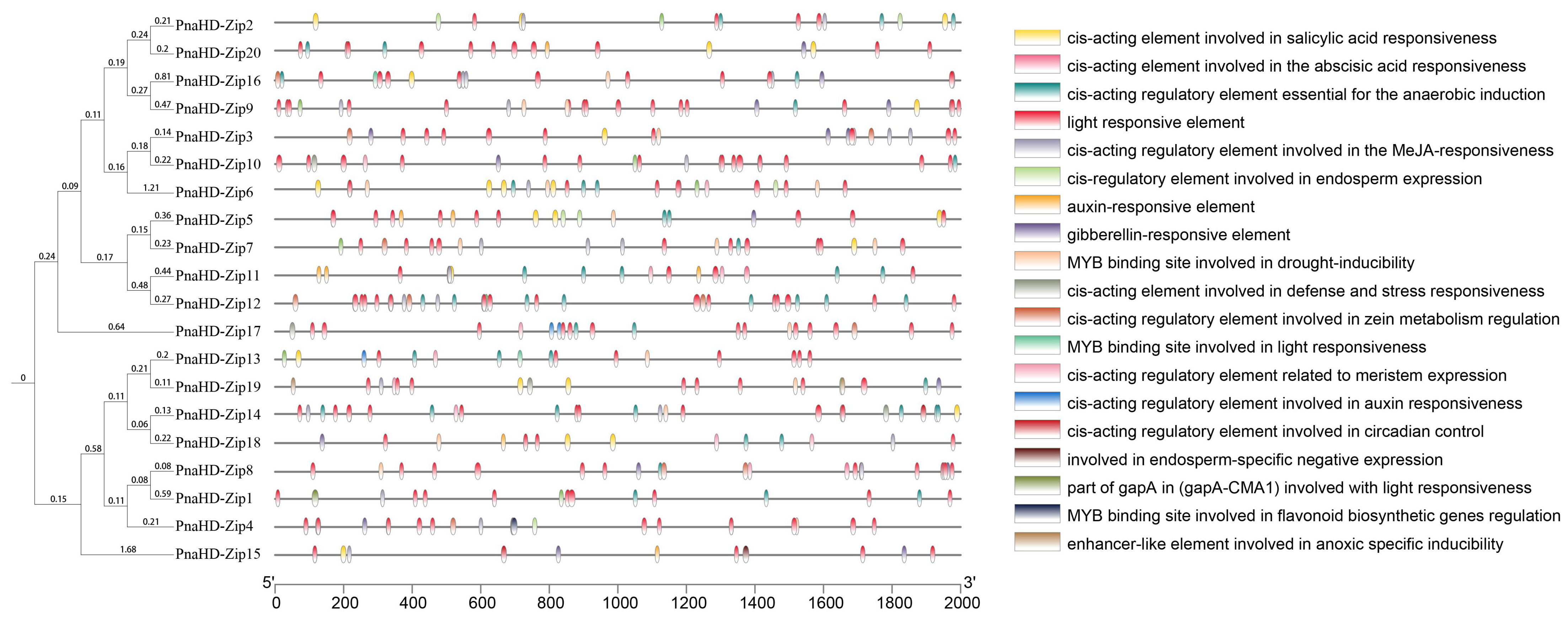
3.4. PnaHD-Zip Gene Promoter Analyses
To further explore the functions and regulation of these
PnaHD-Zip genes, the 2000 bp region upstream of the start codon for each gene was extracted as the putative promoter (
Figure 4). All 30
PnaHD-Zips were found to harbor light-responsive elements, while 15 harbored low-temperature responsive (LTR) elements. MYB binding site and anoxic-induction elements and salicylic acid responsive elements were also respectively detected in 19 and 17 of these
PnaHD-Zips. Endosperm expression elements were identified in 6
PnaHD-Zips and tentatively associated with endosperm-specific expression patterns. Overall, these analyses of cis-regulatory elements suggest that the identified
PnaHD-Zips are likely to be involved in diverse biological processes in
P. nana.
3.5. Analyses of the Chromosomal Distributions of PnaHD-Zip Genes
PnaHD-Zip genes were distributed across seven chromosomes in the
P. nana genome, including seven on chromosome 1, three, three and four on chromosomes 2, 6, and 8, and five each on chromosomes 3 and 5 , while none were present on chromosomes 7. These genes were designated
PnaHD-Zip1—
PnaHD-Zip30 based on their chromosomal locations (
Figure 5). The uneven chromosomal distributions of these
PnaHD-Zips reflect the complexity and diversification of this gene family.
