Submitted:
01 May 2023
Posted:
02 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Demographic Characteristics of Patients.
2.2. Total IgE and blood eosinophils.
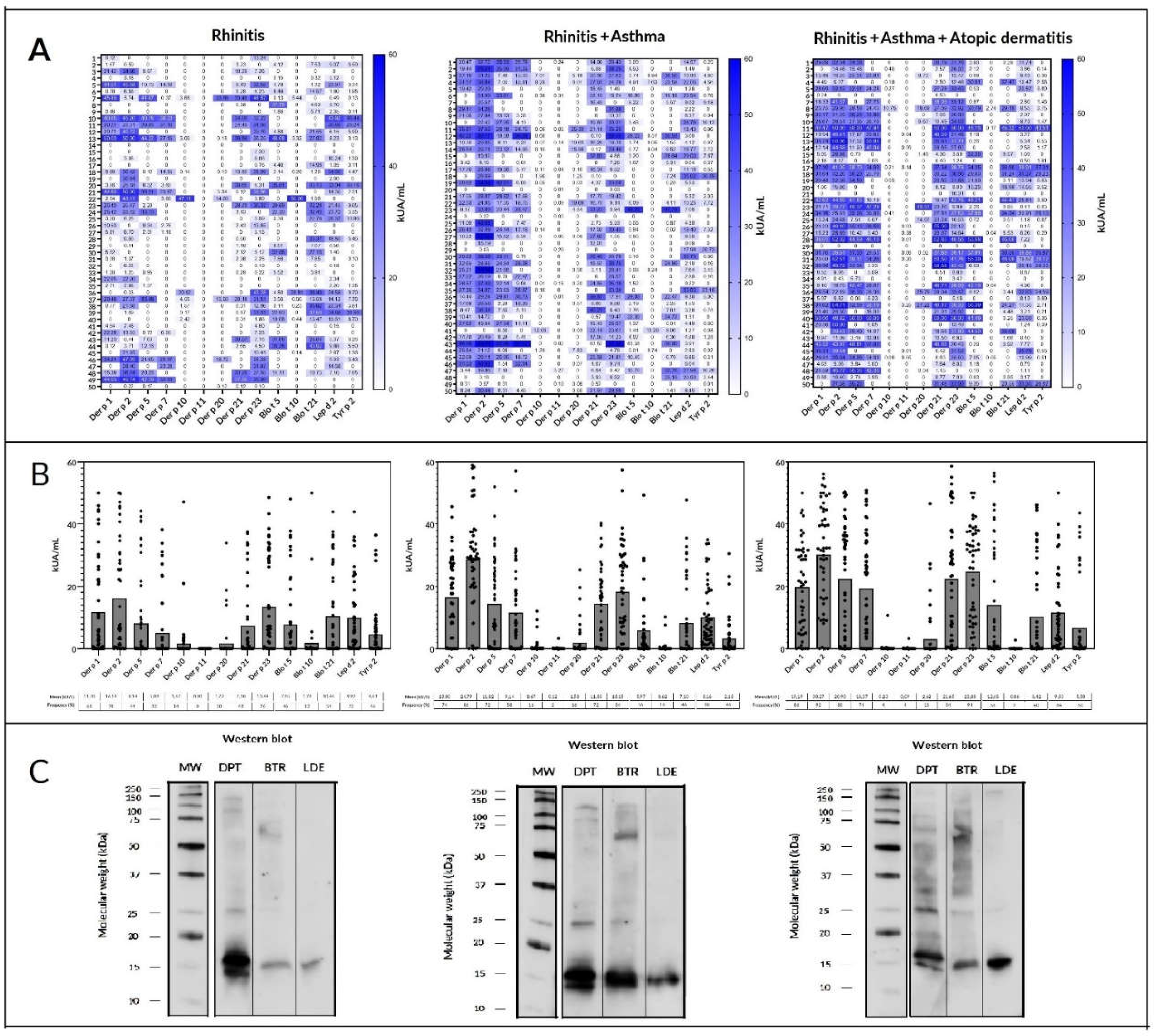
2.3. Prevalence, sIgE reactivity and individual molecular profile according to atopic disease.
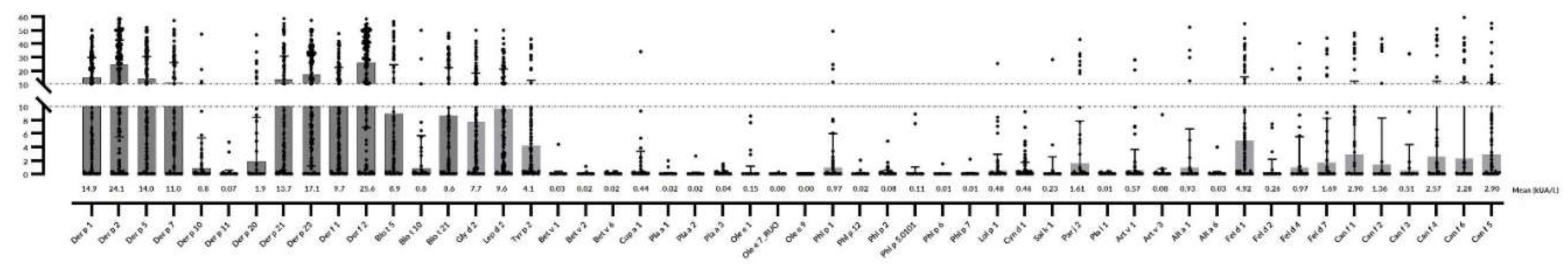
2.4. Mites
2.5. Cat and dog epithelia
| n=84 | % | Number of molecules | Fel d 1 | Fel d 2 | Fel d 4 | Fel d 7 | Can f 1 | Can f 2 | Can f 3 | Can f 4 | Can f 5 | Can f 6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | 22.6 | 1 | * | |||||||||
| 1 | 1.1 | 1 | * | |||||||||
| 13 | 15.4 | 1 | * | |||||||||
| 2 | 2.3 | 2 | * | * | ||||||||
| 11 | 13.0 | 2 | * | * | ||||||||
| 3 | 3.5 | 2 | * | * | ||||||||
| 4 | 4.7 | 2 | * | * | ||||||||
| 1 | 1.1 | 3 | * | * | * | |||||||
| 1 | 1.1 | 3 | * | * | * | |||||||
| 1 | 1.1 | 3 | * | * | * | |||||||
| 2 | 2.3 | 3 | * | * | * | |||||||
| 1 | 1.1 | 3 | * | * | * | |||||||
| 2 | 2.3 | 3 | * | * | * | |||||||
| 1 | 1.1 | 3 | * | * | * | |||||||
| 1 | 1.1 | 3 | * | * | * | |||||||
| 3 | 3.5 | 4 | * | * | * | * | ||||||
| 1 | 1.1 | 4 | * | * | * | * | ||||||
| 1 | 1.1 | 4 | * | * | * | * | ||||||
| 2 | 2.3 | 4 | * | * | * | * | ||||||
| 1 | 1.1 | 4 | * | * | * | * | ||||||
| 1 | 1.1 | 4 | * | * | * | * | ||||||
| 1 | 1.1 | 5 | * | * | * | * | * | |||||
| 1 | 1.1 | 5 | * | * | * | * | * | |||||
| 1 | 1.1 | 6 | * | * | * | * | * | * | ||||
| 1 | 1.1 | 6 | * | * | * | * | * | * | ||||
| 1 | 1.1 | 7 | * | * | * | * | * | * | * | |||
| 1 | 1.1 | 7 | * | * | * | * | * | * | * | |||
| 2 | 2.3 | 8 | * | * | * | * | * | * | * | * | ||
| 1 | 1.5 | 8 | * | * | * | * | * | * | * | * | ||
| 1 | 1.1 | 8 | * | * | * | * | * | * | * | * | ||
