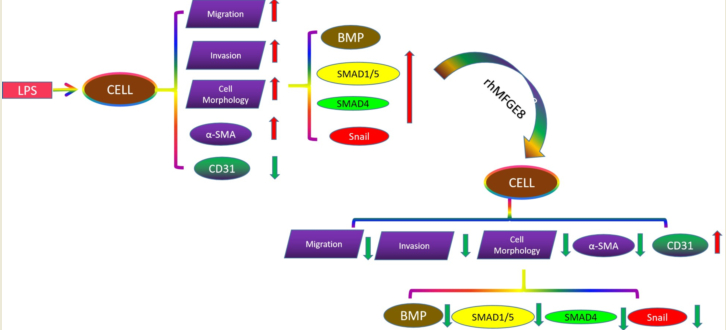
1. Introduction
Acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) are important causes of acute respiratory failure[
1,
2,
3], and their pathological features are acute diffuse inflammatory injury accompanied by a lack of surfactant and acute respiratory distress. Traditionally, the pathological process of ALI is a continuous process that includes exudation, proliferation and fibrosis[
3,
4,
5]. However, the process of fibrosis in ALI was recently shown to be initiated during the early stage. Ameliorating pulmonary fibrosis is beneficial for improving refractory hypoxemia, decreasing the use of ventilators and improving patient survival[
3,
6]. The current research aim is to alleviate pulmonary fibrosis[
7,
8].
Many studies have shown that pulmonary fibrosis is the result of the deposition of extracellular matrix (ECM), which is generated by myofibroblasts[
9,
10]. Myofibroblasts from endothelial cells (ECs) undergo a process known as endothelial-to-mesenchymal transition (EndoMT), and through this process, the cells acquire a mesenchymal phenotype[
11]. Under specific physiological and pathological conditions, the polarity of EndoMT ECs is weakened, the tight junctions between ECs are lost, and the cells acquire the movement and contraction characteristics of mesenchymal cells. After these changes, the cells detach from the endothelial layer, and their migration and invasion are significantly enhanced after the cells migrate to the interstitium and transform into myofibroblasts. During this transformation, the cells change from cobblestone-like to elongated spindle-shaped cells, lose specific molecular markers of ECs [CD31/PECAM-1, von Willebrand factor (vWF), and VE-cadherin], and acquire specific molecular markers of mesenchymal cells (α-SMA, vimentin, and type I collagen)[
12].
Several studies have demonstrated that the generation of myofibroblasts depends on the transforming growth factor beta (TGF-β) signalling pathway[
13,
14,
15]. Bone morphogenetic proteins (BMPs) belong to the TGF-β family and can bind serine-threonine kinase receptors, including type II and type I receptors[
10]. Studies have shown that BMP type I receptors can induce phosphorylation of Smad1, Smad5 and Smad8, and these receptors can also associate with phosphorylated Smad4, form a complex in the nucleus and cooperate with other transcription factors to regulate the transcriptional responses of target genes, which results in regulation of the EndoMT process[
16,
17,
18].
Milk fat globule epidermal growth factor 8 (MFGE8) is an important molecule that mediates phagocytosis and is expressed in the alveolar epithelium, vascular ECs and macrophages[
19]. It has been reported that this protein has crucial physiological and pathophysiological functions in various diseases, including liver, pulmonary and cardiovascular diseases. Laura W. Hansen found that MFGE8 deficiency leads to increased inflammation, tissue damage, neutrophil infiltration, and apoptosis, which causes sepsis in neonatal mice and results in elevated morbidity and mortality[
20]. The Monowar Aziz study clearly indicated the importance of MFGE8 in improving neutrophil infiltration and demonstrated the potential of MFGE8 as a novel treatment for ALI[
21]. Furthermore, in recent years, an increasing number of studies have highlighted the potential role of MFGE8 in organ fibrosis, as revealed by the findings that MFGE8 slows the degree of skin and renal fibrosis in mice and reduces tissue fibrosis in mouse models of liver and cardiac fibrosis[
19,
20,
22]. However, the molecular mechanisms of MFGE8 and EndoMT remain unclear and need to be examined. We hypothesized that rhMFGE8 exerts a protective effect against EndoMT induced by lipopolysaccharide (LPS) through the BMP/Smad1/5/Smad4 signalling pathway.
2. Materials and methods
2.1. Clinical study and data collection
In this study, 44 patients with ARDS and healthy volunteers who were admitted to the Second Affiliated Hospital of Chongqing Medical University (Chongqing, China) between April 1 and August 12, 2021, were enrolled. Hospital nonsurvival was defined as death in the hospital (n = 11), and hospital survival was defined as discharge home with unassisted breathing (n = 33). The inclusion criteria were in accordance with the 2012 Berlin definition of ARDS. The exclusion criteria included congestive heart failure, interstitial lung disease, connective-tissue diseases such as polymyositis, diffuse alveolar haemorrhage due to vasculitis or Goodpasture syndrome, drug-induced lung diseases, cancer, and endobronchial tuberculosis. Blood serum samples were obtained from the patients and volunteers and cryopreserved at −80°C for subsequent experiments. All patients and volunteers provided written informed consent, and the Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University approved this study.
2.2. Cell culture
Human lung microvascular endothelial cells (HLMECs) were purchased from ScienCell (ScienCell Research Laboratories, Inc., CA, USA, cat. no. 3000) and stored at the College of Life Science. We cultured the cells with DMEM/F-12 (ScienCell Research Laboratories, Inc., CA, USA, cat. no. 09421), 10% standard foetal bovine serum (HyClone, Waltham, MA, USA, cat. no. SH30084.03) and 1% penicillin–streptomycin (ScienCell Research Laboratories, Inc., CA, USA, cat. no. 0513) in a 5% CO2 incubator at 37°C.
2.3. Mouse model of ALI
Mice (C57BL/6, aged 7-8 weeks, male) were purchased from the Laboratory Animal Center of Chongqing Medical University. In this study, all experimental animals were housed in the school’s accredited animal facility, which was certified by the Association of Assessment and Accreditation of Laboratory Animal Care International (AAALAC), under a comfortable temperature (24°C) and light cycles (12-h light/12-h dark) and provided sterile mouse chow and water ad libitum. The experimental procedure was implemented in a pathogen-free manner. Forty-five mice were divided into three groups according to a completely randomized design. The mice in the control group were intraperitoneally administered physiological saline. Those in the LPS group were intraperitoneally administered 5 mg/kg LPS dissolved in physiological saline, and those in the LPS+rhMFGE8 group were intraperitoneally administered 5 mg/kg LPS and 20 µg/kg rhMFGE8. At the endpoint of the experiment, the experimental mice were anaesthetized with sodium pentobarbital (60 mg/kg) and euthanised with potassium chloride (100 mg/kg) on days 7, 14 and 28. The Animal Ethics Committee of Chongqing Medical University authorized all animal experimental protocols.
2.4. Antibodies and chemicals
LPS (from Escherichia coli 055:B5, 5 mg/ml, cat. no. L8880) was purchased from Solarbio Limited Company. LDN-2128549 (BMP receptor inhibitor, cat. no.1432597-26-6) was procured from APEXBIO Company. rh-MFGE8 (cat. no. 2767-MF) was purchased from R&D Systems, a biotech company. The MFGE8 ELISA kit (cat. no. SEB286Hu) was obtained from Cloud-Clone Limited Company. siRNA-MFGE8-1 (lot number: 430019983), siRNA-MFGE8-2 (lot number: 430084101), and siRNA-MFGE8-3 (lot number: 430048100) were obtained from Qiagen Limited Company. AB Antibody Technology provided primary antibodies, such as CD31 (1:1,000, cat. no. ab9498) and BMP4 (1:1,000, cat. no. ab39973) antibodies, and Cell Signaling Technology (Danvers, CT, USA) provided other antibodies, including α -SMA (1:1,000, cat. no. 19245), Snail (1:1,000, cat. no. 3879), Smad1 (1:1,000, cat. no. 6499), Smad4 (1:1,000, cat. no.46535), and Smad5 (1:1,000, cat. no.12534) antibodies. Fluorochrome-conjugated antibody (goat anti-rabbit IgG H&L/AF488 antibody, bs-0295G-AF488) was purchased from Bioss Antibodies Limited Company. The Beyotime Institute of Biotechnology (Shanghai, China) provided secondary antibodies, including horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (cat. no. A0208) and anti-mouse IgG (cat. no. A0192).