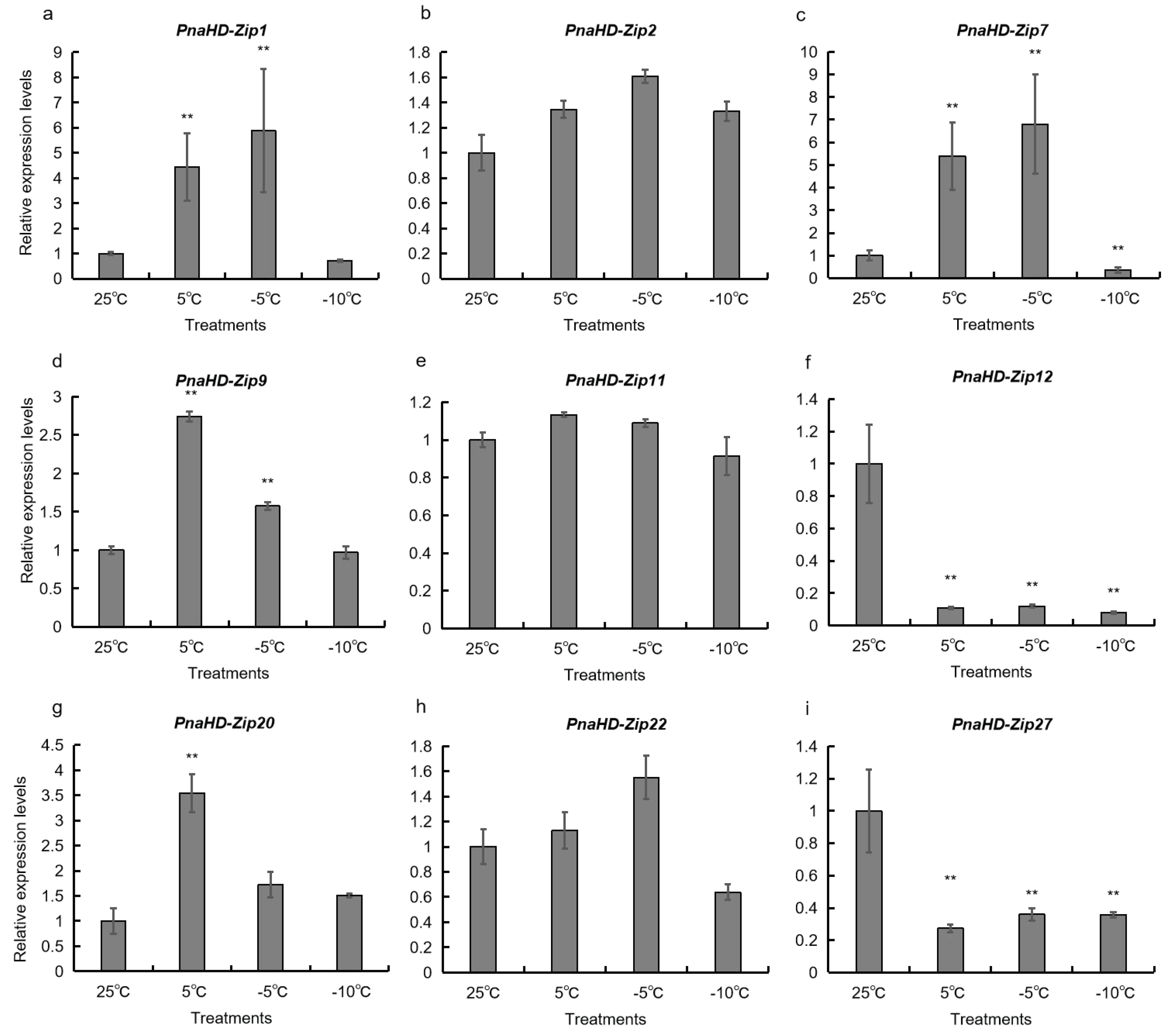
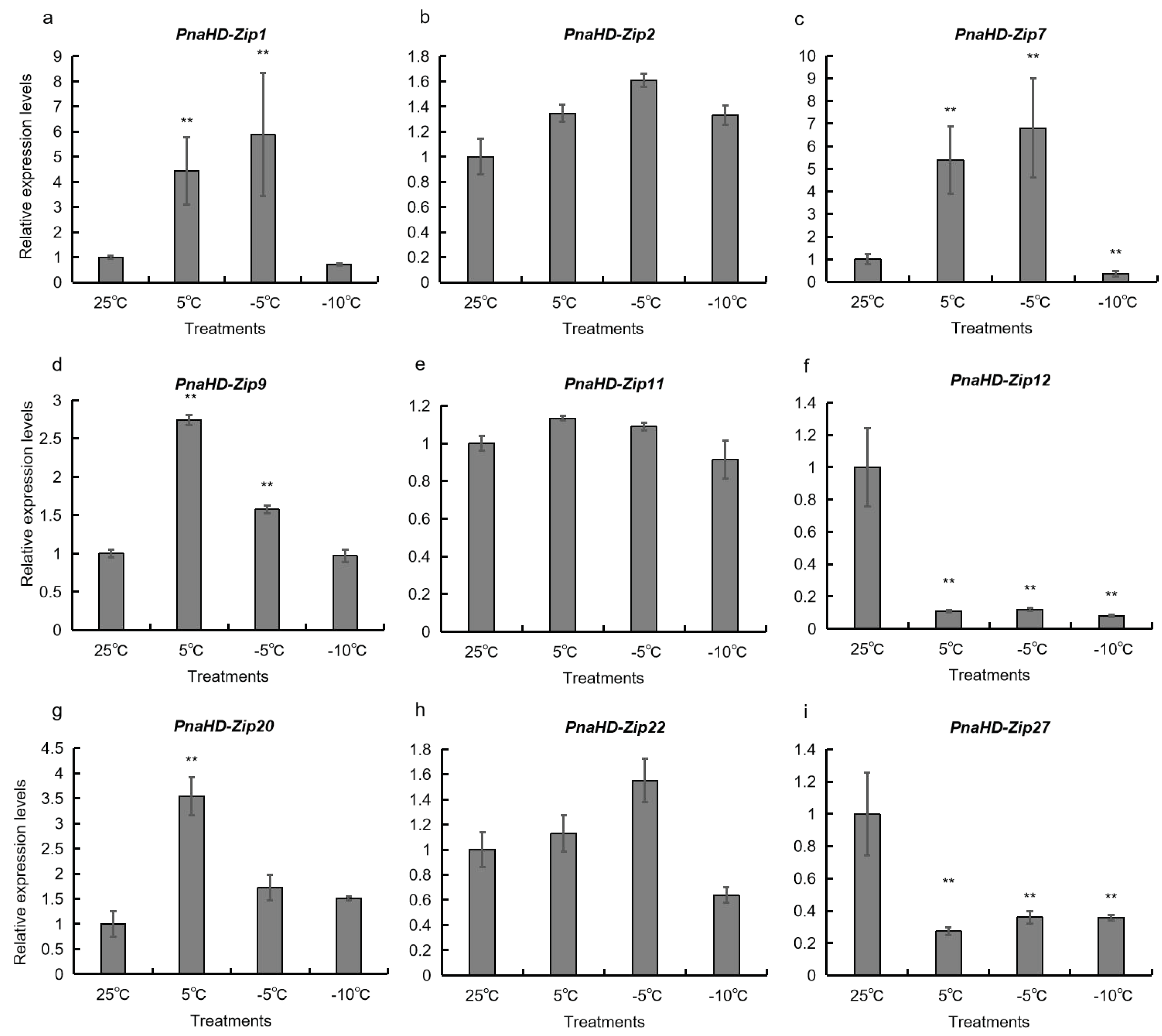
3.6. Changes in PnaHD-Zip Expression Patterns in Response to Cold Stress
To better understand the potential functional roles of the identified
PnaHD-Zip genes, a qPCR approach was next used to evaluate the expression of the 9
PnaHD-Zips harboring cold-responsive elements in response to different temperatures (25°C/5°C/-5°C/-10°C) (
Figure 6).
