Submitted:
05 May 2023
Posted:
10 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Synucleins Structure and Homology
3. Synucleins Expression and Physiological Roles
4. Synucleins Are Cancer-Related Proteins
5. Synucleins Regulation and Posttranslational Modifications
6. Synucleins Aggregation and Cancer
7. Synucleins Controlled Pathways in Cancer
8. Synucleins as Cancer Biomarkers
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dunker, A.K.; Oldfield, C.J.; Meng, J.; Romero, P.; Yang, J.Y.; Chen, J.W.; Vacic, V.; Obradovic, Z.; Uversky, V.N. The Unfoldomics Decade: An Update on Intrinsically Disordered Proteins. BMC Genomics, 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Maroteaux, L.; Campanelli, J.T.; Scheller, R.H. Synuclein: A Neuron-Specific Protein Localized to the Nucleus and Presynaptic Nerve Terminal. J Neurosci. 1988, 8, 2804–2815. [Google Scholar] [CrossRef] [PubMed]
- Nakajo, S.; Omata, K.; Aiuchi, T.; Shibayama, T.; Okahashi, I.; Ochiai, H.; Nakai, Y.; Nakaya, K.; Nakamura, Y. Purification and Characterization of a Novel Brain-Specific 14-kDa Protein. J Neurochem 1990, 55, 2031–2038. [Google Scholar] [CrossRef] [PubMed]
- Nakajo, S.; Tsukada, K.; Omata, K.; Nakamura, Y.; Nakaya, K. A New Brain-specific 14-kDa Protein Is a Phosphoprotein: Its Complete Amino Acid Sequence and Evidence for Phosphorylation. Eur J Biochem 1993, 217, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Jakes, R.; Spillantini, M.G.; Goedert, M. Identification of Two Distinct Synucleins from Human Brain. FEBS Lett 1994, 345, 27–32. [Google Scholar] [CrossRef]
- Ji, H.; Liu, Y.E.; Jia, T.; Wang, M.; Liu, J.; Xiao, G.; Joseph, B.K.; Rosen, C.; Shi, Y.E. Identification of a Breast Cancer-Specific Gene, BCSG1, by Direct Differential CDNA Sequencing. Cancer Res 1997, 57, 759–764. [Google Scholar]
- Buchman, V.L. Persyn, a Member of the Synuclein Family, Has a Distinct Pattern of Expression in the Developing Nervous System. J Neurosci. 1998, 18, 9335–9341. [Google Scholar] [CrossRef]
- Ninkina, N.N.; Alimova-Kost, M. V.; Paterson, J.W.E.; Delaney, L.; Cohen, B.B.; Imreh, S.; Gnuchev, N. V.; Davies, A.M.; Buchman, V.L. Organization, Expression and Polymorphism of the Human Persyn Gene. Hum Mol Genet. 1998, 7, 1417–1424. [Google Scholar] [CrossRef]
- Lavedan, C.; Leroy, E.; Dehejia, A.; Buchholtz, S.; Dutra, A.; Nussbaum, R.L.; Polymeropoulos, M.H. Identification, Localization and Characterization of the Human γ-Synuclein Gene. Hum Genet 1998, 103, 106–112. [Google Scholar] [CrossRef]
- Alberti, S.; Hyman, A.A. Are Aberrant Phase Transitions a Driver of Cellular Aging? Bioessays 2016, 38, 959–968. [Google Scholar] [CrossRef]
- Berlow, R.B.; Dyson, H.J.; Wright, P.E. Functional Advantages of Dynamic Protein Disorder. FEBS Lett. 2015, 589, 2433–2440. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat Rev Mol Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins in Human Diseases: Introducing the D 2 Concept. Annu Rev Biophys 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Pirman, N.L.; Milshteyn, E.; Galiano, L.; Hwelett, J.C.; Fanucci, G.E. Characterization of the disordered-to-α-helical transition of IA₃ by SDSL-EPR spectroscopy. Protein Sci. 2011, 20(1), 150–159. [Google Scholar] [CrossRef] [PubMed]
- Carnazza, K.E.; Komer, L.E.; Xie, Y.X.; Pineda, A.; Briano, J.A.; Gao, V.; Na, Y.; Ramlall, T.; Buchman, V.L.; Eliezer, D. ; Sharma, M,.; Burré, J. Synaptic vesicle binding of α-synuclein is modulated by β- and γ-synucleins. Cell Rep 2022, 39, 110675. [Google Scholar] [CrossRef]
- Li, H.T.; Du, H.N.; Tang, L.; Hu, J.; Hu, H.Y. Structural transformation and aggregation of human α-synuclein in trifluoroethanol: Non-amyloid component sequence is essential and β-sheet formation is prerequisite to aggregation. Biopolymers 2002, 64(4), 221–226. [Google Scholar] [CrossRef]