2.5. Cell morphology and imaging
HLMECs were seeded in culture bottles after the cells reached logarithmic growth. The control cells were treated with administered physiological saline, the cells in the LPS group was treated with 20 μg/ml LPS dissolved in physiological saline, and those in the LPS+rhMFGE8 group were treated with 20 μg/ml LPS and 80 μg/ml rhMFGE8. The cells in the LPS+siRNA group were treated with 20 μg/ml LPS and 100 nM siRNA, and those in the LPS+siRNA+rhMFGE8 group were treated with 20 μg/ml LPS, 80 μg/ml rhMFGE8 and 100 nM siRNA. The cells were incubated for 96 h. The cell morphology was observed using an inverted system microscope provided by Olympus Company and photographed.
2.6. Transwell migration assay
Cell migration was assessed using a Transwell chamber assay (8 μm pore size, Corning, 3413) according to the manufacturer’s instructions. A total of 1 × 105 HLMECs were treated with LPS (20 μg/ml), siRNA and/or rhMFGE8 and resuspended in serum-free medium, and the cells were added to the upper layer of the Transwell chambers. DMEM/F12 medium containing 10% foetal bovine serum was added to the lower layer, and the cells were incubated for 24 h. The chambers were washed, and cotton buds were used to remove the cells on the upper surface. The cells on the lower surface were fixed with 4% paraformaldehyde for 30 min and then stained with crystal violet for 30 min. The cell numbers in five randomly selected areas were counted to evaluate cell migration.
2.7. Scratch assay
Cells were seeded in 6-well plates at a density of 5.0 × 105 cells/ml. When the confluence of the cells reached 90%, the cell monolayer was scratched with a sterile 200 μL pipette tip, and the wells were washed twice with PBS. Images were taken using an inverted system microscope provided by Olympus Company, and the migration distance was analysed using Image-Pro Plus 6.0
2.8. Cellular immunofluorescence
Cells were added to 24-well plates with cover slips, cultured for 96 h with stimulation, fixed in 4% paraformaldehyde for 15 min, permeabilised with 0.5% Triton X-100 for 20 min and blocked for 30 min. Subsequently, primary antibodies against CD31 and/or α-SMA were added, and the cells were incubated overnight at 4°C. Secondary antibodies conjugated to Alexa Fluor 488 were added, and the cells were incubated for 1 h. DAPI was then added for visualization. ImageJ was used to quantify the fluorescence intensity in five randomly selected areas.
2.9. Flow cytometry
After stimulation, the cells were fixed in 4% paraformaldehyde for 15 min and permeabilized with Triton X-100 (0.5%). The cells were then incubated with anti-CD31 and α-SMA antibodies, and cell fluorescence was measured with a FACSCanto II instrument (Becton Dickinson, San Jose, CA, USA) and analysed using Flow Jo software (Tree Star, San Carlos, CA, USA).
2.10. RNA interference
Small interfering RNAs (siRNAs) targeting human MFGE8 were obtained from Qiagen. According to the manufacturer’s instructions, we transiently transfected the cultured cells using Lipofectamine™ 8000 (Beyotime Institute of Biotechnology) and 100 nM siRNA. We performed western blotting and q-PCR to measure the efficiency and to obtain specific siRNAs, which were used for future experiments. The targeting sequences of the siRNAs were (a) AGTGGAGAACACGAAT, (b) TCACTCCGGAATAAAT, and (c) TGGCTGTCAGGAATTG.
2.11. Quantitative real-time PCR
Total RNA from the cells was extracted using TRIzol reagent (Takara, Code No. 9108) and transcribed into complementary DNA (cDNA) using the PrimeScript RT kit (Takara, RR047A). Quantitative real-time polymerase chain reaction (qRT‒PCR) was performed in a 50 μL reaction volume using SYBR Green (Takara, RR820A). β-Actin was used as the reference gene, and the change in mRNA was quantified using the 2
-ΔΔCq method. The primer pairs are listed in
Table 1.
2.12. Western blotting
Total protein was isolated from the cells, and the protein concentration was measured with a BCA protein assay kit. Equal amounts of protein (40 µg) were separated using 10-15% SDS‒PAGE. The separated proteins were subsequently transferred to PVDF (0.22 μm or 0.45 μm) membranes. The membranes were then blocked, incubated with primary antibodies overnight at 4°C, washed with TBST, and incubated with HRP-conjugated goat anti-rabbit IgG or goat anti-mouse IgG secondary antibodies. ECL reagents were used to visualize the signals. Image Lab software was used for densitometry analysis of the immunoblots.
2.13. H&E staining
Lung tissue blocks were embedded in paraffin, cut into specimens and deparaffinized with xylene. Ethanol was then used for dehydration. The sections were dyed with haematoxylin for 5 min and differentiated with hydrochloric acid in ethanol. The samples were then soaked in warm water for 15 min, transferred into eosin solution for 2 min and dehydrated with ethanol. The sections were then sealed with neutral resin.
2.14. Masson staining
The degree of lung fibrosis was determined by Masson staining. Paraffin sections of lung tissues were routinely prepared and stained with Masson’s trichrome blue. The samples were sequentially stained with haematoxylin for at least 6 min, Masson complex oil for 5 min, 5% phosphomolybdic acid for 5 min, and 2% aniline blue for 10 min. The samples were routinely dehydrated and placed on slides, and the sections were observed with an optical microscope.
2.15. Statistical analysis
All data were calculated and analysed using PSS 22.0 statistical software and GraphPad Prism 6.0. The statistical analyses were performed using two-tailed Student’s t test. Differences between the control and treated samples were considered statistically significant if P <0.01.
4. Discussion
In this study, we first demonstrated that LPS could induce EndoMT in HLMECs. Our previous studies showed that LPS, which is an important inflammatory inducer, could be used in cell and animal models to investigate the mechanism and feasible treatments of ALI[
7,
8,
23]. Toshio Suzuki et al. found that EndoMT induced by LPS is dependent on the ROS levels
in vitro, and transient EndoMT was observed in mice with septic ALI, which could be repaired[
24]. Several environmental factors, including inflammation, hypoxia, radiation, hyperglycaemia and TGF-β, have been shown to trigger EndoMT[
25,
26,
27]. During initiation of EndoMT, the molecular and structural properties of ECs are altered by various stimulating factors. Cells lose their endothelial properties (reduced expression of surface EC markers such as CD31 and vWF) and cell adhesion abilities and acquire high migratory potential and fibroblast-like phenotypes[
11]. Our results are in agreement with previous research. This study demonstrated that LPS greatly inhibited the level of CD31 protein and increased the level of α-SMA protein, which showed that LPS could induce EndoMT.