PnaHD-Zip1, and
PnaHD-Zip7 were significantly up regulated at -5°C and were also expressed at 5°C. In contrast, no significant changes in
PnaHD-Zip2, PnaHD-Zip11, or PnaHD-Zip22 expression were observed across these different temperature treatment conditions, and PnaHD-Zip12 and PnaHD-Zip27 expression levels were continuously suppressed under cold stress conditions. Together these results indicated that these different PnaHD-Zips exhibit distinct and varied responses to cold stress exposure.
4. Discussion
Here, a systematic genome-wide analysis of members of the
P. nana HD-ZIP gene family was conducted with a focus on the phylogenetic relationships, structural characteristics, conserved motifs, and expression profiles for these different genes. Phylogenic analyses enabled the classification of these
P. nana HD-ZIP proteins into four distinct subgroups (I-IV), in line with what has been reported in
A. thaliana, poplar (
Populus trichocarpa), and rice (
Oryza sativa) [
3,
5,
11,
14,
29,
30]. High levels of similarity with respect to conserved protein motifs and intron-exon numbers were observed within each of these subgroups, validating these phylogenetic relationships. A qPCR-based analysis of the expression dynamics of
PnaHD-Zip genes in response to cold stress further highlighted the potential utility of these genes as candidates for efforts to enhance
P. nana abiotic stress resistance. Specifically,
PnaHD-Zip2 and
PnaHD-Zip20 were continuous upregulated under cold-stress conditions inhibited during cryogenic treatment, suggesting the value of these genes as candidate target regulators of cold stress resistance in
P. nana.
P. nana is a member of the
Rosaceae genus. Wild almonds have been established as a drought-resistant, cold-tolerant, and highly adaptable species of fast-growing trees with high levels of economic and medicinal importance [
31]. The average annual temperatures to which wild almonds are exposed are approximately 5°C lower than for cultivated almonds. Specific genes involved in adaptation to ambient temperatures are thus likely to have undergone changes over the course of almond domestication [
31]. To better characterize these genes, a genome-wide analysis of
P. nana was thus performed to provide a foundation for future efforts to breed hardier almond varieties. Overall, the evolutionary history of TFs is complex, and the present results offer new insight into the molecular evolution of the
PnaHD-Zip gene family, enabling predicting analyses of the characteristics and functions of these proteins based on their sequences.
Different plant species express varying numbers of HD-ZIP genes, with the encoded proteins serving as key TFs that coordinate growth and developmental processes [
30,
32]. Here, genes encoding these HD-ZIP proteins were systematically analyzed in
P. nana, leading to the identification of 30
PnaHD-Zips. These results were then combined with previously published HD-ZIP gene family data from other plants [
2,
5,
33,
34,
35], thus enabling comparisons of these genes with those from other species including
Arabidopsis [
3,
11,
14,
29].
P. nana was found to encode fewer HD-ZIP family proteins (n=30) as compared to
A. thaliana (n=47) [
28,
36] ,
Prunus mume (n=32)[
37] or
O. sativa (n=38) [
38,
39,
40]. The greater numbers of HD-ZIP-encoding genes in
A. thaliana [
41] and
V. vinifera (n=63) [
42,
43] suggest that they play complex regulatory roles that are at least partially species-specific.