- Uéda, K.; Fukushima, H.; Masliah, E.; Xia, Y.; Iwai, A.; Yoshimoto, M.; Otero, D.A.; Kondo, J.; Ihara, Y.; Saitoh, T. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A 1993, 90, 11282–11286. [Google Scholar] [CrossRef]
- Golebiewska, U.; Zurawsky, C.; Scarlata, S. Defining the oligomerization state of γ-synuclein in solution and in cells. Biochemistry 2014, 53(2), 293–299. [Google Scholar] [CrossRef]
- Surgucheva, I.; Sharov, V.S.; Surguchov, A. γ-Synuclein: Seeding of α-synuclein aggregation and transmission between cells. Biochemistry 2012, 51(23), 4743–4754. [Google Scholar] [CrossRef]
- Eliezer, D. The mysterious C-terminal tail of alpha-synuclein: Nanobody’s guess. J Mol Biol. 2013, 425(14), 2393–2396. [Google Scholar] [CrossRef]
- Bussell, R.; Eliezer, D. A structural and functional role for 11-mer repeats in α-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003, 329(4), 763–778. [Google Scholar] [CrossRef] [PubMed]
- George, J.M. The Synucleins. Genome Biol. 2002, 3. [Google Scholar]
- Vargas, K.J.; Makani, S.; Davis, T.; Westphal, C.H.; Castillo, P.E.; Chandra, S.S. Synucleins regulate the kinetics of synaptic vesicle endocytosis. J Neurosci. 2014, 34(28), 9364–9376. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.H.; Eliezer, D. Secondary structure and dynamics of micelle bound β- and γ-synuclein. Protein Sci. 2006, 15(5), 1162–1174. [Google Scholar] [CrossRef] [PubMed]
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef] [PubMed]
- Cabin, D.E.; Shimazu, K.; Murphy, D.; Cole, N.B.; Gottschalk, W.; McIlwain, K.L.; Orrison, B.; Chen, A.; Ellis, C.E.; Paylor, R.; Lu, B.; Nussbaum, R.L. Synaptic vesicle depletion correlates with attenuated synaptic α-synuclein. J Neurosci. 2002, 22, 8797–8807. [Google Scholar] [CrossRef] [PubMed]
- Peters, O.M.; Millership, S.; Shelkovnikova, T.A.; Soto, I.; Keeling, L.; Hann, A.; Marsh-Armstrong, N.; Buchman, V.L.; Ninkina, N. Selective pattern of motor system damage in gamma-synuclein transgenic mice mirrors the respective pathology in amyotrophic lateral sclerosis. Neurobiol Dis. 2012, 48(1), 124–131. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, K.; Wider, C.; Vilariño-Güell, C.; Soto-Ortolaza, A.I.; Lincoln, S.J.; Kachergus, J.M.; Jasinska-Myga, B.; Ross, O.A.; Rajput, A.; Robinson, C.A.; Ferman, T.J.; Wszolek, K.; Dickson, D.W.; Farrer, M.J. Association of alpha-, beta-, and gamma-Synuclein with diffuse lewy body disease. Arch Neurol 2010, 67, 970–975. [Google Scholar] [CrossRef]
- Galvin, J.E.; Uryu, K.; Lee, V.M.; Trojanowski, J.Q. Axon pathology in Parkinson’s disease and Lewy body dementia hippocampus contains alpha-, beta-, and gamma-synuclein. Proc Natl Acad Sci U S A 1999, 96(23), 13450–13455. [Google Scholar] [CrossRef]
- Brown, J.W.; Buell, A.K.; Michaels, T.C.; Meisl, G.; Carozza, J.; Flagmeier, P.; Vendruscolo, M.; Knowles, T.P.; Dobson, C.M.; Galvagnion, C. β-Synuclein suppresses both the initiation and amplification steps of α-synuclein aggregation via competitive binding to surfaces. Sci Rep. 2016, 6, 36010. [Google Scholar] [CrossRef]
- Hayashi, J.; Carver, J.A. β-Synuclein: An Enigmatic Protein with Diverse Functionality. Biomolecules 2022, 12, 142. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Bar-On, P.; Ho, G.; Takenouchi, T.; Rockenstein, E.; Crews, L.; Masliah, E. β-Synuclein Regulates Akt Activity in Neuronal Cells: A Possible Mechanism for Neuroprotection in Parkinson’s Disease. J Biol Chem 2004, 279, 23622–23629. [Google Scholar] [CrossRef] [PubMed]
- Surgucheva, I.; Ninkina, N.; Buchman, V.L.; Grasing, K.; Surguchov, A. Protein Aggregation in Retinal Cells and Approaches to Cell Protection. Cell Mol Neurobiol 2005, 25, 1051–1066. [Google Scholar] [CrossRef] [PubMed]