MFGE8 is derived from macrophages and plays an important role in reducing inflammation and maintaining tissue homeostasis. Xiao Wang et al. found that LPS suppresses MFGE8 gene expression in macrophages[
28]. Atabai et al. discovered that the expression of MFGE8 was decreased, which could exacerbate the severity of lung injury[
29]. We confirmed these reports and also showed that MFGE8 was decreased by approximately 40% in HLMECs after LPS stimulation and in ARDS patients. In contrast to previous findings, Jun Wang et al. showed that the protein expression of MFGE8 was elevated in the serum of patients with chronic pulmonary hypertension[
19]. The differences in the results of these studies are likely attributable to the differences in pathophysiological processes between ALI and pulmonary hypertension. Monowar Aziz et al. showed that exogenous supplementation with MFGE8 could reduce neutrophil and cellular infiltration induced by LPS by upregulating GRK2 expression and downregulating CXCR2 expression, which reduces lung injury[
21]. A previous study revealed that MFGE8 is secreted by mesenchymal stem cells (MSCs) and acts as an autocrine factor that suppresses TGF-β signalling by binding to αvβ3 integrin and decreases ECM deposition and liver fibrosis in mice[
30]. Our results that endogenous supplementation with rhMFGE8 could attenuate LPS-induced EndoMT
in vitro are in accordance with Bo Wang’s study, who showed that MFGE8 could exert a protective effect on TGF-β1-induced EndoMT and may be a therapeutic strategy for cardiac fibrosis[
30].
Previous studies have indicated that the transforming growth factor (TGF)-β and BMP families of growth factors act as mediators that promote EndoMT, including downstream Smad-dependent and Smad-independent proteins. After their phosphorylation, Smad1 and Smad5 recruit Smad4 and then create a complex that plays a crucial role in enhancing signal cascade conduction[
13]. After translocation from the cytoplasm to the nucleus, the complexes interact with target transcription factors, such as Zeb1, Twist, Slug and Snail. We found that the expression of BMP, Smad1/5 and Smad4 was elevated in response to stimulation with LPS, and LDN-212854, a BMP/Smad1/5/4 signalling pathway inhibitor, was used to verify the involvement of this signalling pathway. The results showed that knockdown of MFGE8 increased the activity of the BMP signalling pathway, whereas the administration of rhMFGE8 reversed this effect.
In this study, we found that LPS induced the deposition of blue-stained ECM in the alveolar interstitial space and around mouse pulmonary arteries at the early stage and that the level of deposition increased over time. Kun Xiao found that proinflammatory cytokines are the most effective inducers of EndoMT, which leads to inflammatory conditions that transform ECs into myofibroblasts and thereby facilitates fibrotic diseases[
31]. In the lungs, fibrosis may occur as a result of abnormal remodelling following ALI and inflammatory diseases, in the context of systemic autoimmune diseases, or as an idiopathic process[
8]. Pulmonary tissue architecture is determined by the balance among collagen production, deposition and removal, which is a dynamic process. When sustained collagen production exceeds or overwhelms the mechanisms of collagen removal, excess collagen is deposited in the ECM, leading to tissue fibrosis. Atabai et al demonstrated that mice with deficiency in the MFGE8 gene exhibit collagen accumulation in pulmonary tissues after bleomycin treatment and that rhMFGE8 is effective in binding and internalizing collagen and accelerating the removal of collagen. We also found that rhMFGE8 ameliorated ECM deposition induced by LPS. Wayne W. Chaung et al. reported that rhMFGE8 sharply attenuates sepsis in rats with acute alcohol exposure and should be further developed as a safe and effective therapy for sepsis in alcohol abusers[
32]. Laura W. Hansen et al. used male and female newborn mice and showed that a lack of MFGE8 exacerbates lung injury and mortality in neonatal sepsis and that rhMFGE8 has a protective effect in neonatal sepsis[
20].
However, the present study has several limitations. We did not obtain patient lung tissue and were therefore unable to compare this tissue with mouse lung tissue. We detected the serum expression of MFGE8 in patients only when they were diagnosed with ARDS, and we did not detect the serum expression of MFGE8 after the patients had recovered; thus, we will conduct these experiments in future studies. We administered rhMFGE8 to mice for only six days; if we had administered rhMFGE8 to the mice for additional days, the difference between each group would have been more obvious.
In conclusion, the results obtained in this study suggested that rhMFGE8 exerted a protective effect at the early stage of LPS-induced EndoMT through the BMP/Smad1/5/Smad4 signalling pathway and could be a therapeutic target in ALI.
Table 1.
1. Clinical characteristics.
Table 1.
1. Clinical characteristics.
| Variables |
Healthy control (n=44) |
ARDS (n=44) |
| Age (years) |
68.11±9.38 |
72.02±9.26 |
| Gender (male/female) |
20/24 |
23/21 |
| BMI(kg/cm2) |
23.59±1.13 |
23.38±1.37 |
| PaO2/FiO2 (mmHg) |
431.85±22.67 |
93.61±14.64* |
| WBC counts (109/L) |
7.43±1.68 |
16.69±5.34* |
| C-reactive protein (mg/L) |
6.08±2.47 |
30.64±11.16* |
| Procalcitonin(ng/mL) |
0.54±0.29 |
2.83±1.27* |
| MFGE8(pg/ml) |
79.36±14.90 |
60.11±22.77* |
Table 1.
2. Demographic characteristics and clinical data of 44 patients with ARDS.
Table 1.
2. Demographic characteristics and clinical data of 44 patients with ARDS.
| |
Survivors (n=33) |
Nonsurvivors (n=11) |
| Age (years) |
67.03±9.30 |
71.36±9.29 |
| Gender (male/female) |
20/13 |
3/8 |
| BMI(kg/cm2) |
23.4±1.32 |
23.34±1.58 |
| PaO2/FiO2 (mmHg) |
95.30±14.32 |
88.54±15.11 |
| WBC counts (109/L) |
17.44±5.23 |
14.43±5.26 |
| C-reactive protein (mg/L) |
30.51±11.75 |
31.02±1.02 |
| Procalcitonin (ng/mL) |
2.75±1.35 |
3.06±1.27 |
| MFGE8 (pg/ml) |
65.6±18.94 |
45.09±10.79* |
| Comorbidity |
|
|
| Obstructive airway disease |
10 |
6 |
| Hypertension |
14 |
4 |
| Diabetes |
6 |
3 |
| Cardiovascular disease |
6 |
3 |
| Cerebrovascular accident |
1 |
0 |
| Haematological disease |
1 |
1 |
| Chronic renal failure |
0 |
2 |
| Cancer history |
5 |
2 |
| Aetiology of ARDS, n (%) |
|
|
| Pulmonary aetiology |
20(60.60%) |
7(63.63%) |
| Pulmonary infection |
13(39.39%) |
5(45.45%) |
| Aspiration |
7(21.21%) |
2(18.18%) |
| Extrapulmonary aetiology |
13(39.39%) |
4(36.36%) |
| Sepsis |
8(24.24%) |
2(18.18%) |
| Pancreatitis |
5(15.15%) |
2(18.18%) |
Figure 1.