Members of the PnaHD-Zip gene family were classified into four subgroups, as has also been performed for members of the HD-ZIP families in A. thaliana, O. sativa, and V. vinifera. As these four subfamilies of HD-ZIP TFs are evident in both monocots and dicots, they presumably predate the monocot-dicot evolutionary divergence. A high degree of HD-ZIP gene family evolutionary conservation was evident across species. While variations in the total numbers of genes in each HD-ZIP gene family were evident among species, the HD-ZIP III subfamily tended to be the smallest of these families while the HD-ZIP I subfamily tended to be largest, with these differences likely being attributable to specific gene replication events. Övernäs et al. (2010) determined that HD-ZIP I proteins encoded by multiple plant species may serve as important components of the sucrose and abscisic acid signaling machinery, functioning as important regulators of plant embryogenesis and de-etiolation. The relatively large numbers of HD-ZIP I proteins may thus be the result of plants evolving such that they can adapt to a wide range of environmental stressors.
Cold stress exposure can directly impact enzymatic kinetics and the fluidity of the cell membrane in plant cells, contributing to deleterious reductions in photosynthetic activity, metabolic dysregulation, and impaired material transport [
44,
45]. Plants engage various signaling pathways to respond to low-temperature stress, and the effective identification of the associated genes represents a promising means of enhancing the ability of crops to tolerate cold winter weather [
46,
47]. The sunflower HD-Zip I gene homolog HaHB1 and the
AtHB13 gene encoded by
Arabidopsis thaliana have both been shown to play a role in cold tolerance in plants [
48,
49], with both being upregulated upon exposure to low temperatures. Transgenic
A. thaliana plants overexpressing
HaHB1 and
AtHB13 exhibited improved membrane stability and were better able to tolerate exposure to cold temperatures. Five HD-Zip I subfamily genes (
TaHDZipI-1 to
TaHDZipI-5) have also been reported in wheat [
50,
51,
52]. In the present analysis, 15
PnaHD-Zip genes harboring cold-responsive cis-acting regulatory elements, of which
PnaHD-Zip1 and
PnaHD-Zip7 were significantly upregulated upon exposure to -5°C conditions, while
PnaHD-Zip9,
PnaHD-Zip20 were significantly expressed at 5°C, and
PnaHD-Zip2,
PnaHD-Zip11, and
PnaHD-Zip22 were not expressed at significant levels under these low-temperature conditions. Of note,
PnaHD-Zip12 and
PnaHD-Zip27 expression was continuously suppressed under low-temperature conditions. Together, these findings suggest that
PnaHD-Zip1 and
PnaHD-Zip7 may serve as important regulators of cold stress responses in
P. nana.
A phylogenetic tree established based on the amino acid sequences of these 30 PnaHD-Zips indicated that they share a high degree of sequence homology, suggesting that they may engage in similar functional roles in line with those of HD-ZIP proteins encoded by A. thaliana. These results may help guide future proteomic research focused on these HD-ZIP proteins in P. nana, providing an opportunity to better clarify the processes that govern P. nana development and responsivity to environmental stressors.
5. Conclusions
The present study represents the first systematic bioinformatics-based genome-wide analysis of the HD-ZIP gene family in P. nana. In total, 30 PnaHD-Zips that were classified into 4 subgroups were identified herein and subjected to in silico analyses of their phylogenetic relationships, physicochemical properties, conserved motifs, and promoter sequence composition. The PnaHD-Zips within each subgroup were found to exhibit conserved domain structures. A multi-species phylogenetic tree incorporating HD-ZIP genes suggested that they play roles in the process of secondary wall synthesis. Chromosomal localization and duplication event analyses further suggested that both segmental and tandem duplication events are likely to have contributed to the expansion of the PnaHD-Zip gene family. In addition, promoter analyses and qPCR-based analyses of the expression profiles for these different genes suggested that PnaHD-Zip1 and PnaHD-Zip7 exhibit specific expression patterns. Overall, these findings provide valuable new insights into the evolutionary and functional differences among members of this PnaHD-zip gene family, offering a robust foundation for the further study and characterization of these important TFs.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Table S1: The primer sequences of genes used in qRT-PCR analysis; Table S2: Protein sequences of HD-Zip family in
A. thaliana,
O. sativa and
P.nana.