- Buchman, V.L.; Adu, J.; Pinõn, L.G.P.; Ninkina, N.N.; Davies, A.M. Persyn, a Member of the Synuclein Family, Influences Neurofilament Network Integrity. Nat Neurosci 1998, 1, 101–103. [Google Scholar] [CrossRef] [PubMed]
- Barba, L.; Paolini Paoletti, F.; Bellomo, G.; Gaetani, L.; Halbgebauer, S.; Oeckl, P.; Otto, M.; Parnetti, L. Alpha and Beta Synucleins: From Pathophysiology to Clinical Application as Biomarkers. Mov Disord. 2022, 37, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Pinho, R.; Paiva, I.; Jerčić, K.G.; Fonseca-Ornelas, L.; Gerhardt, E.; Fahlbusch, C.; Garcia-Esparcia, P.; Kerimoglu, C.; Pavlou, M.A.S.; Villar-Piqué, A.; Szego, É.; Lopes da Fonseca, T.; Odoardi, F.; Soeroes, S.; Rego, A.C.; Fischle, W.; Schwamborn, J.C.; Meyer, T.; Kügler, S.; Ferrer, I.; Attems, J.; Fischer, A.; Becker, S.; Zweckstetter, M.; Borovecki. F.; Outeiro, T.F. Nuclear Localization and Phosphorylation Modulate Pathological Effects of Alpha-Synuclein. Hum Mol Genet 2019, 28, 31–50. [Google Scholar] [CrossRef] [PubMed]
- Sousa, V.L.; Bellani, S.; Giannandrea, M.; Yousuf, M.; Valtorta, F.; Meldolesi, J.; Chieregatti, E. α-Synuclein and Its A30P Mutant Affect Actin Cytoskeletal Structure and Dynamics. Mol Biol Cell 2009, 20, 3725–3739. [Google Scholar] [CrossRef]
- Araki, K.; Sugawara, K.; Hayakawa, E.H.; Ubukawa, K.; Kobayashi, I.; Wakui, H.; Takahashi, N.; Sawada, K.; Mochizuki, H.; Nunomura, W. The Localization of α-Synuclein in the Process of Differentiation of Human Erythroid Cells. Int J Hematol 2018, 108, 130–138. [Google Scholar] [CrossRef]
- Liu, H.; Liu, W.; Wu, Y.; Zhou, Y.; Xue, R.; Luo, C.; Wang, L.; Zhao, W.; Jiang, J.D.; Liu, J. Loss of Epigenetic Control of Synuclein-γ Gene as a Molecular Indicator of Metastasis in a Wide Range of Human Cancers. Cancer Res 2005, 65, 7635–7643. [Google Scholar] [CrossRef]
- Goh, K.W.; Say, Y.H. γ-Synuclein Confers Both pro-Invasive and Doxorubicin-Mediated pro-Apoptotic Properties to the Colon Adenocarcinoma LS 174T Cell Line. Tumor Biol. 2015, 36, 7947–7960. [Google Scholar] [CrossRef]
- Jia, T.; Liu, Y.E.; Liu, J.; Shi, Y.E. Stimulation of Breast Cancer Invasion and Metastasis by Synuclein gamma. Cancer Res 1999, 59, 742–747. [Google Scholar] [PubMed]
- Bruening, W.; Giasson, B.I.; Klein-Szanto, A.J.P.; Lee, V.M.Y.; Trojanowski, J.Q.; Godwin, A.K. Synucleins Are Expressed in the Majority of Breast and Ovarian Carcinomas and in Preneoplastic Lesions of the Ovary. Cancer 2000, 88, 2154–2163. [Google Scholar] [CrossRef]
- Raghavan, R.; White, C.L.; Rogers, B.; Coimbra, C.; Rushing, E.J. Alpha-Synuclein Expression in Central Nervous System Tumors Showing Neuronal or Mixed Neuronal/Glial Differentiation. J Neuropathol Exp Neurol 2000, 59, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, M.; Suzuki, S.O.; Doh-Ura, K.; Iwaki, T. α-Synuclein Is Expressed in a Variety of Brain Tumors Showing Neuronal Differentiation. Acta Neuropathol 2000, 99, 154–160. [Google Scholar] [CrossRef]
- Surguchov, A.; Palazzo, R.E.; Surgucheva, I. Gamma Synuclein: Subcellular Localization in Neuronal and Non-Neuronal Cells and Effect on Signal Transduction. Cell Motil Cytoskeleton 2001, 49, 218–228. [Google Scholar] [CrossRef]
- Pan, Z.Z.; Bruening, W.; Giasson, B.I.; Lee, V.M.Y.; Godwin, A.K. γ-Synuclein Promotes Cancer Cell Survival and Inhibits Stress- and Chemotherapy Drug-Induced Apoptosis by Modulating MAPK Pathways. J Biol Chem. 2002, 277, 35050–35060. [Google Scholar] [CrossRef]
- Zhou, C.Q.; Liu, S.; Xue, L.Y.; Wang, Y.H.; Zhu, H.X.; Lu, N.; Xu, N.Z. Down-Regulation of γ-Synuclein in Human Esophageal Squamous Cell Carcinoma. World J Gastroenterol 2003, 9, 1900–1903. [Google Scholar] [CrossRef]