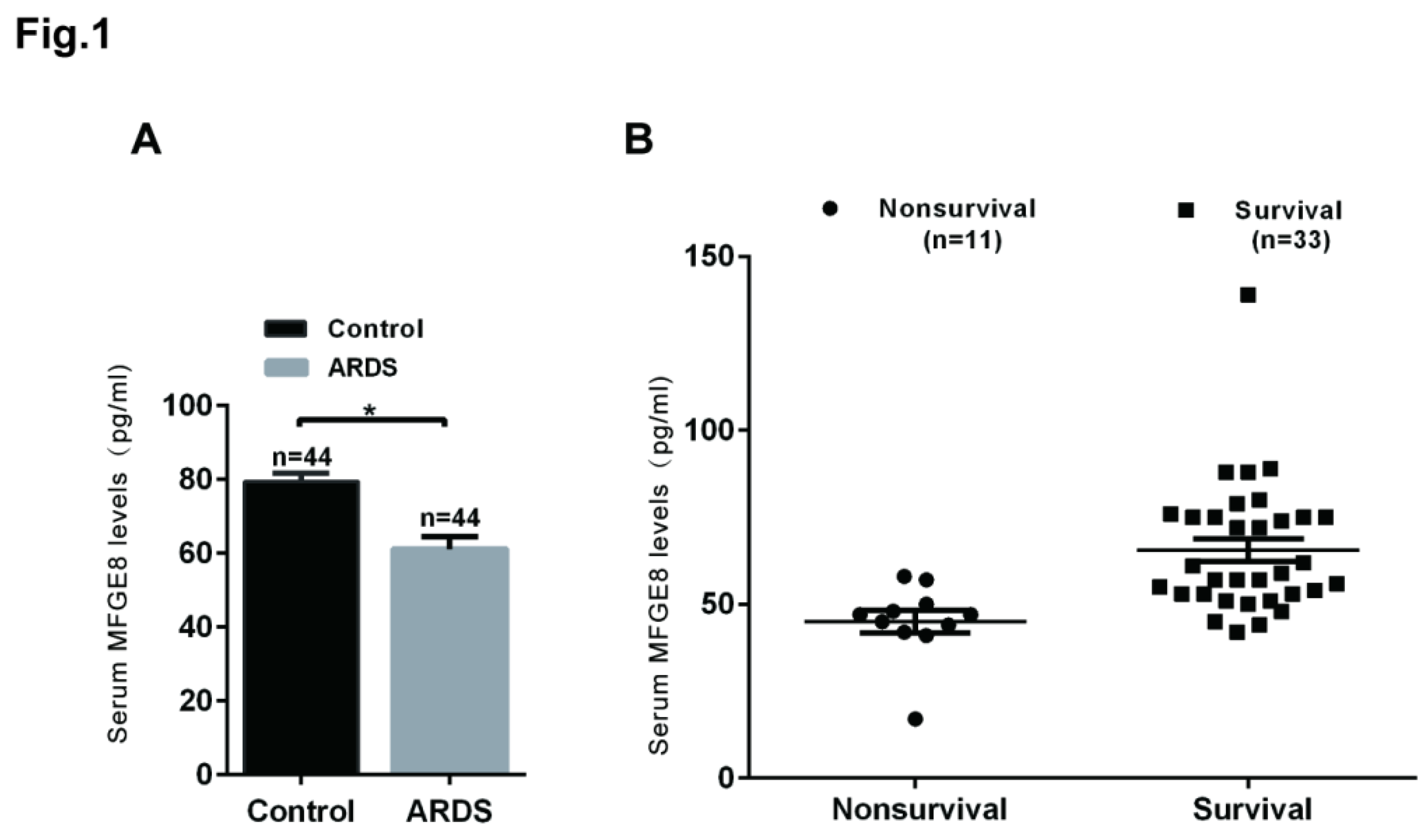
MFGE8 was a protective factor in ARDS patients. An ELISA kit was used to determine the MFGE8 serum concentrations in ARDS patients (n=44) and healthy volunteers (n=44). (B) MFGE8 levels in patients who survived (n=33) and in nonsurvivors (n=11). The statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the ARDS group and the control group.
Figure 1.
MFGE8 was a protective factor in ARDS patients. An ELISA kit was used to determine the MFGE8 serum concentrations in ARDS patients (n=44) and healthy volunteers (n=44). (B) MFGE8 levels in patients who survived (n=33) and in nonsurvivors (n=11). The statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the ARDS group and the control group.
Figure 2.
HLMECs treated with LPS underwent EndoMT. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into two groups: control group and LPS group. (A) The cells were incubated with LPS (20 μg/mL) for 96 h, and microscopy was used to observe their morphology (magnification, x100, scale bar = 50 μm). (B) The cells were incubated with LPS (20 μg/mL) for 48 h, and a Transwell migration assay was performed to examine cell invasion (magnification, x100, scale bar = 50 μm). (C) The cells were incubated with LPS (20 μg/mL) for 24 h, and a scratch assay was performed to examine cell migration (magnification, x100, scale bar = 50 μm). (D) The cells were incubated with LPS (20 μg/mL) for 96 h, and fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA (magnification, x200, scale bar = 20 μm). (E) The cells were incubated with LPS (20 μg/mL) for 96 h, and the protein expression of CD31 and α-SMA was examined by western blotting. β-Actin was used as a reference protein. (F) The cells were incubated with LPS (20 μg/mL) for 96 h, and the expression levels of the CD31 and α-SMA genes were examined by qRT‒PCR. (G) The cells were incubated with LPS (20 μg/mL) for 96 h, and FCM was used to examine the protein expression of CD31 and α-SMA. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the LPS group and the control group.
Figure 2.
HLMECs treated with LPS underwent EndoMT. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into two groups: control group and LPS group. (A) The cells were incubated with LPS (20 μg/mL) for 96 h, and microscopy was used to observe their morphology (magnification, x100, scale bar = 50 μm). (B) The cells were incubated with LPS (20 μg/mL) for 48 h, and a Transwell migration assay was performed to examine cell invasion (magnification, x100, scale bar = 50 μm). (C) The cells were incubated with LPS (20 μg/mL) for 24 h, and a scratch assay was performed to examine cell migration (magnification, x100, scale bar = 50 μm). (D) The cells were incubated with LPS (20 μg/mL) for 96 h, and fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA (magnification, x200, scale bar = 20 μm). (E) The cells were incubated with LPS (20 μg/mL) for 96 h, and the protein expression of CD31 and α-SMA was examined by western blotting. β-Actin was used as a reference protein. (F) The cells were incubated with LPS (20 μg/mL) for 96 h, and the expression levels of the CD31 and α-SMA genes were examined by qRT‒PCR. (G) The cells were incubated with LPS (20 μg/mL) for 96 h, and FCM was used to examine the protein expression of CD31 and α-SMA. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the LPS group and the control group.