Author Contributions
Conceptualization, H.Z and Y.Q.; Resources, H.Z.; Writing—original draft preparation, W.H., C.Q. and L.L.; Investigation, W.L. and Y.Q.; Writing—review and editing, W.H. and C.Q.; Data curatio, L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the “National Key R&D Program of China (2022YFD2200400)”
Data Availability Statement
Data supporting the present study are available from the corresponding author upon reasonable request.
Acknowledgments
The author thanks all members of the research team for their help and inspiration. Thank all the reviewers who participated in the review for their language assistance in preparing this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gehring, W.J. Homeo Boxes in the Study of Development. Science 1987, 236, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Elhiti, M.; Stasolla, C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009, 4, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, E.; Olsson, A.S.; Johannesson, H.; Hanson, J.; Engström, P.; Söderman, E. Homeodomain Leucine Zipper Class I Genes in Arabidopsis. Expression Patterns and Phylogenetic Relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Davis, R.W. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. USA 1992, 89, 3894–3898. [Google Scholar] [CrossRef]
- Hu, R.; Chi, X.; Chai, G.; Kong, Y.; He, G.; Wang, X.; Shi, D.; Zhang, D.; Zhou, G. Genome-Wide Identification, Evolutionary Expansion, and Expression Profile of Homeodomain-Leucine Zipper Gene Family in Poplar (Populus trichocarpa). PLOS ONE 2012, 7, e31149. [Google Scholar] [CrossRef]
- Zhang, P.; Duo, T.; Wang, F.; Zhang, X.; Yang, Z.; Hu, G. De novo transcriptome in roots of switchgrass (Panicum virgatum L.) reveals gene expression dynamic and act network under alkaline salt stress. BMC Genom. 2021, 22, 1–14. [Google Scholar] [CrossRef]
- Olsson, A.; Engström, P.; Söderman, E. The homeobox genes ATHB12 and ATHB7encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004, 55, 663–677. [Google Scholar] [CrossRef]
- Carlsbecker Annelie,Lee Ji-Young,Roberts Christina J,Dettmer Jan,Lehesranta Satu,Zhou Jing,Lindgren Ove,Moreno-Risueno Miguel A,Vatén Anne,Thitamadee Siripong,Campilho Ana,Sebastian Jose,Bowman John L,Helariutta Ykä,Benfey Philip N. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate.. Nature,2010,465(7296). [CrossRef]
- Shen, W. , Plant HD-Zip Transcription Factor. Chemistry of life, 2003. 23(5): p. 387-389.
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Ciarbelli, A.R.; Ciolfi, A.; Salvucci, S.; Ruzza, V.; Possenti, M.; Carabelli, M.; Fruscalzo, A.; Sessa, G.; Morelli, G.; Ruberti, I. The Arabidopsis Homeodomain-leucine Zipper II gene family: diversity and redundancy. Plant Mol. Biol. 2008, 68, 465–478. [Google Scholar] [CrossRef]
- Harris, J.C.; Hrmova, M.; Lopato, S.; Langridge, P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011, 190, 823–837. [Google Scholar] [CrossRef]
- Ruberti, I.; Sessa, G.; Ciolfi, A.; Possenti, M.; Carabelli, M.; Morelli, G. Plant adaptation to dynamically changing environment: The shade avoidance response. Biotechnol. Adv. 2012, 30, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M. J. , Kadakia, N., Greenham, K., & Estelle, M. (2019). Members of the arabidopsis auxin receptor gene family are essential early in embryogenesis and have broadly overlapping functions.