- Fung, K.M.; Rorke, L.B.; Giasson, B.; Lee, V.M.Y.; Trojanowski, J.Q. Expression of alpha-, beta-, and gamma-Synuclein in Glial Tumors and Medulloblastomas. Acta Neuropathol. 2003, 106, 167–175. [Google Scholar] [CrossRef]
- Li, Z.; Sclabas, G.M.; Peng, B.; Hess, K.R.; Abbruzzese, J.L.; Evans, D.B.; Chiao, P.J. Overexpression of Synuclein-γ in Pancreatic Adenocarcinoma. Cancer 2004, 101, 58–65. [Google Scholar] [CrossRef]
- Yanagawa, N.; Tamura, G.; Honda, T.; Endoh, M.; Nishizuka, S.; Motoyama, T. Demethylation of the Synuclein γ Gene CpG Island in Primary Gastric Cancers and Gastric Cancer Cell Lines. Clinical Cancer Research 2004, 10, 2447–2451. [Google Scholar] [CrossRef]
- Iwaki, H.; Kageyama, S.; Isono, T.; Wakabayashi, Y.; Okada, Y.; Yoshimura, K.; Terai, A.; Arai, Y.; Iwamura, H.; Kawakita, M.; Yoshiki, T. Diagnostic Potential in Bladder Cancer of a Panel of Tumor Markers (Calreticulin, γ-Synuclein, and Catechol-o-Methyltransferase) Identified by Proteomic Analysis. Cancer Sci 2004, 95, 955–956. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Zheng, M.H.; Cai, Q.; Feng, B.; Chen, X.H.; Yu, B.Q.; Gao, Y.B.; Ji, J.; Lu, A.G.; Li, J.W.; Wang, M.L.; Liu, B.Y. Aberrant Expression and Demethylation of γ-Synuclein in Colorectal Cancer, Correlated with Progression of the Disease. Cancer Sci 2008, 99, 1924–1932. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wang, T.F.; Peng, Y.F.; Xie, J.; Feng, B.; Qiu, M.Y.; Li, L.H.; Lu, A.G.; Liu, B.Y.; Zheng, M.H. Expression of α-, β- and γ-Synuclein in Colorectal Cancer, and Potential Clinical Significance in Progression of the Disease. Oncol Rep 2010, 23, 429–436. [Google Scholar] [PubMed]
- Morgan, J.; Hoekstra, A. V.; Chapman-Davis, E.; Hardt, J.L.; Kim, J.J.; Buttin, B.M. Synuclein-γ (SNCG) May Be a Novel Prognostic Biomarker in Uterine Papillary Serous Carcinoma. Gynecol Oncol 2009, 114, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Kamitani, T. Parkinson’s Disease-Related Protein, α-Synuclein, in Malignant Melanoma. PLoS One 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Maitta, R.W.; Wolgast, L.R.; Wang, Q.; Zhang, H.; Bhattacharyya, P.; Gong, J.Z.; Sunkara, J.; Albanese, J.M.; Pizzolo, J.G.; A. Cannizzaro, L.; Ramesh, K.H.; Ratech, H. Alpha- and Beta-Synucleins Are New Diagnostic Tools for Acute Erythroid Leukemia and Acute Megakaryoblastic Leukemia. Am J Hematol. 2011, 86, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Mhawech-Fauceglia, P.; Wang, D.; Syriac, S.; Godoy, H.; Dupont, N.; Liu, S.; Odunsi, K. Synuclein-γ (SNCG) Protein Expression Is Associated with Poor Outcome in Endometrial Adenocarcinoma. Gynecol Oncol. 2012, 124, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; She, F.; Wang, D.; Yao, X.; Jiang, L.; Chen, Y. SNCG Gene Silencing in Gallbladder Cancer Cells Inhibits Key Tumorigenic Activities. Front Biosci. 2012, 17, 1589–1598. [Google Scholar] [CrossRef]
- Cheng, J.C.; Chiang, M.T.; Lee, C.H.; Liu, S.Y.; Chiu, K.C.; Chou, Y.T.; Huang, R.Y.; Huang, S.M.; Shieh, Y.S. γ-Synuclein Expression Is a Malignant Index in Oral Squamous Cell Carcinoma. J Dent Res 2016, 95, 439–445. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Turriani, E.; Lázaro, D.F.; Ryazanov, S.; Leonov, A.; Giese, A.; Schön, M.; Schön, M.P.; Griesinger, C.; Outeiro, T.F.; Arndt-Jovin, D.J.; Becker, D. Treatment with Diphenyl-Pyrazole Compound Anle138b/c Reveals That α-Synuclein Protects Melanoma Cells from Autophagic Cell Death. Proc Natl Acad Sci U S A 2017, 114, E4971–E4977. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, M.; Giambelluca, M.; Scavuzzo, M.C.; Di Franco, G.; Guadagni, S.; Palmeri, M.; Furbetta, N.; Gianardi, D.; Costa, A.; Gentiluomo, M.; Gaeta, R.; Pollina, L.E.; Falcone. A.; Vivaldi, C.; Di Candio, G.; Biagioni, F.; Busceti, C.L.; Soldani, P.; Puglisi-Allegra, S.; Morelli, L.; Fornai, F. In Pancreatic Adenocarcinoma Alpha-Synuclein Increases and Marks Peri-Neural Infiltration. Int J Mol Sci. 2022, 23, 3775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, Z.; Ma, K. SNCA Correlates with Immune Infiltration and Serves as a Prognostic Biomarker in Lung Adenocarcinoma. BMC Cancer 2022, 22, 406. [Google Scholar]