| 2 | 2.3 | 9 | * | * | * | * | * | * | * | * | * |
2.6. Pollen
| n=64 | % | Number of molecules | Cyn d 1 | Par j 2 | Phl p 1 | Phl p 2 | Phl p 12 | Lol p 1 | Art v 1 | Art v 3 | Cup a 1 | Sal k 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4.6 | 1 | * | |||||||||
| 3 | 4.6 | 1 | * | |||||||||
| 2 | 3.1 | 1 | * | |||||||||
| 3 | 4.6 | 1 | * | |||||||||
| 17 | 26.5 | 1 | * | |||||||||
| 2 | 3.1 | 1 | * | |||||||||
| 3 | 4.6 | 1 | * | |||||||||
| 3 | 4.6 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 2 | 3.1 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 2 | 3.1 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 1 | 1.5 | 2 | * | * | ||||||||
| 1 | 1.5 | 3 | * | * | * | |||||||
| 2 | 3.1 | 3 | * | * | * | |||||||
| 1 | 1.5 | 3 | * | * | * | |||||||
| 1 | 1.5 | 3 | * | * | * | |||||||
| 1 | 1.5 | 3 | * | * | * | |||||||
| 1 | 1.5 | 3 | * | * | * | |||||||
| 3 | 4.6 | 4 | * | * | * | * | ||||||
| 1 | 1.5 | 4 | * | * | * | * | ||||||
| 1 | 1.5 | 4 | * | * | * | * | ||||||
| 1 | 1.5 | 4 | * | * | * | * | ||||||
| 1 | 1.5 | 4 | * | * | * | * | ||||||
| 1 | 1.5 | 5 | * | * | * | * | * |
2.7. Mold and cockroach allergens
3. Discussion
4. Materials and Methods
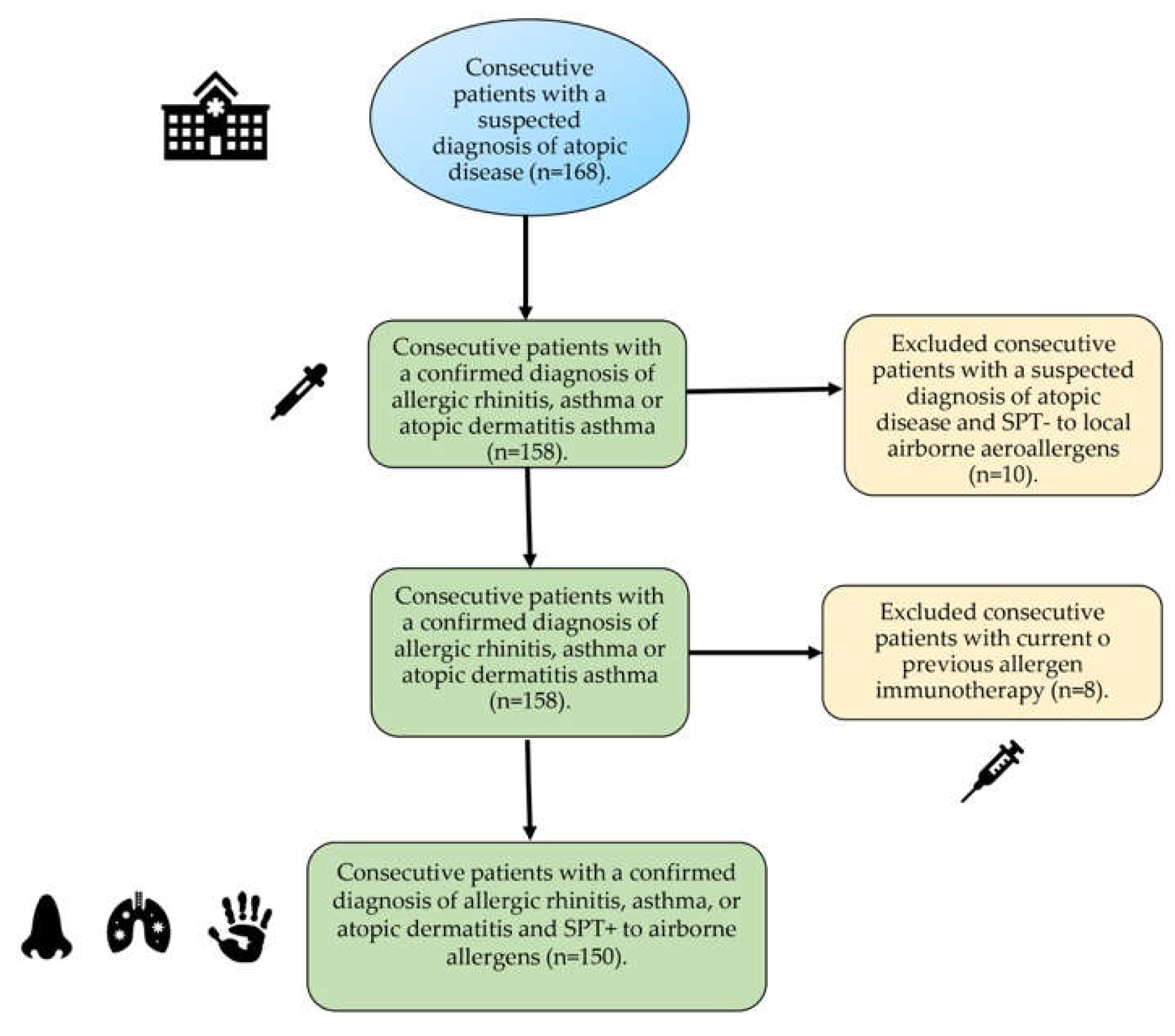
4.1. Subjects
4.2. Skin Prick Test.
4.3. Mite allergenic extracts.
4.4. SDS PAGE and IgE Western Blot.
4.5. Serological analysis.
4.6. Statistical analysis.
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wild, C.P. Complementing the genome with an "exposome": the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P. The exposome: from concept to utility. Int J Epidemiol. 2012, 41, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Renz, H.; Holt, P.G.; Inouye, M.; Logan, A.C.; Prescott, S.L.; Sly, P.D. An exposome perspective: early-life events and immune development in a changing world. J Allergy Clin Immunol. 