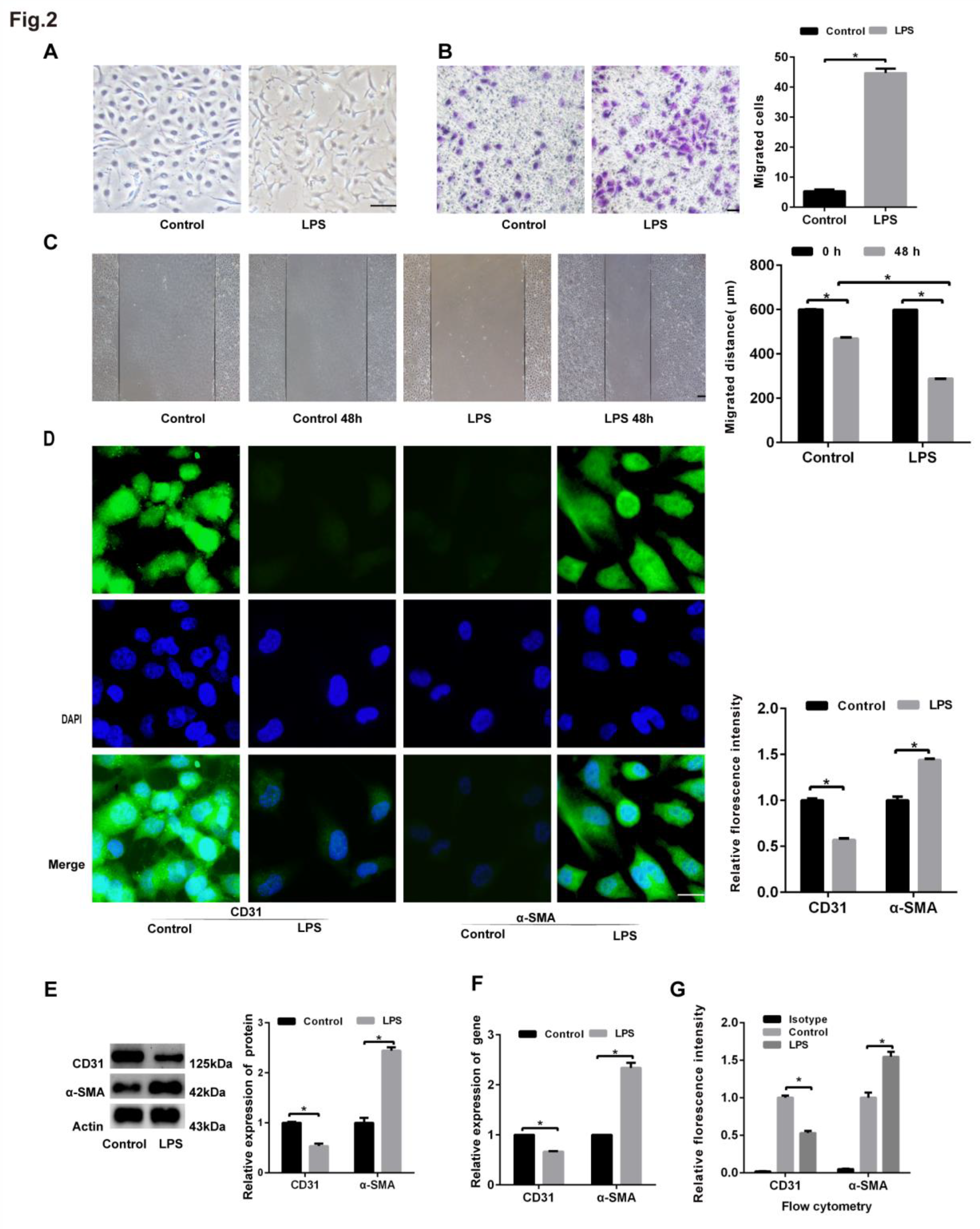
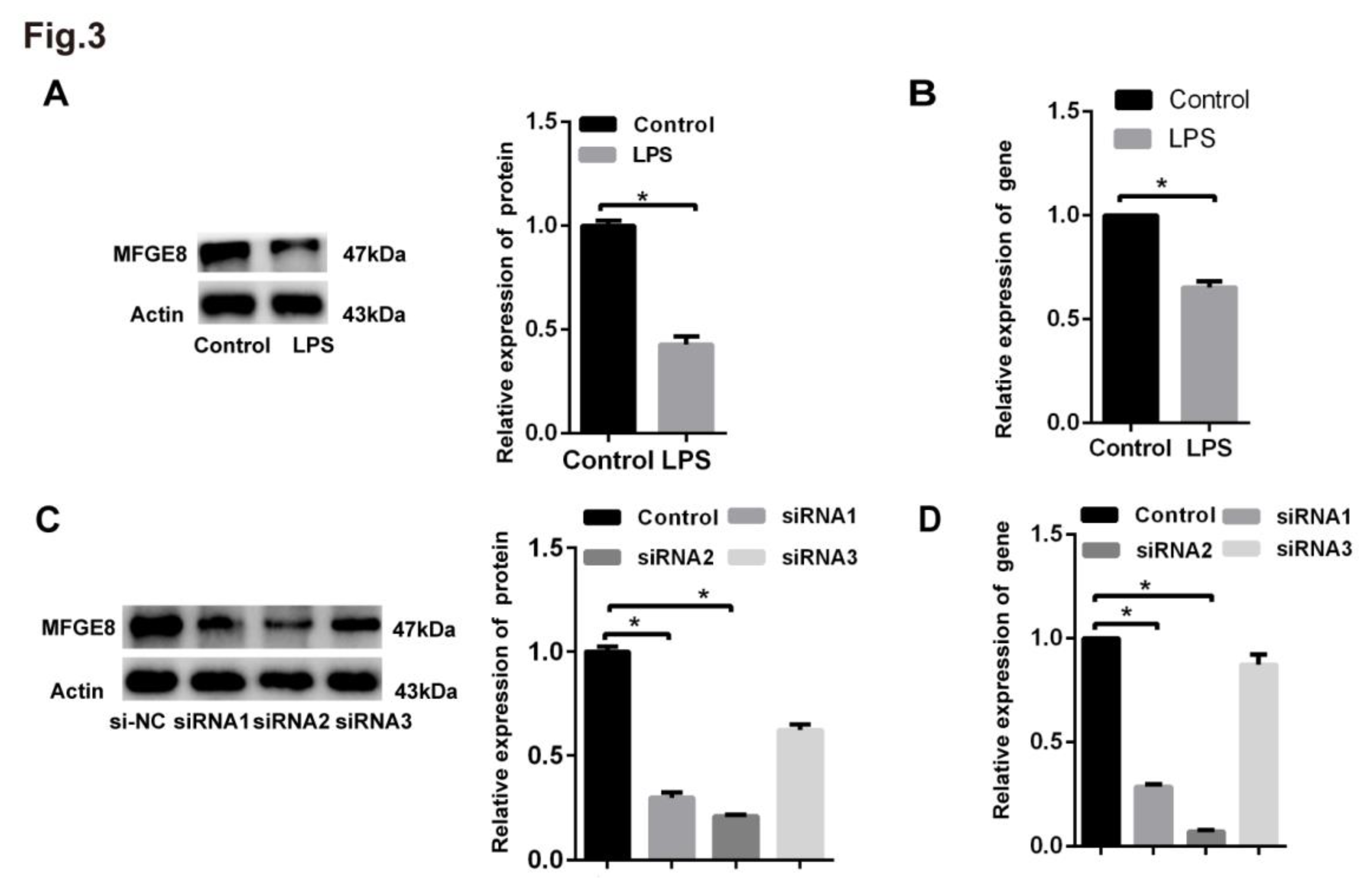
Figure 3.
Analysis of the knockdown efficiency of MFGE8. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum. (A) Western blotting was used to examine the expression of MFGE8. β-Actin was used as a reference protein. (B) qRT‒PCR was used to determine the expression of MFGE8. (C, D) Three specific siRNAs (siRNA-MFGE8-1, siRNA-MFGE8-2, and siRNA-MFGE8-3) were transfected into HLMECs for 24 h to downregulate MFGE8 expression, and the knockdown efficiency was analysed by qRT‒PCR and western blotting. β-Actin was used as a reference protein. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the treated group and the control group.
Figure 3.
Analysis of the knockdown efficiency of MFGE8. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum. (A) Western blotting was used to examine the expression of MFGE8. β-Actin was used as a reference protein. (B) qRT‒PCR was used to determine the expression of MFGE8. (C, D) Three specific siRNAs (siRNA-MFGE8-1, siRNA-MFGE8-2, and siRNA-MFGE8-3) were transfected into HLMECs for 24 h to downregulate MFGE8 expression, and the knockdown efficiency was analysed by qRT‒PCR and western blotting. β-Actin was used as a reference protein. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the treated group and the control group.
Figure 4.
The siRNA-mediated knockdown of MFGE8 exacerbated LPS-induced EndoMT in HLMECs. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into three groups: control group, siRNA group, and LPS+siRNA group. (A) Microscopy was used to observe the cell morphology (magnification, x100, scale bar = 50 μm). (B) A Transwell migration assay was performed to examine cell invasion (magnification, x100, scale bar = 50 μm). (C) A scratch assay was performed to examine cell migration (magnification, x100, scale bar = 50 μm). (D) Fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA (magnification, x200, scale bar = 20 μm). (E) The protein expression of CD31 and α-SMA was examined by western blotting. β-Actin was used as a reference protein. (F) The expression levels of the CD31 and α-SMA genes were examined by qRT‒PCR. (G) FCM was used to examine the protein expression of CD31 and α-SMA. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk indicates a significant difference (*P<0.01) between the siRNA group and the control group and between the LPS+siRNA group and the control group.
Figure 4.