- Becker, C.; Brobert, G.P.; Johansson, S.; Jick, S.S.; Meier, C.R. Cancer Risk in Association with Parkinson Disease: A Population-Based Study. Parkinsonism Relat Disord. 2010, 16, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Shou, C.; Meng, L.; Jiang, B.; Dong, B.; Yao, L.; Xie, Y.; Zhang, J.; Chen, Y.; Budman, D.R.; Shi, Y.E. Neuronal Protein Synuclein γ Predicts Poor Clinical Outcome in Breast Cancer. Int J Cancer 2007, 121, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Dong, B.; Lu, A.; Qu, L.; Xing, X.; Meng, L.; Wu, J.; Eric Shi, Y.; Shou, C. Synuclein Gamma Predicts Poor Clinical Outcome in Colon Cancer with Normal Levels of Carcinoembryonic Antigen. BMC Cancer 2010, 10, 359. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Qu, L.; Dong, B.; Xing, X.; Ren, T.; Zeng, Y.; Jiang, B.; Meng, L.; Wu, J.; Shou, C. Combined Phenotype of 4 Markers Improves Prognostic Value of Patients with Colon Cancer. Am J Med Sci. 2012, 343, 295–302. [Google Scholar] [CrossRef]
- Taguchi, K.; Watanabe, Y.; Tsujimura, A.; Tanaka, M. Expression of α-Synuclein Is Regulated in a Neuronal Cell Type-Dependent Manner. Anat Sci Int. 2019, 94, 11–22. [Google Scholar] [CrossRef]
- Lee Clough, R.; Stefanis, L. A Novel Pathway for Transcriptional Regulation of alpha-synuclein. FASEB J. 2007, 21, 596–607. [Google Scholar] [CrossRef]
- Mittal, S.; Bjørnevik, K.; Im, D.S.; Flierl, A.; Dong, X.; Locascio, J.J.; Abo, K.M.; Long, E.; Jin, M.; Xu, B.; Xiang, Y.K.; Rochet, J.C.; Engeland, A.; Rizzu, P.; Heutink, P.; Bartels, T.; Selkoe, D.J.; Caldarone, B.J.; Glicksman, M.A.; Khurana, V.; Schüle, B.; Park, D.S.; Riise, T.; Scherzer, C.R. Β2-Adrenoreceptor Is a Regulator of the α-Synuclein Gene Driving Risk of Parkinson’s Disease. Science (1979) 2017, 357, 891–898. [Google Scholar] [CrossRef]
- Gómez-Santos, C.; Barrachina, M.; Giménez-Xavier, P.; Dalfó, E.; Ferrer, I.; Ambrosio, S. Induction of C/EBPβ and GADD153 Expression by Dopamine in Human Neuroblastoma Cells: Relationship with α-Synuclein Increase and Cell Damage. Brain Res Bull. 2005, 65, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Surguchov, A. Molecular and Cellular Biology of Synucleins. Int Rev Cell Mol Biol. 2008, 270, 225–317. [Google Scholar] [PubMed]
- McHugh, P.C.; Wright, J.A.; Brown, D.R. Transcriptional Regulation of the Beta-Synuclein 5′-Promoter Metal Response Element by Metal Transcription Factor-1. PLoS One 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yin, Y.; Hua, H.; Sun, X.; Luo, T.; Wang, J.; Jiang, Y. The Reciprocal Regulation of γ-Synuclein and IGF-I Receptor Expression Creates a Circuit That Modulates IGF-I Signaling. J Biol Chem. 2010, 285, 30480–30488. [Google Scholar] [CrossRef]
- Lu, A.; Zhang, F.; Gupta, A.; Liu, J. Blockade of AP1 Transactivation Abrogates the Abnormal Expression of Breast Cancer-Specific Gene 1 in Breast Cancer Cells. J Biol Chem. 2002, 277, 31364–31372. [Google Scholar] [CrossRef]
- Shao, T.; Song, P.; Hua, H.; Zhang, H.; Sun, X.; Kong, Q.; Wang, J.; Luo, T.; Jiang, Y. Gamma Synuclein Is a Novel Twist1 Target That Promotes TGF-β-Induced Cancer Cell Migration and Invasion Article. Cell Death Dis. 2018, 9, 625. [Google Scholar] [CrossRef]