2017, 140, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Celebi Sozener, Z.; Ozdel Ozturk, B.; Cerci, P.; Turk, M.; Gorgulu Akin, B.; Akdis, M.; et al. Epithelial barrier hypothesis: Effect of the external exposome on the microbiome and epithelial barriers in allergic disease. Allergy 2022, 77, 1418–1449. [Google Scholar] [CrossRef] [PubMed]
- Custovic, A. To what extent is allergen exposure a risk factor for the development of allergic disease? Clin Exp Allergy 2015, 45, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Putcha, N.; Woo, H.; McCormack, M.C.; Fawzy, A.; Romero, K.; Davis, M.F.; et al. Home Dust Allergen Exposure Is Associated with Outcomes among Sensitized Individuals with Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2022, 205, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Ellwood, P.; Asher, M.I.; Billo, N.E.; Bissell, K.; Chiang, C.Y.; Ellwood, E.M.; et al. The Global Asthma Network rationale and methods for phase I global surveillance: prevalence, severity, management and risk factors. Eur Respir J. 2017, 49, 1601605. [Google Scholar] [CrossRef] [PubMed]
- Pawankar, R. Allergic diseases and asthma: a global public health concern and a call to action. World Allergy Organ J. 2014, 7, 12. [Google Scholar] [CrossRef]
- Kuruvilla, M.E.; Lee, F.E.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin Rev Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef]
- Kim, J.; Ahn, K. Atopic dermatitis endotypes: knowledge for personalized medicine. Curr Opin Allergy Clin Immunol. 2022, 22, 153–159. [Google Scholar] [CrossRef]