- Sessa, G.; Carabelli, M.; Possenti, M.; Morelli, G.; Ruberti, I. Multiple Links between HD-Zip Proteins and Hormone Networks. Int. J. Mol. Sci. 2018, 19, 4047. [Google Scholar] [CrossRef] [PubMed]
- Hawker, N.P.; Bowman, J.L. Roles for Class III HD-Zip and KANADI Genes in Arabidopsis Root Development. Plant Physiol. 2004, 135, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Itoh, J.-I.; Hibara, K.-I.; Sato, Y.; Nagato, Y. Developmental Role and Auxin Responsiveness of Class III Homeodomain Leucine Zipper Gene Family Members in Rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef]
- Zhu, Y. , Yu, L., Wang, X., & Li, L. (2013). Hd-zip iii transcription factor and cell differentiation in plants. CHINESE BULLETIN OF BOTANY, 48(2), 199-209. [CrossRef]
- Yip, H.K.; Floyd, S.K.; Sakakibara, K.; Bowman, J.L. Class III HD-Zip activity coordinates leaf development in Physcomitrella patens. Dev. Biol. 2016, 419, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Côté, C.L.; Boileau, F.; Roy, V.; Ouellet, M.; Levasseur, C.; Morency, M.-J.; Cooke, J.E.; Séguin, A.; MacKay, J.J. Gene family structure, expression and functional analysis of HD-Zip III genes in angiosperm and gymnosperm forest trees. BMC Plant Biol. 2010, 10, 273–273. [Google Scholar] [CrossRef]
- Petricka, J.J.; Schauer, M.A.; Megraw, M.; Breakfield, N.W.; Thompson, J.W.; Georgiev, S.; Soderblom, E.J.; Ohler, U.; Moseley, M.A.; Grossniklaus, U.; et al. The protein expression landscape of the Arabidopsis root. Proc. Natl. Acad. Sci. 2012, 109, 6811–6818. [Google Scholar] [CrossRef]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996 Sep;10(3):393-402. [CrossRef]
- Bou-Torrent, J.; Salla-Martret, M.; Brandt, R.; Musielak, T.; Palauqui, J.-C.; Martínez-García, J.F.; Wenkel, S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 2012, 7, 1382–1387. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Wei, J. Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 2012, 39, 8465–8473. [Google Scholar] [CrossRef]
- Wei, Y. , Studies on cold-resistance of several Chinese almond species. Journal of Northwest A&F University (Nat. Sci. Ed.), 2012. 40(6): p. 99-106.
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chun-Cheng, W.; Yun-Ling, Z.; Song-Mei, M.; Gang, H.; Dan, Z.; Han, Y.; Wang, C.-C.; Zhang, Y.-L.; Ma, S.-M.; Huang, G.; et al. Phylogeny and species differentiation of four wild almond species of subgen. Amygdalus in China. Chin Jour Plan Ecolo 2021, 45, 987–995. [Google Scholar] [CrossRef]
- Huala, Dickerman, Allan, W., Garcia-Hernandez, Margarita, & Weems. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant.
- Nakamura, M.; Katsumata, H.; Abe, M.; Yabe, N.; Komeda, Y.; Yamamoto, K.T.; Takahashi, T. Characterization of the Class IV Homeodomain-Leucine Zipper Gene Family in Arabidopsis. Plant Physiol. 2006, 141, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Agalou, A.; Purwantomo, S.; Övernäs, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engström, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2007, 66, 87–103. [Google Scholar] [CrossRef]
- Li Peng, Luo Shuping, Li Jiang. Morphological differences and pollen germination rate analysis of four cultivars of cultivars and wild almonds. Journal of Tarim University,2016,28(03):74-79.
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-ZIP Gene Family: Potential Roles in Improving Plant Growth and Regulating Stress-Responsive Mechanisms in Plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Aso, K.; Kato, M.; Banks, J.A.; Hasebe, M. Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Mol. Biol. Evol. 1999, 16, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Clark, S.E. Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 2006, 8, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhou, Y.; Jiang, H.; Li, X.; Gan, D.; Peng, X.; Zhu, S.; Cheng, B. Systematic Analysis of Sequences and Expression Patterns of Drought-Responsive Members of the HD-Zip Gene Family in Maize. PLOS ONE 2011, 6, e28488. [Google Scholar] [CrossRef]
- Söderman, E.; Hjellström, M.; Fahleson, J.; Engström, P. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol. Biol. 1999, 40, 1073–1083. [Google Scholar] [CrossRef]
- Li Lulu, Zheng Tangchun, Zhuo Xiaokang, Li Suzhen, Qiu Like, Wang Jia, Cheng Tangren, Zhang Qixiang. Genome-wide identification, characterization and expression analysis of the HD-Zip gene family in the stem development of the woody plant Prunus mume.. PeerJ,2019,7.