The siRNA-mediated knockdown of MFGE8 exacerbated LPS-induced EndoMT in HLMECs. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into three groups: control group, siRNA group, and LPS+siRNA group. (A) Microscopy was used to observe the cell morphology (magnification, x100, scale bar = 50 μm). (B) A Transwell migration assay was performed to examine cell invasion (magnification, x100, scale bar = 50 μm). (C) A scratch assay was performed to examine cell migration (magnification, x100, scale bar = 50 μm). (D) Fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA (magnification, x200, scale bar = 20 μm). (E) The protein expression of CD31 and α-SMA was examined by western blotting. β-Actin was used as a reference protein. (F) The expression levels of the CD31 and α-SMA genes were examined by qRT‒PCR. (G) FCM was used to examine the protein expression of CD31 and α-SMA. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk indicates a significant difference (*P<0.01) between the siRNA group and the control group and between the LPS+siRNA group and the control group.
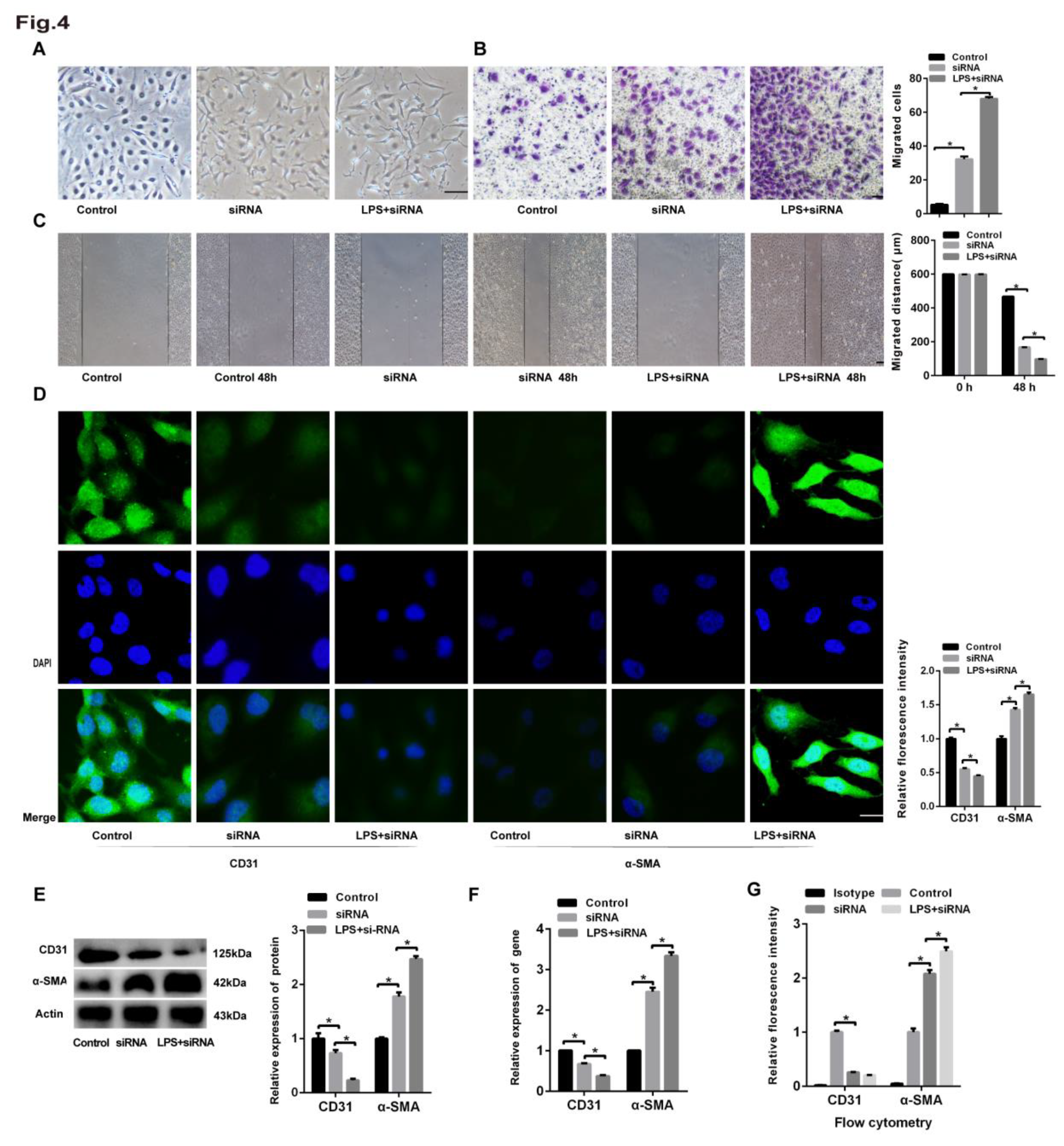
Figure 5.
rhMFGE8 attenuated LPS-induced EndoMT in HLMECs. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into five groups: control group, LPS group, LPS+siRNA group, LPS+rhMFGE8 group, and LPS+siRNA+rhMFGE8 group. (A) The cells were incubated for 96 h. (A) Microscopy was used to observe the cell morphology (magnification, x100, scale bar = 50 μm). (B) Transwell migration assays were performed to examine cell invasion (magnification, x100, scale bar = 50 μm). (C) A scratch assay was performed to examine cell migration (magnification, x100, scale bar = 50 μm). (D) Fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA (magnification, x200, scale bar = 20 μm). (E) The protein expression of CD31 and α-SMA was examined by western blotting. β-Actin was used as a reference protein. (F) The expression levels of the CD31 and α-SMA genes were examined by qRT‒PCR. (G) FCM was used to examine the protein expression of CD31 and α-SMA. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the LPS group and the LPS+rhMFGE8 group and between the LPS+siRNA group and the LPS+siRNA+rhMFGE8 group.
Figure 5.
rhMFGE8 attenuated LPS-induced EndoMT in HLMECs. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into five groups: control group, LPS group, LPS+siRNA group, LPS+rhMFGE8 group, and LPS+siRNA+rhMFGE8 group. (A) The cells were incubated for 96 h. (A) Microscopy was used to observe the cell morphology (magnification, x100, scale bar = 50 μm). (B) Transwell migration assays were performed to examine cell invasion (magnification, x100, scale bar = 50 μm). (C) A scratch assay was performed to examine cell migration (magnification, x100, scale bar = 50 μm). (D) Fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA (magnification, x200, scale bar = 20 μm). (E) The protein expression of CD31 and α-SMA was examined by western blotting. β-Actin was used as a reference protein. (F) The expression levels of the CD31 and α-SMA genes were examined by qRT‒PCR. (G) FCM was used to examine the protein expression of CD31 and α-SMA. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference (P<0.01) between the LPS group and the LPS+rhMFGE8 group and between the LPS+siRNA group and the LPS+siRNA+rhMFGE8 group.