- Perera, F.; Nadeau, K. Climate Change, Fossil-Fuel Pollution, and Children's Health. N Engl J Med. 2022, 386, 2303–2314. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Khatri, S.B. The Exposome and Asthma. Clin Chest Med. 2019, 40, 107–23. [Google Scholar]
- Huang, C.F.; Chie, W.C.; Wang, I.J. Effect of environmental exposures on allergen sensitization and the development of childhood allergic diseases: A large-scale population-based study. World Allergy Organ J. 2021, 14, 100495. [Google Scholar] [CrossRef] [PubMed]
- Schoos, A.M.; Chawes, B.L.; Jelding-Dannemand, E.; Elfman, L.B.; Bisgaard, H. Early indoor aeroallergen exposure is not associated with development of sensitization or allergic rhinitis in high-risk children. Allergy 2016, 71, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Weghofer, M.; Thomas, W.R.; Kronqvist, M.; Mari, A.; Purohit, A.; Pauli, G.; et al. Variability of IgE reactivity profiles among European mite allergic patients. Eur J Clin Invest. 2008, 38, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Stemeseder, T.; Schweidler, B.; Doppler, P.; Klinglmayr, E.; Moser, S.; Lueftenegger, L.; et al. Exposure to Indoor Allergens in Different Residential Settings and Its Influence on IgE Sensitization in a Geographically Confined Austrian Cohort. PLoS One 2017, 12, e0168686. [Google Scholar] [CrossRef]
- D'Amato, G.; Chong-Neto, H.J.; Monge Ortega, O.P.; Vitale, C.; Ansotegui, I.; Rosario, N.; et al. The effects of climate change on respiratory allergy and asthma induced by pollen and mold allergens. Allergy 2020, 75, 2219–2228. [Google Scholar] [CrossRef]
- Paudel, B.; Chu, T.; Chen, M.; Sampath, V.; Prunicki, M.; Nadeau, K.C. Increased duration of pollen and mold exposure are linked to climate change. Sci Rep. 2021, 11, 12816. [Google Scholar] [CrossRef]