- Foulds, P.G.; Mitchell, J.D.; Parker, A.; Turner, R.; Green, G.; Diggle, P.; Hasegawa, M.; Taylor, M.; Mann, D.; Allsop, D. Phosphorylated A-synuclein Can Be Detected in Blood Plasma and Is Potentially a Useful Biomarker for Parkinson’s Disease. FASEB J. 2011, 25, 4127–4137. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Li, J. D. The Roles of Post-Translational Modifications on α-Synuclein in the Pathogenesis of Parkinson’s Diseases. Front Neurosci. 2019, 13, 381. [Google Scholar] [CrossRef]
- Cole, R.N.; Hart, G.W. Cytosolic O-Glycosylation Is Abundant in Nerve Terminals. J Neurochem. 2001, 79, 1080–1089. [Google Scholar] [CrossRef]
- Surgucheva, I.; Newell, K.L.; Burns, J.; Surguchov, A. New α- and γ-Synuclein Immunopathological Lesions in Human Brain. Acta Neuropathol Commun. 2013, 2, 132. [Google Scholar] [CrossRef]
- Gong, H.; Yang, X.; Zhao, Y.; Petersen, R.; Liu, X.; Liu, Y.; Huang, K. Amyloidogenicity of P53: A Hidden Link Between Protein Misfolding and Cancer. Curr Protein Pept Sci. 2014, 16, 135–146. [Google Scholar] [CrossRef]
- Ano Bom, A.P.D.; Rangel, L.P.; Costa, D.C.F.; de Oliveira, G.A.P.; Sanches, D.; Braga, C.A.; Gava, L.M.; Ramos, C.H.I.; Cepeda, A.O.T.; Stumbo, A.C.; De Moura Gallo, C.V.; Cordeiro, Y.; Silva, J.L. Mutant P53 Aggregates into Prion-like Amyloid Oligomers and Fibrils. Journal of Biological Chemistry 2012, 287, 28152–28162. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, N.; Ikezaki, M.; Nishikawa, T.; Namba, N.; Ohgita, T.; Saito, H.; Ihara, Y.; Shimanouchi, T.; Ino, K.; Uchimura, K.; Nishitsuji, K. Sulfated Glycosaminoglycans Mediate Prion-like Behavior of P53 Aggregates. Proc Natl Acad Sci U S A 2020, 117, 33225–33234. [Google Scholar] [CrossRef] [PubMed]
- Claes, F.; Maritschnegg, E.; Baets, G. De; Siekierska, A.; Rubio, M.S.; Ramakers, M.; Michiels, E.; Smet, F. De; Depreeuw, J.; Vergote, I.; Vanderstichele, A.; van den Broeck, A.; Olbrecht, S.; Hermans, E.; Amant, F.; Lambrechts, D.; Nilsson, P. R.; Rousseau, F.; Schymkowitz, J. The Tumor Suppressor Protein PTEN Undergoes Amyloid-like Aggregation in Tumor Cells. bioRxiv. 2020, 11, 402115. [Google Scholar]
- Kehrloesser, S.; Osterburg, C.; Tuppi, M.; Schäfer, B.; Vousden, K.H.; Dötsch, V. Intrinsic Aggregation Propensity of the P63 and P73 TI Domains Correlates with P53R175H Interaction and Suggests Further Significance of Aggregation Events in the P53 Family. Cell Death Differ. 2016, 23, 1952–1960. [Google Scholar] [CrossRef]
- Direito, I.; Monteiro, L.; Melo, T.; Figueira, D.; Lobo, J.; Enes, V.; Moura, G.; Henrique, R.; Santos, M.A.S.; Jerónimo, C.; Amado F, Fardilha M, Helguero LA. Protein Aggregation Patterns Inform about Breast Cancer Response to Antiestrogens and Reveal the Rna Ligase Rtcb as Mediator of Acquired Tamoxifen Resistance. Cancers (Basel) 2021, 13, 3195. [Google Scholar] [CrossRef]
- Matafora, V.; Farris, F.; Restuccia, U.; Tamburri, S.; Martano, G.; Bernardelli, C.; Sofia, A.; Pisati, F.; Casagrande, F.; Lazzari, L.; Marsoni, S.; Bonoldi, E.; Bachi, A. Amyloid Aggregates Accumulate in Melanoma Metastasis Modulating YAP Activity. EMBO Rep. 2020, 21. [Google Scholar] [CrossRef]
- Genovese, I.; Fornetti, E.; Ruocco, G. Mitochondria Inter-Organelle Relationships in Cancer Protein Aggregation. Front Cell Dev Biol. 2022, 10. [Google Scholar] [CrossRef]
- Shekoohi, S.; Rajasekaran, S.; Patel, D.; Yang, S.; Liu, W.; Huang, S.; Yu, X.; Witt, S.N. Knocking out Alpha-Synuclein in Melanoma Cells Dysregulates Cellular Iron Metabolism and Suppresses Tumor Growth. Sci Rep. 2021, 11, 5267. [Google Scholar] [CrossRef]
- Dean, D.N.; Lee, J.C. Linking Parkinson’s Disease and Melanoma: Interplay Between α-Synuclein and Pmel17 Amyloid Formation. Mov Disord. 2021, 36, 1489–1498. [Google Scholar] [CrossRef]