- Goff S, A. Erratum: A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) (Science (, 2002) (92)). 2005. 5 April.
- Bang, S.W.; Lee, D.; Jung, H.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Suh, J.; Kim, J. Overexpression of OsTF1L, a rice HD-Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 2018, 17, 118–131. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014, 15, 1–25. [Google Scholar] [CrossRef]
- Giovanna Sessa, Monica Carabelli and Ida Ruberti,. (1994). Identification of Distinct Families of HD-ZIP Proteins in Arabidopsis thaliana. Plant Molecular Biology.1994, 411-426. [CrossRef]
- Li, Z.; Zhang, C.; Guo, Y.; Niu, W.; Wang, Y.; Xu, Y. Evolution and expression analysis reveal the potential role of the HD-Zip gene family in regulation of embryo abortion in grapes (Vitis vinifera L.). BMC Genom. 2017, 18, 744. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H. , Jin, H., Liu, H., Dong, Q. Yan, H., Gan, D., Zhang, W., Zhu, S. Genome-wide analysis of HD-zip genes in grape (Vitis vinifera). Tree genetics & genomes, 2014. 11: p. 1-11. [CrossRef]
- Janská A, Marsík P, Zelenková S, Ovesná J. Cold stress and acclimation—what is important for metabolic adjustment? Plant Biol (Stuttg). 2010 ;12(3):395-405. 1 May. [CrossRef] [PubMed]
- Jiang, F. , Li,Y., Wen, B. Review on physiology of chilling stress and chilling resistance of plants. Fujian Journal of Agricultural Sciences, 2002. 17(3): p. 190-195.
- Su, J. , Dong, Y., Chai, S. Progress of Studies on Detection of Alien Chromatin in Wheat. Crops, 2008. 24(3): p. 17-19.
- Ji, S. , D, S., L, W. Research progress on the mechanism of plant response to low temperature stress. Life Science, 2010, 22(10): 1013-1019. [CrossRef]
- Manavella, P.A.; Dezar, C.A.; Bonaventure, G.; Baldwin, I.T.; Chan, R.L. HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 2008, 56, 376–388. [Google Scholar] [CrossRef]
- Dezar, C.A.; Fedrigo, G.V.; Chan, R.L. The promoter of the sunflower HD-Zip protein gene Hahb4 directs tissue-specific expression and is inducible by water stress, high salt concentrations and ABA. Plant Sci. 2005, 169, 447–456. [Google Scholar] [CrossRef]
- Lopato, S.; Borisjuk, L.; Milligan, A.S.; Shirley, N.; Bazanova, N.; Langridge, P. Systematic identification of factors involved in post-transcriptional processes in wheat grain. Plant Mol. Biol. 2006, 62, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.C.; Sornaraj, P.; Taylor, M.; Bazanova, N.; Baumann, U.; Lovell, B.; Langridge, P.; Lopato, S.; Hrmova, M. Molecular interactions of the γ-clade homeodomain-leucine zipper class I transcription factors during the wheat response to water deficit. Plant Mol. Biol. 2016, 90, 435–452. [Google Scholar] [CrossRef]
- Kovalchuk, N.; Chew, W.; Sornaraj, P.; Borisjuk, N.; Yang, N.; Singh, R.; Bazanova, N.; Shavrukov, Y.; Guendel, A.; Munz, E.; et al. The homeodomain transcription factor TaHDZipI-2 from wheat regulates frost tolerance, flowering time and spike development in transgenic barley. New Phytol. 2016, 211, 671–687. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).