- Miller, J.D. The Role of Dust Mites in Allergy. Clin Rev Allergy Immunol. 2019, 57, 312–329. [Google Scholar] [CrossRef]
- Andiappan, AK; Puan, KJ; Lee, B; Nardin, A; Poidinger, M; Connolly, J; et al. Allergic airway diseases in a tropical urban environment are driven by dominant mono-specific sensitization against house dust mites. Allergy 2014, 69, 501–509. [Google Scholar]
- Anto, J.M.; Bousquet, J.; Akdis, M.; Auffray, C.; Keil, T.; Momas, I.; et al. Mechanisms of the Development of Allergy (MeDALL): Introducing novel concepts in allergy phenotypes. J Allergy Clin Immunol. 2017, 139, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, J.; Melén, E.; Haahtela, T.; Koppelman, G.H.; Togias, A.; Valenta, R.; et al. Rhinitis associated with asthma is distinct from rhinitis alone: The ARIA-MeDALL hypothesis. Allergy 2023. [CrossRef] [PubMed]
- Dramburg, S.; Hilger, C.; Santos, A.; et al. EAACI Molecular Allergology User's Guide 2.0. Pediatr Allergy Immunol. 2023, 34 (Suppl. 28), e13854. [Google Scholar] [CrossRef] [PubMed]
- https://www.aemet.es/documentos/es/conocermas/recursos_en_linea/publicaciones_y_estudios/publicaciones/Aerobiologia/Aerobiologia.pdf Last accessed, April 14, 2023.
- Bousquet, J.; Schünemann, H.J.; Togias, A.; Bachert, C.; Erhola, M.; Hellings, P.W.; et al. Allergic Rhinitis and Its Impact on Asthma Working Group. Next-generation Allergic Rhinitis and Its Impact on Asthma (ARIA) guidelines for allergic rhinitis based on Grading of Recommendations Assessment, Development and Evaluation (GRADE) and real-world evidence. J Allergy Clin Immunol. 2020, 145, 70–80. [Google Scholar] [PubMed]
- https://ginasthma.org/gina-reports/ Last accessed March 15, 2023.