- Pavlenko, T.A.; Roman, A.Y.; Lytkina, O.A.; Pukaeva, N.E.; Everett, M.W.; Sukhanova, I.S.; Soldatov, V.O.; Davidova, N.G.; Chesnokova, N.B.; Ovchinnikov, R.K.; et al. Gamma-Synuclein Dysfunction Causes Autoantibody Formation in Glaucoma Patients and Dysregulation of Intraocular Pressure in Mice. Biomedicines 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Sugama, S.; Nakai, M.; Takenouchi, T.; Wei, J.; Urano, T.; Inoue, S.; Hashimoto, M. α-Synuclein Stimulates Differentiation of Osteosarcoma Cells: Relevance to down-Regulation of Proteasome Activity. J Biol Chem. 2007, 282, 5736–5748. [Google Scholar] [CrossRef] [PubMed]
- Israeli, E.; Yakunin, E.; Zarbiv, Y.; Hacohen-Solovich, A.; Kisos, H.; Loeb, V.; Lichtenstein, M.; Ben-Gedalya, T.; Sabag, O.; Pikarsky, E.; et al. α-Synuclein Expression Selectively Affects Tumorigenesis in Mice Modeling Parkinson’s Disease. PLoS One 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Xu, K. Alpha-Synuclein Contributes to Malignant Progression of Human Meningioma via the Akt/MTOR Pathway. Cancer Cell Int. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chen, X.; Huang, S.; Li, G.; Mo, M.; Zhang, L.; Chen, C.; Guo, W.; Zhou, M.; Wu, Z.; Cen, L.; Long, S.; Li, S.; Yang, X.; Qu, S.; Pei, Z.; Xu, P. Iron Promotes α-Synuclein Aggregation and Transmission by Inhibiting TFEB-Mediated Autophagosome-Lysosome Fusion. J Neurochem 2018, 145, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Kim, Y.M.; Junn, E.; Lee, G.; Park, K.-H.; Tanaka, M.; Ronchetti, R.D.; Quezado, M.M.; Mouradian, M.M. Cell Cycle Aberrations by α-Synuclein over-Expression and Cyclin B Immunoreactivity in Lewy Bodies. Neurobiol Aging 2003, 24, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.M.; Huang, Y.X.; Li, X.L.; Chen, C.; Shi, Q.; Wang, G.R.; Tian, C.; Wang, Z.Y.; Jing, Y.Y.; Gao, C.; Dong, X.P. Molecular Interaction of α-Synuclein with Tubulin Influences on the Polymerization of Microtubule in Vitro and Structure of Microtubule in Cells. Mol Biol Rep 2010, 37, 3183–3192. [Google Scholar] [CrossRef]
- Koss, D.J.; Erskine, D.; Porter, A.; Palmoski, P.; Menon, H.; Todd, O.G.J.; Leite, M.; Attems, J.; Outeiro, T.F. Nuclear Alpha-Synuclein Is Present in the Human Brain and Is Modified in Dementia with Lewy Bodies. Acta Neuropathol Commun. 2022, 10. [Google Scholar] [CrossRef]
- Thorne, N.J.; Tumbarello, D.A. The Relationship of Alpha-Synuclein to Mitochondrial Dynamics and Quality Control. Front Mol Neurosci. 2022, 15. [Google Scholar] [CrossRef]
- Gupta, A.; Inaba, S.; Wong, O.K.; Fang, G.; Liu, J. Breast Cancer-Specific Gene 1 Interacts with the Mitotic Checkpoint Kinase BubR1. Oncogene 2003, 22, 7593–7599. [Google Scholar] [CrossRef]
- Chen, J.; Jiao, L.; Xu, C.; Yu, Y.; Zhang, Z.; Chang, Z.; Deng, Z.; Sun, Y. Neural Protein Gamma-Synuclein Interacting with Androgen Receptor Promotes Human Prostate Cancer Progression. BMC Cancer 2012, 12, 593. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Niu, J.; Sun, E.; Rong, X.; Zhang, X.; Ju, Y. Gamma-Synuclein Binds to AKT and Promotes Cancer Cell Survival and Proliferation. Tumor Biol. 2016, 37, 14999–15005. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Liu, J.; Tian, J.; Zhang, Y.; Yan, M.; Zhu, C. SiRNA Targeting of the SNCG Gene Inhibits the Growth of Gastric Carcinoma SGC7901 Cells in Vitro and in Vivo by Downregulating the Phosphorylation of AKT/ERK. Cytogenet Genome Res. 2018, 154, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gu, L.; Li, X.; Wang, J. Silencing of Synuclein-γ Inhibits Human Cervical Cancer through the AKT Signaling Pathway. Cell Mol Biol Lett. 2019, 24, 49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, X.H.; Li, C.; Wu, X.X.; Chen, Y.L.; Li, W.W.; Li, X.; Gong, F.; Tang, Q.; Jiang, D. SNCG Promotes the Progression and Metastasis of High-Grade Serous Ovarian Cancer via Targeting the PI3K/AKT Signaling Pathway. J Exp and Clin Cancer Res. 2020, 39, 79. [Google Scholar] [CrossRef]