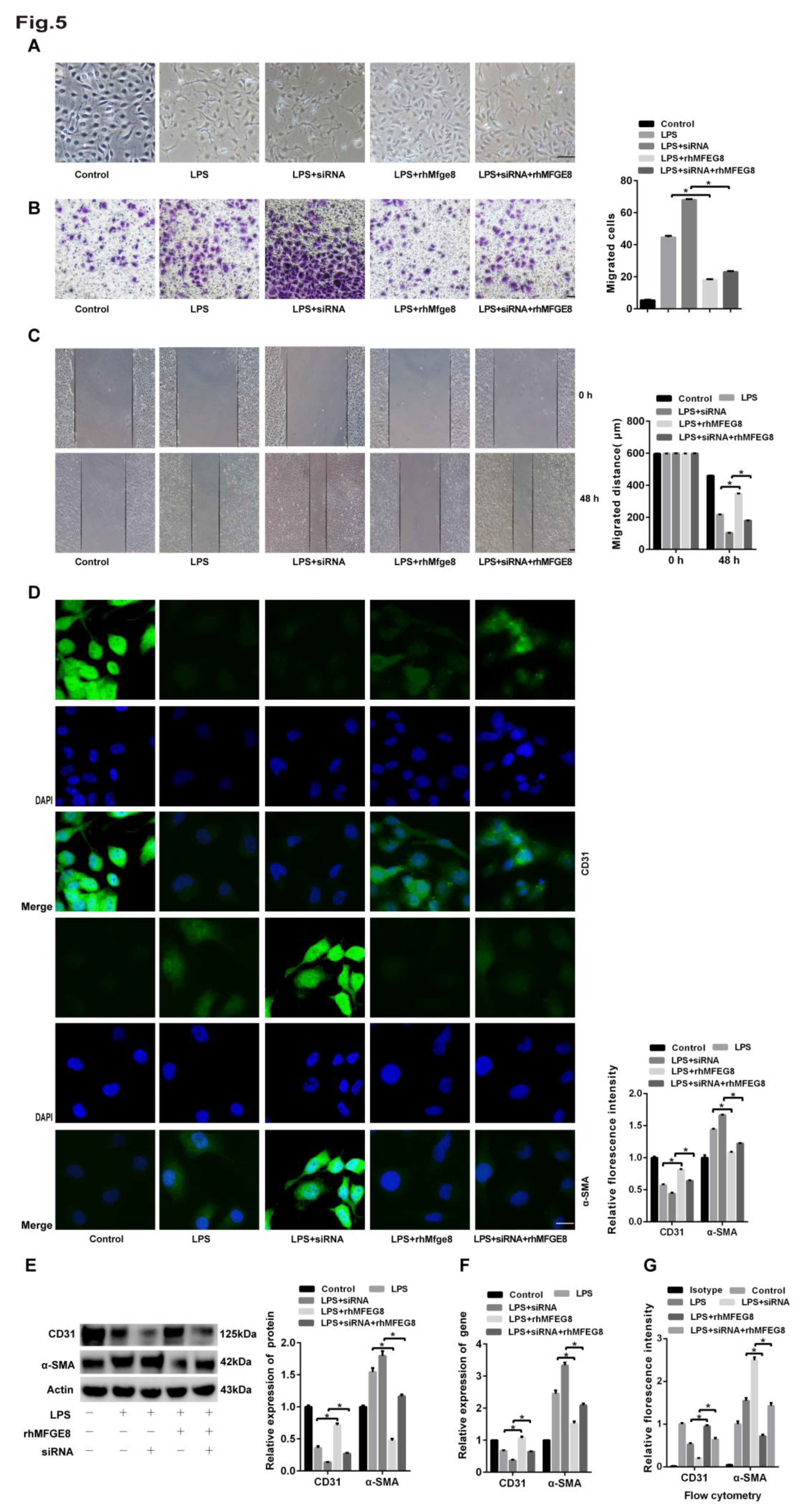
Figure 6.
The BMP/Smad1/5/Smad1/4 signalling pathway was activated by LPS. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into five groups: control group, LPS group, LPS+siRNA group, LPS+rhMFGE8 group, and LPS+siRNA+rhMFGE8 group. (A, C) Western blotting and qRT‒PCR were used to determine the protein and gene expression levels of BMP, Smad1, Smad5, Smad4 and Snail. (B) The BMP inhibitor LDN-212854 was used to verify the relationship between LPS and the BMP signalling pathway. Western blotting was used to examine the protein expression of Smad1, Smad5 and Smad4. Β-Actin was used as a reference protein. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference between the LPS group and the control group and between the LPS group and the LPS+rhMFGE8 group.
Figure 6.
The BMP/Smad1/5/Smad1/4 signalling pathway was activated by LPS. Human lung microvascular endothelial cells (HLMECs) were cultured with DMEM/F-12 and 10% standard foetal bovine serum, and the cells were divided into five groups: control group, LPS group, LPS+siRNA group, LPS+rhMFGE8 group, and LPS+siRNA+rhMFGE8 group. (A, C) Western blotting and qRT‒PCR were used to determine the protein and gene expression levels of BMP, Smad1, Smad5, Smad4 and Snail. (B) The BMP inhibitor LDN-212854 was used to verify the relationship between LPS and the BMP signalling pathway. Western blotting was used to examine the protein expression of Smad1, Smad5 and Smad4. Β-Actin was used as a reference protein. Representative images from three replicated independent experiments are shown. n = 3 per group. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference between the LPS group and the control group and between the LPS group and the LPS+rhMFGE8 group.
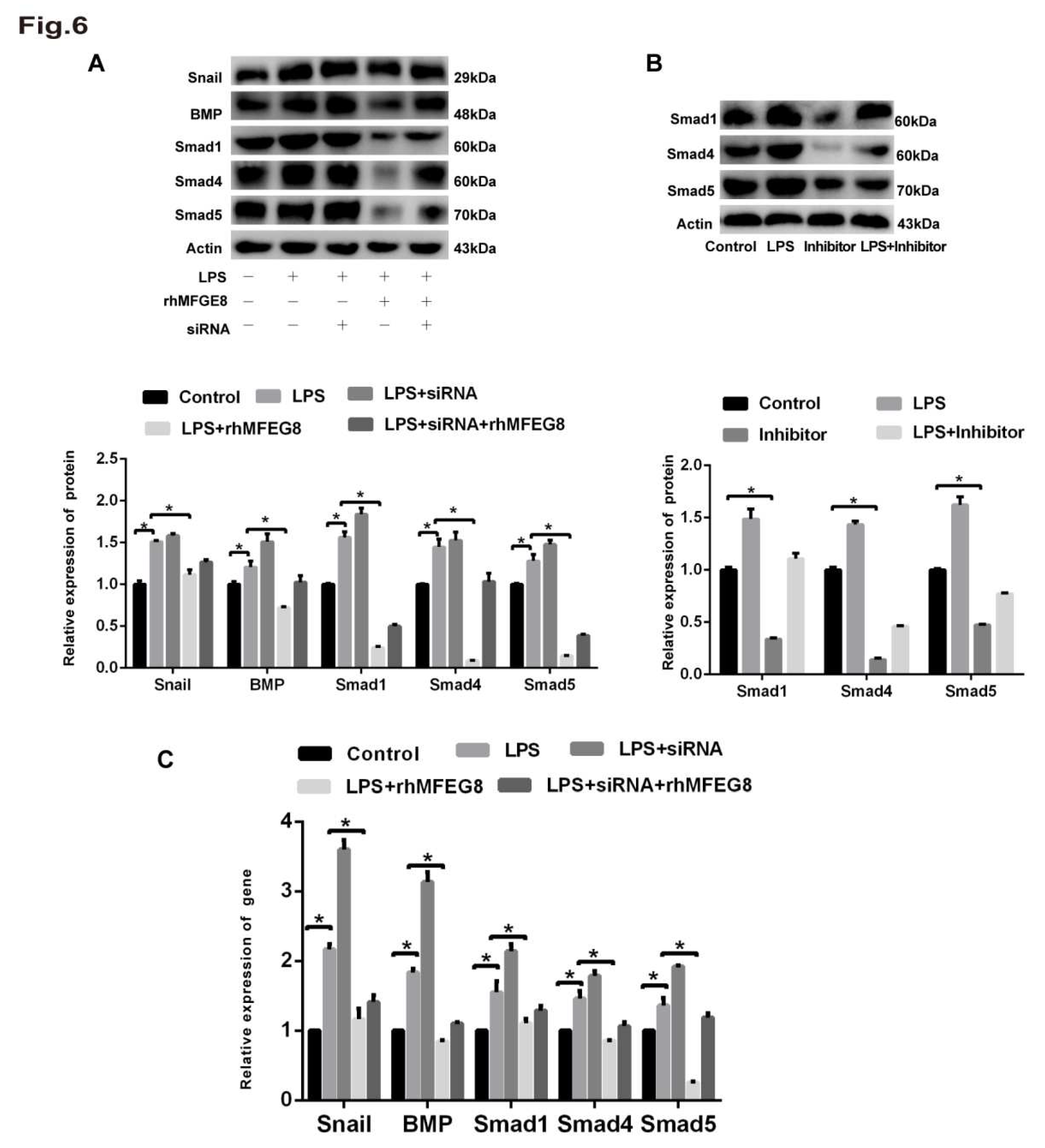
Figure 7.