- Wollenberg, A.; Barbarot, S.; Bieber, T.; Christen-Zaech, S.; Deleuran, M.; Fink-Wagner, A.; et al. Consensus-based European guidelines for treatment of atopic eczema (atopic dermatitis) in adults children: part, I. J Eur Acad Dermatol Venereol. 2019, 32, 657–682. [Google Scholar] [CrossRef]
- Goodess, C.M.; Giorgi, F.; Hamaoui-Laguel, L.; Semenov, M.A.; Solmon, F.; Storkey, J.; Vautard, R.; Epstein, M.M. Climate Change and Future Pollen Allergy in Europe. Environ Health Perspect. 2017, 125, 385–391. [Google Scholar]
- Beggs, P.J.; Clot, B.; Sofiev, M.; Johnston, F.H. Climate change, airborne allergens, and three translational mitigation approaches. EBioMedicine. 2023, 104478. [Google Scholar] [CrossRef]
- Martín, J.L.; Bethencourt, J.; Cuevas-Agulló, E. Assessment of global warming on the island of Tenerife, Canary Islands (Spain). Trends in minimum, maximum and mean temperatures since 1944. Climatic Change 2012, 114, 343–355. [Google Scholar] [CrossRef]
- Dominguez-Rodriguez, A.; Baez-Ferrer, N.; Rodríguez, S.; Avanzas, P.; Abreu-Gonzalez, P.; Terradellas, E.; Cuevas, E.; Basart, S.; Werner, E. Saharan Dust Events in the Dust Belt -Canary Islands- and the Observed Association with in-Hospital Mortality of Patients with Heart Failure. J. Clin. Med. 2020, 9, 376. [Google Scholar] [CrossRef]
- Szefler, S.J.; Wenzel, S.; Brown, R.; Erzurum, S.C.; Fahy, J.V.; Hamilton, R.G.; et al. Asthma outcomes: biomarkers. J Allergy Clin Immunol. 2012, 129 (3 Suppl), S9–23. [Google Scholar] [CrossRef]
- Kidon, M.I.; Chiang, W.C.; Chin, C.W.; Liew, W.K.; Kang, L.W.; Ong, T.C.; et al. Mite componentspecific IgE repertoire and phenotypes of allergic disease in childhood: the tropical perspective. Pediatr Allergy Immunol. 2011, 22, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Oksel, C.; Haider, S.; Fontanella, S.; Frainay, C.; Custovic, A. Classification of pediatric asthma: from phenotype discovery to clinical practice. Front Pediatr. 2018, 6, 258. [Google Scholar] [CrossRef] [PubMed]
- Scala, E.; Caprini, E.; Abeni, D.; Meneguzzi, G.; Buzzulini, F.; Cecchi, L.; Villalta, D.; Asero, R. A qualitative and quantitative comparison of IgE antibody profiles with two multiplex platforms for component-resolved diagnostics in allergic patients. Clin Exp Allergy 2021, 51, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Platteel, A.C.M.; van der Pol, P.; Murk, J.L.; Verbrugge-Bakker, I.; Hack-Steemers, M.; Roovers, T.H.W.M.; Heron, M. A comprehensive comparison between ISAC and ALEX2 multiplex test systems. Clin Chem Lab Med. 2022, 60, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.; Arias, J.; Boquete, M.; Cardona, V.; Carrillo, T.; Gala, G.; et al. Analysis of mite allergic patients in a diverse territory by improved diagnostic tools. Clin Exp Allergy 2012, 42, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- González-Pérez, R.; Pineda, F.; Poza-Guedes, P.; Castillo, M.; Matheu, V.; Sánchez-Machín, I. Molecular Allergen Profiling of Dual Mite Sensitization in Severe Allergic Rhinitis. J Investig Allergol Clin Immunol. 2020, 30, 421–429. [Google Scholar] [CrossRef] [PubMed]