- Sun, D.; Li, W.Y.; Chen, S.H.; Zhi, Z.F.; Lin, H.S.; Fan, J.T.; Fan, Y.J. ShRNA-Mediated Suppression of γ-Synuclein Leading to Downregulation of P38/ERK/JNK Phosphorylation and Cell Cycle Arrest in Endometrial Cancer Cells. Mol Biol. 2020, 54, 1006–1017. [Google Scholar] [CrossRef]
- Chen, J.; Huang, S.; Wu, K. jin; Wang, Y. kun; Jia, Y. jun; Lu, Y. shu; Weng, Z. yi [The Correlation of Synuclein-γ and Matrix Metalloproteinase 9 in Breast Cancer]. Zhonghua Wai Ke Za Zhi 2013, 51, 641–644. [Google Scholar]
- Surgucheva, I.G.; Sivak, J.M.; Fini, M.E.; Palazzo, R.E.; Surguchov, A.P. Effect of γ-Synuclein Overexpression on Matrix Metalloproteinases in Retinoblastoma Y79 Cells. Arch Biochem Biophys. 2003, 410, 167–176. [Google Scholar] [CrossRef]
- Pan, Z.Z.; Bruening, W.; Godwin, A.K. Involvement of RHO GTPases and ERK in Synuclein-γ Enhanced Cancer Cell Motility. Int J Oncol. 2006, 29, 1201–1205. [Google Scholar] [CrossRef]
- Liu, C.; Qu, L.; Lian, S.; Tian, Z.; Zhao, C.; Meng, L.; Shou, C. Unconventional Secretion of Synuclein-γ Promotes Tumor Cell Invasion. FEBS J. 2014, 281, 5159–5171. [Google Scholar] [CrossRef]
- Zhao, J.; Xing, N. Identification of γ-Synuclein as a Stage-Specific Marker in Bladder Cancer by Immunohistochemistry. Med Sci Monit. 2014, 20, 2550–2555. [Google Scholar] [PubMed]
- Chen, Z.; Ji, Z.; Wang, Q.; Shi, B.; Shou, C.; Liu, C.; Fan, H.; Li, H.; Davidson, K.T.; Wakefield, M.R.; Ball, T.W.; Fang, Y. Expression of γ-Synuclein in Bladder Carcinoma: A Possible Marker for Prognosis. Anticancer Res. 2016, 36, 951–956. [Google Scholar] [PubMed]
- Takemura, Y.; Ojima, H.; Oshima, G.; Shinoda, M.; Hasegawa, Y.; Kitago, M.; Yagi, H.; Abe, Y.; Hori, S.; Fujii-Nishimura, Y.; Kubota, N.; Masuda, Y.; Hibi, T.; Sakamoto, M.; Kitagawa, Y. Gamma-synuclein is a Novel Prognostic Marker that Promotes Tumor Cell Migration in Biliary Tract Carcinoma. Cancer Med. 2021, 10, 5599–5613. [Google Scholar] [CrossRef] [PubMed]
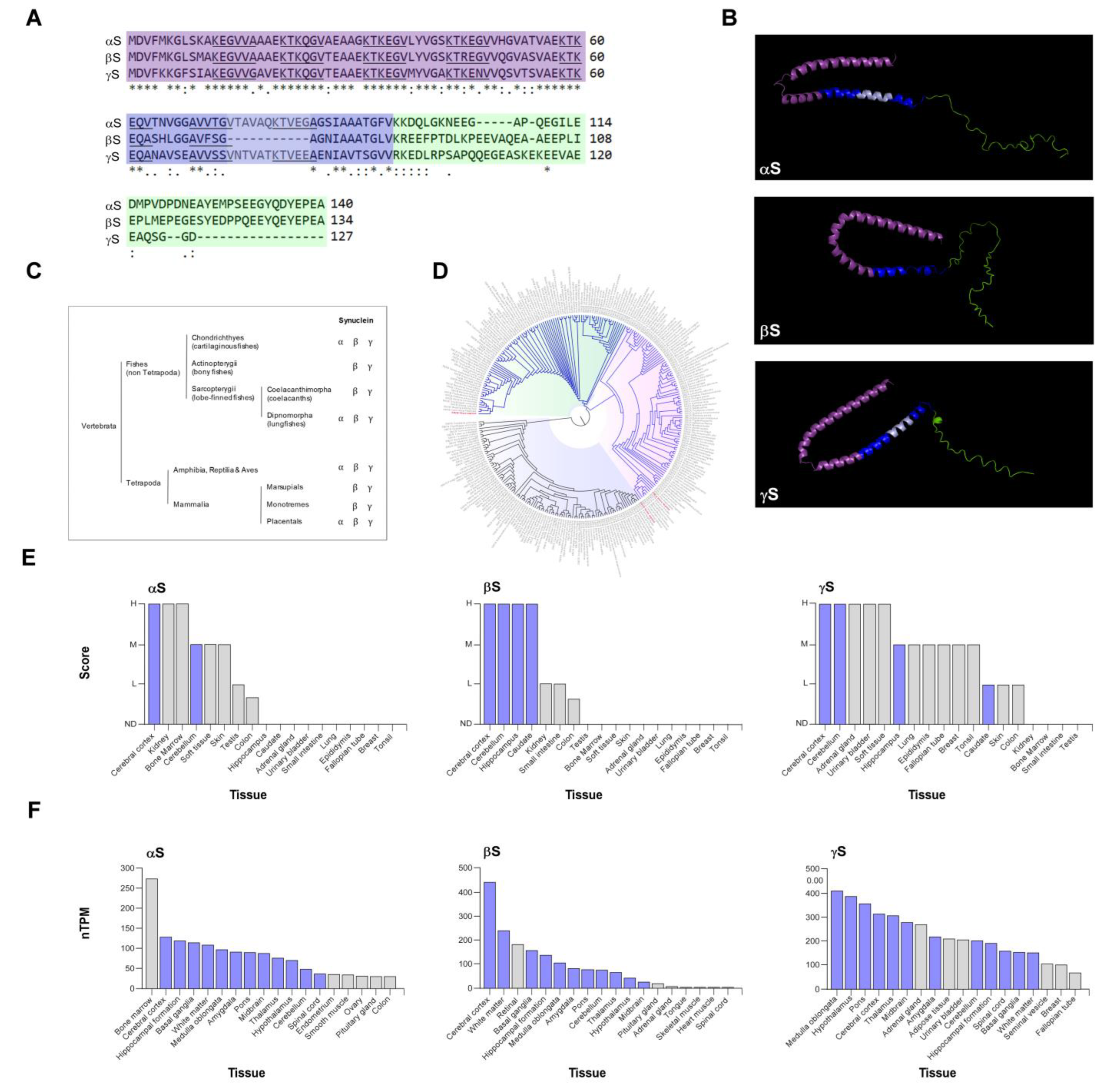
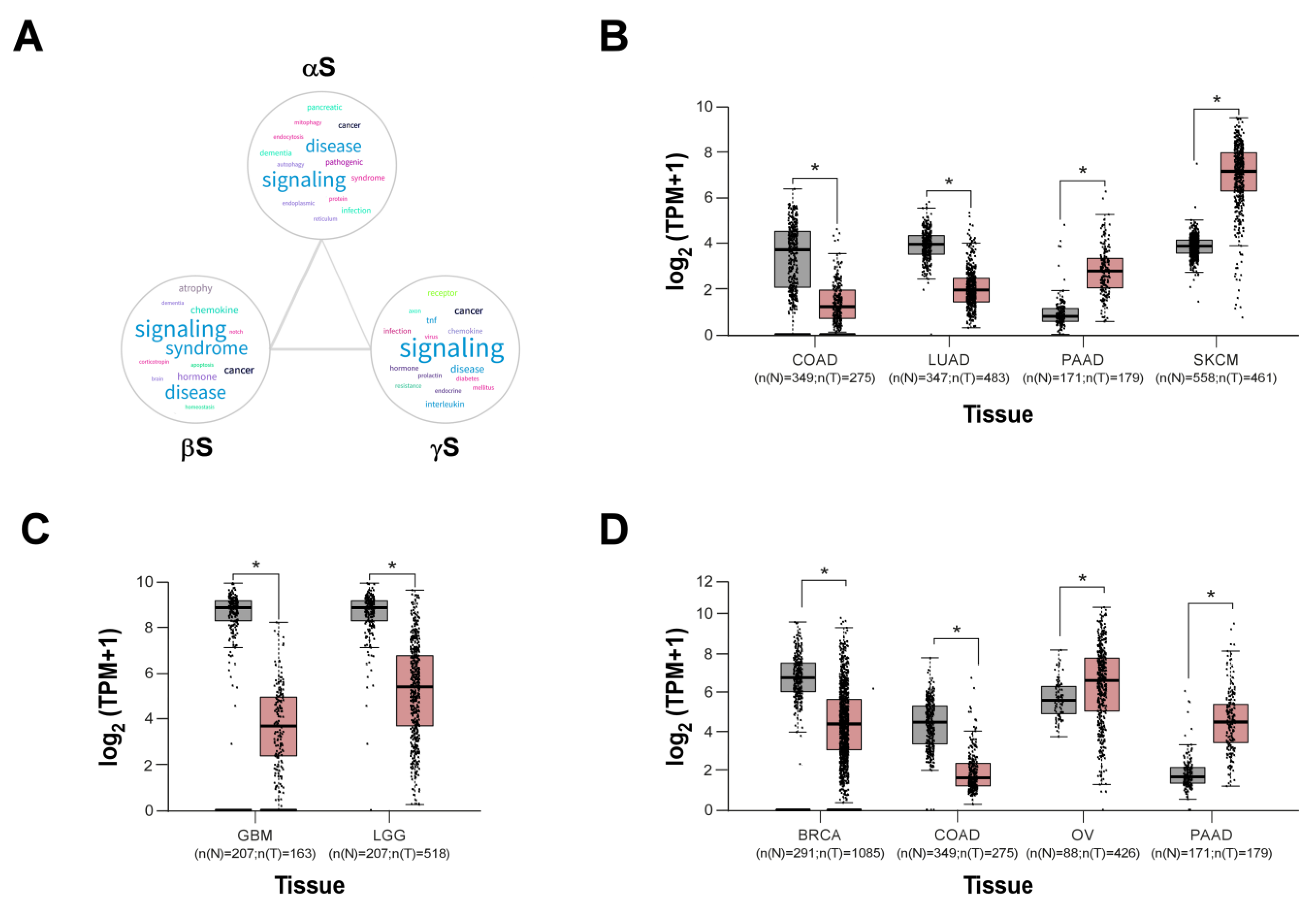
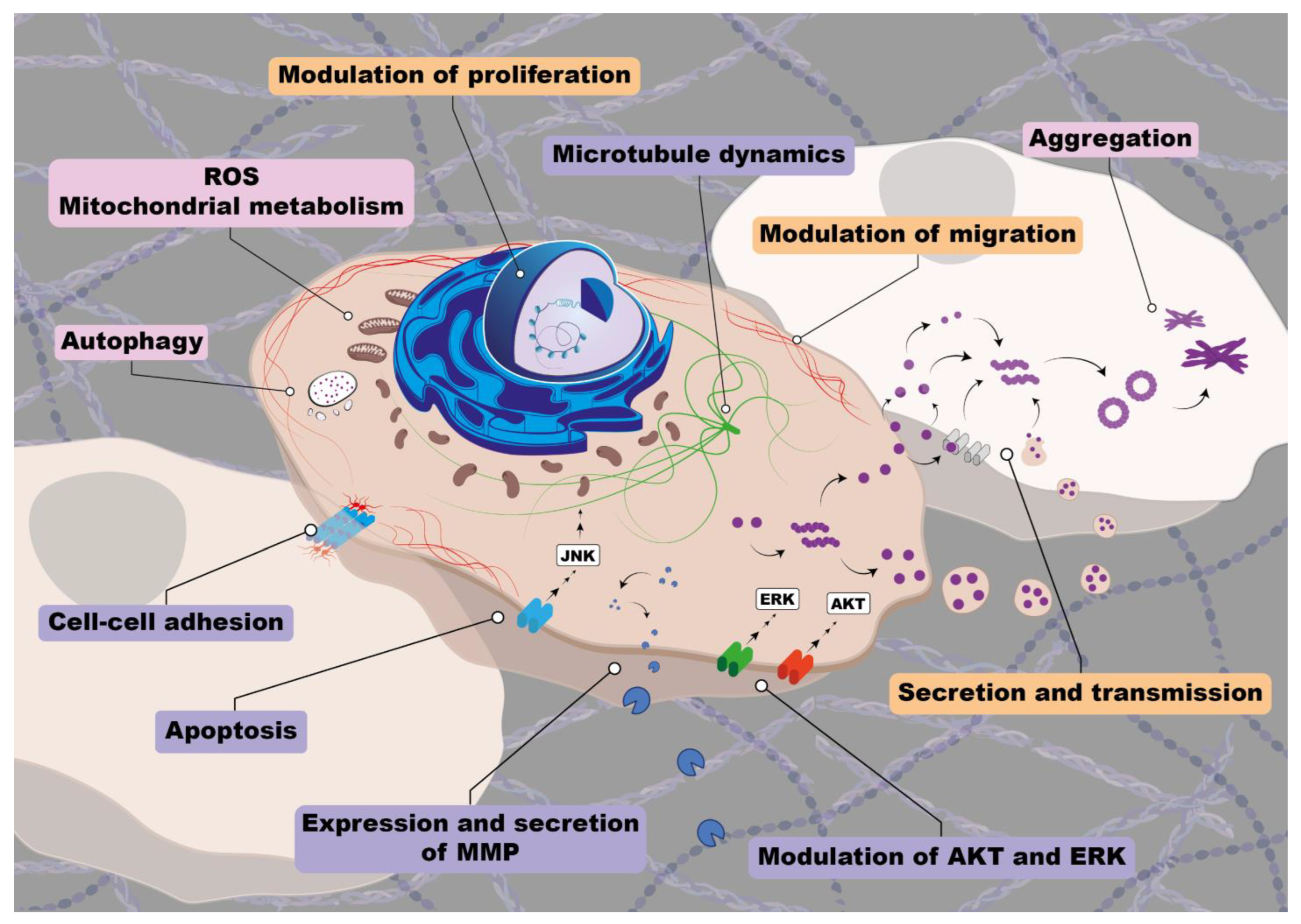
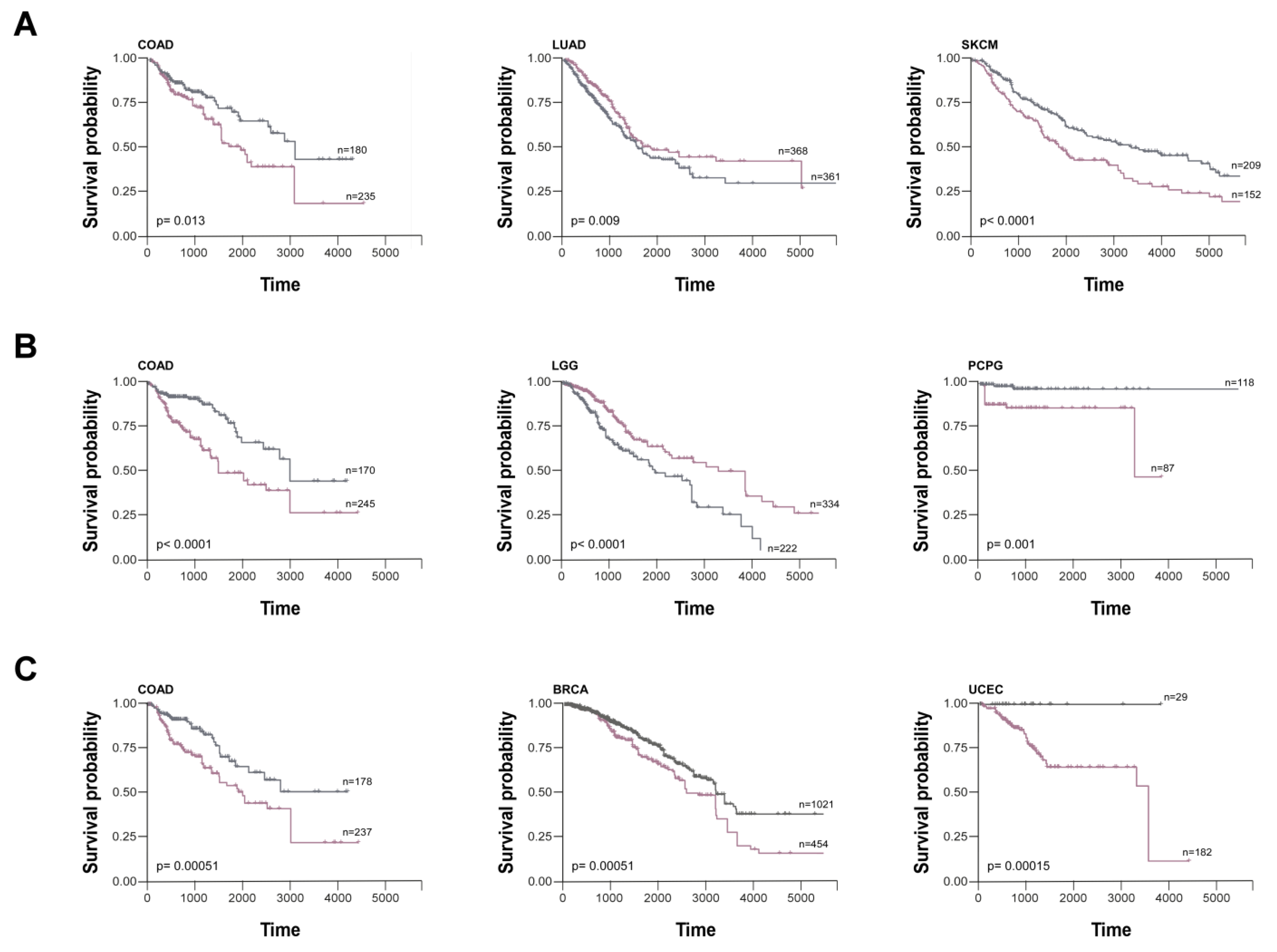
| Synucleins | Year | Tumor type | Type of study | Reference |
| γS | 1997 | Breast | Patients | [6] |
| γS | 1999 | Breast | Cell lines | [41] |
| βS, γS | 2000 | Breast, Ovarian | Cell lines and patients | [42] |
| αS | 2000 | Ganglioglioma, Ganglioneuroblastoma | Patients | [43] |
| αS | 2000 | Medulloblastoma, Pineocytoma, Pineoblastoma | Patients | [44] |
| γS | 2001 | Retinoblastoma | Cell lines | [45] |
| γS | 2001 | Ovarian | Cell lines | [46] |
| γS | 2003 | Esophageal | Cell lines and patients | [47] |
| αS, βS | 2003 | Glial, Medulloblastoma | Patients | [48] |
| γS | 2004 | Pancreatic | Cell lines and patients | [49] |
| γS | 2004 | Gastric | Cell lines and patients | [50] |
| γS | 2004 | Bladder | Patients | [51] |
| γS | 2005 | Prostate, Cervical, Colon, Lung | Patients | [39] |
| αS | 2007 | Osteosarcoma | Cell lines | [92] |
| γS | 2008 | Colon | Cell lines and patients | [52] |
| γS | 2009 | Uterine serous papillary carcinoma | Cell lines and patients | [54] |
| αS, βS, γS | 2010 | Colon | Cell lines and patients | [53] |
| αS | 2010 | Melanoma | Cell lines and patients | [55] |
| αS, βS | 2011 | Leukemia | Cell lines and patients | [56] |
| αS | 2011 | Melanoma, Breast, Lung | Cell lines and mice | [93] |
| γS | 2012 | Endometrium | Patients | [57] |
| γS | 2016 | Squamous cell carcinoma of the oral cavity | Cell lines and patients | [59] |
| αS | 2016 | Meningioma | Cell lines and patients | [94] |
| γS | 2021 | Biliary tract | Cell lines and patients | [113] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