rhMFGE8 attenuated ECM deposition and played a protective role in the early stage of EndoMT. Forty-five mice were divided into three groups according to a completely randomized design: control group, LPS group and LPS+rhMFGE8 group. The control group was intraperitoneally administered physiological saline. The LPS group was intraperitoneally administered 20 mg/kg LPS dissolved in physiological saline. The LPS+rhMFGE8 group was intraperitoneally administered 20 mg/kg LPS and 20 µg/kg rhMFGE8. At the endpoint of the experiment, the mice were anaesthetised with sodium pentobarbital (60 mg/kg) and euthanised with potassium chloride (100 mg/kg) on days 7, 14 and 28. (A) H&E staining was used to observe the LPS-induced changes in cell morphology and the alveolar septum in the lung (magnification, x100, scale bar = 100 μm). (B) Masson staining was used to examine ECM deposition in lung tissue (magnification, x100, scale bar = 100 μm). (C) Fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA in lung tissue. CD31 is shown by red fluorescence, and α-SMA is shown by green fluorescence (magnification, x200, scale bar = 20 μm). Western blotting was used to examine the protein expression of CD31 and α-SMA. n = 5 mice per group were analysed in the experiment. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference between the LPS group and the control group and between the LPS group and the LPS+rhMFGE8 group.
Figure 7.
rhMFGE8 attenuated ECM deposition and played a protective role in the early stage of EndoMT. Forty-five mice were divided into three groups according to a completely randomized design: control group, LPS group and LPS+rhMFGE8 group. The control group was intraperitoneally administered physiological saline. The LPS group was intraperitoneally administered 20 mg/kg LPS dissolved in physiological saline. The LPS+rhMFGE8 group was intraperitoneally administered 20 mg/kg LPS and 20 µg/kg rhMFGE8. At the endpoint of the experiment, the mice were anaesthetised with sodium pentobarbital (60 mg/kg) and euthanised with potassium chloride (100 mg/kg) on days 7, 14 and 28. (A) H&E staining was used to observe the LPS-induced changes in cell morphology and the alveolar septum in the lung (magnification, x100, scale bar = 100 μm). (B) Masson staining was used to examine ECM deposition in lung tissue (magnification, x100, scale bar = 100 μm). (C) Fluorescence microscopy was used to examine the protein expression of CD31 and α-SMA in lung tissue. CD31 is shown by red fluorescence, and α-SMA is shown by green fluorescence (magnification, x200, scale bar = 20 μm). Western blotting was used to examine the protein expression of CD31 and α-SMA. n = 5 mice per group were analysed in the experiment. The data are presented as the means ± SDs. Statistical analysis was performed using Student’s t test, and an asterisk (*) indicates a significant difference between the LPS group and the control group and between the LPS group and the LPS+rhMFGE8 group.
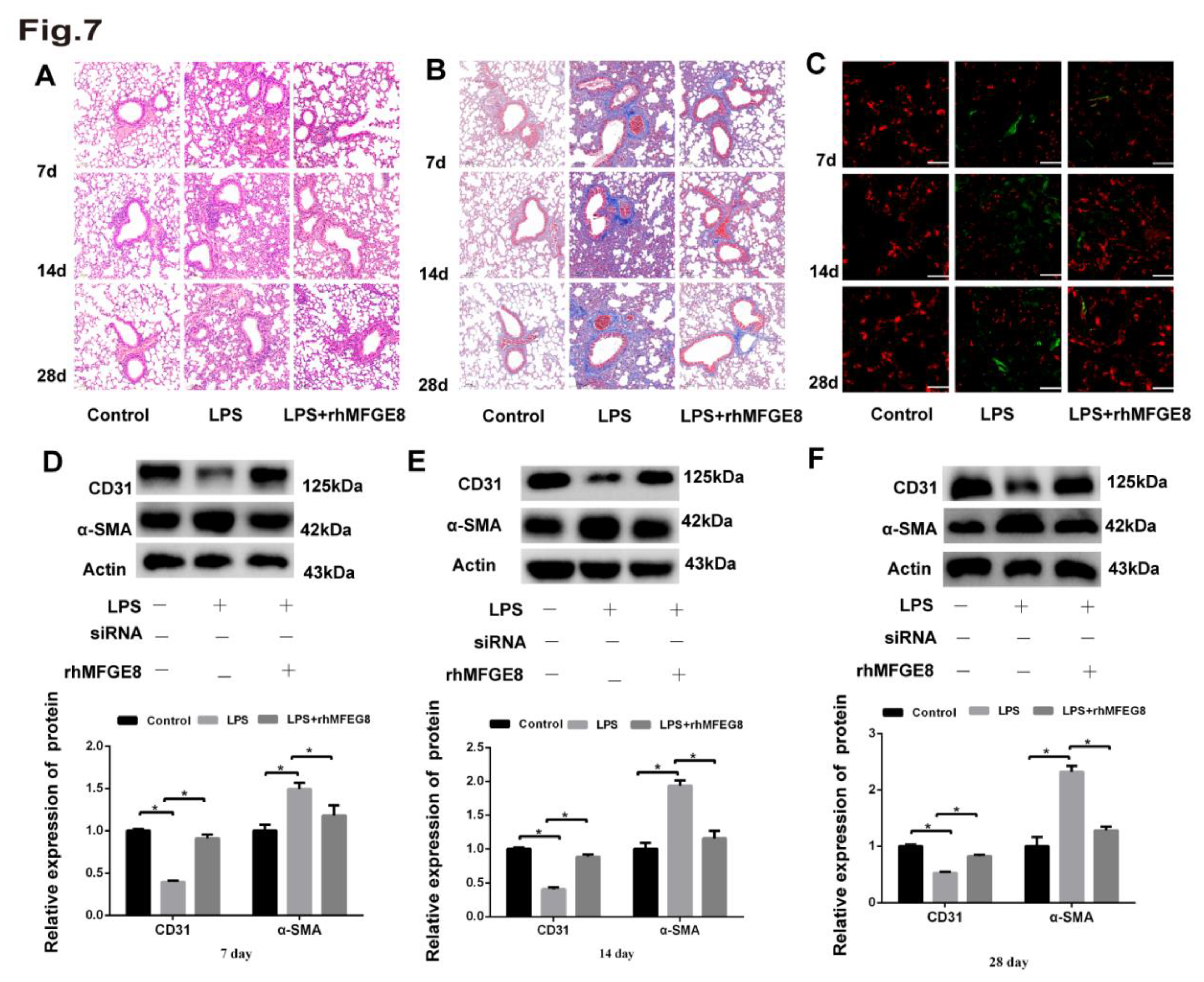
Table 1.
Primer sequences used for qRT‒PCR.
Table 1.
Primer sequences used for qRT‒PCR.
| |
Gene |
Forward primer |
Reverse primer |
| human |
MFGE8 |
atgggaacattgccaactcac |
ggggttatcgtcattgctgc |
| human |
BMP4 |
tagcccctaagcatcactcacag |
cacatcgctgaagtccacatag |
| human |
CD31 |
ccaagatagcctcaaagtcgg |
ctgggcatcataagaaatcctg |
| human |
Smad1 |
tctcagccgatggacacaaac |
agcaaccgcctgaacatctc |
| human |
Smad4 |
tgagggacagccatcgttg |
cacaggaatgttgggaaacttg |
| human |
Smad5 |
cctccagtattagtgcctcgtc |
ctgtggttcattgtggctcag |
| human |
Snail |
aatcggaagctaactacagcg |
atgagcattggcagcgagg |
| human |
α -SMA |
ggctgttttcccatccattg |
cttttgctctgtgcttcgtca |