- González-Pérez, R.; Poza-Guedes, P.; Pineda, F.; Forstenlechner, P.; Castillo, M.; Mederos-Luís, E.; Aumayr, M.; Matheu, V.; Alava-Cruz, C.; Sánchez-Machín, I. Mite Molecular Profile in the Th2-Polarized Moderate-to-Severe Persistent Asthma Endotype Subjected to High Allergen Exposure. Int Arch Allergy Immunol. 2021, 182, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Vidal, C.; Lojo, S.; Juangorena, M.; Gonzalez-Quintela, A. Association Between Asthma and Sensitization to Allergens of Dermatophagoides pteronyssinus. J Investig Allergol Clin Immunol. 2016, 26, 304–309. [Google Scholar] [CrossRef]
- López-Rodríguez, R.; Rial, M.J.; Esteban-Gorgojo, I.; Veleiro Pérez, B.; López-Araújo, G.A.; Pérez-Quintero, O.; Carballas, C.; Parra, A.; Pineda, F. Serodominance Profile in a Dust Mite Complex Region. Int Arch Allergy Immunol. 2022, 183, 843–851. [Google Scholar] [CrossRef]
- Zeng, G.; Luo, W.; Zheng, P.; Wei, N.; Huang, H.; Sun, B.; Zhao, X. Component-Resolved Diagnostic Study of Dermatophagoides Pteronyssinus Major Allergen Molecules in a Southern Chinese Cohort. J Investig Allergol Clin Immunol. 2015, 25, 343–351. [Google Scholar]
- Rodinkova, V.V.; Yuriev, S.D.; Kryvopustova, M.V.; Mokin, V.B.; Kryzhanovskyi, Y.M.; Kurchenko, A.I. Molecular Profile Sensitization to House Dust Mites as an Important Aspect for Predicting the Efficiency of Allergen Immunotherapy. Front Immunol. 2022, 13, 848616. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Barnes, K.C.; Naidu, R.P.; Levett, P.N.; Beaty, T.H.; Wilson, A.F. Correlates of sensitization to Blomia tropicalis and Dermatophagoides pteronyssinus in asthma in Barbados. Int Arch Allergy Immunol 2003, 131, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Zhou, Q.; Prabhu, N.; Furuhashi, K.; Chua, Y.L.; Grotenbreg, G.M.; et al. Blomia tropicalis allergen 5 (Blo t 5) T-cell epitopes and their ability to suppress the allergic immune response. Immunology. 2017, 152, 344–355. [Google Scholar] [CrossRef] [PubMed]
- González-Pérez, R.; Poza-Guedes, P.; Pineda, F.; Castillo, M.; Sánchez-Machín, I. House Dust Mite Precision Allergy Molecular Diagnosis (PAMD@) in the Th2-prone Atopic Dermatitis Endotype. Life (Basel). 2021, 11, 1418. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, N.; Zakzuk, J.; Caraballo, L. House Dust Mite Allergy Under Changing Environments. Allergy Asthma Immunol Res. 2019, 11, 450–469. [Google Scholar] [CrossRef] [PubMed]
- Iraola Calvo, V.; Fernández-Caldas, E. Mapa acarológico de España; Laboratorios Leti: Barcelona, 2009; ISBN 978-84-69173336. [Google Scholar]
- Gafvelin, G.; Johansson, E.; Lundin, A.; Smith, A.M.; Chapman, M.D.; Benjamin, D.C.; et al. Cross-reactivity studies of a new group 2 allergen from the dust mite Glycyphagus domesticus, Gly d 2, and group 2 allergens from Dermatophagoides pteronyssinus, Lepidoglyphus destructor, and Tyrophagus putrescentiae with recombinant allergens. J Allergy Clin Immunol. 2001, 107, 511–518. [Google Scholar] [CrossRef]
- Osterlund, C.; Grönlund, H.; Gafvelin, G.; Bucht, A. Non-proteolytic aeroallergens from mites, cat and dog exert adjuvant-like activation of bronchial epithelial cells. Int Arch Allergy Immunol. 2011, 155, 111–118. [Google Scholar] [CrossRef]
- Gruchalla, R.S.; Pongracic, J.; Plaut, M.; et al. Inner City Asthma Study: relationships among sensitivity, allergen exposure, and asthma morbidity. J Allergy Clin Immunol 2005, 115, 478–485. [Google Scholar] [CrossRef]
- Subramanian, A.; Khatri, S.B. The Exposome and Asthma. Clin Chest Med. 2019, 40, 107–123. [Google Scholar] [CrossRef]
- Available online: https://izana.aemet.es/wp-content/docs/Izana_Report_2017_2018.pdf Last accessed March 3, 2023.
- Izquierdo, R.; Belmonte, J.; Avila, A.; Alarcón, M.; Cuevas, E.; Alonso-Pérez, S. Source areas and long-range transport of pollen from continental land to Tenerife (Canary Islands). Int J Biometeorol. 2011, 55, 67–85. [Google Scholar]
- Posa, D.; Perna, S.; Resch, Y.; Lupinek, C.; Panetta, V.; Hofmaier, S.; et al. Evolution and predictive value of IgE responses toward a comprehensive panel of house dust mite allergens during the first 2 decades of life. J Allergy Clin Immunol. 2017, 139, 541–549.e8. [Google Scholar] [CrossRef] [PubMed]
- Custovic, A.; Murray, C.S. The effect of allergen exposure in early childhood on the development of atopy. Curr Allergy Asthma Rep. 2002, 2, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, N.; Flohr, C.; Irvine, A.D. The exposome in atopic dermatitis. Allergy 2020, 75, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Bronnert, M.; Mancini, J.; Birnbaum, J.; Agabriel, C.; Liabeuf, V.; Porri, F.; et al. Component-resolved diagnosis with commercially available D. pteronyssinus Der p 1, Der p 2 and Der p 10: relevant markers for house dust mite allergy. Clin Exp Allergy 2012, 42, 1406–1415. [Google Scholar]
- Celi, G.; Brusca, I.; Scala, E.; Villalta, D.; Pastorello, E.; Farioli, L.; et al. House dust mite allergy in Italy-Diagnostic and clinical relevance of Der p 23 (and of minor allergens): A real-life, multicenter study. Allergy 2019, 74, 1787–1789. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, A.; Utsumi, D.; Lund, K.; Okano, M.; Ohashi-Doi, K.; Okubo, K. Correlation between sensitization to house dust mite major allergens, age, and symptoms in Japanese house dust mite allergic subjects. Int Immunopharmacol. 2022, 107, 108640. [Google Scholar] [CrossRef] [PubMed]
- Walsemann, T.; Böttger, M.; Traidl, S.; Schwager, C.; Gülsen, A.; Freimooser, S.; Roesner, L.M.; Werfel, T.; Jappe, U. Specific IgE against the house dust mite allergens Der p 5, 20 and 21 influences the phenotype and severity of atopic diseases. Allergy 2023, 78, 731–742. [Google Scholar] [CrossRef]
- Caraballo, L.; Valenta, R.; Puerta, L.; Pomés, A.; Zakzuk, J.; Fernandez-Caldas, E.; et al. The allergenic activity and clinical impact of individual IgE-antibody binding molecules from indoor allergen sources. World Allergy Organ J. 2020, 13, 100118. [Google Scholar] [CrossRef]
- Heinzerling, L.; Mari, A.; Bergmann, K.C.; Bresciani, M.; Burbach, G.; Darsow, U.; et al. The skin prick test - European standards. Clin Transl Allergy 2013, 3, 3. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685. [CrossRef] [PubMed]
- Bojcukova, J.; Vlas, T.; Forstenlechner, P.; Panzner, P. Comparison of two multiplex arrays in the diagnostics of allergy. Clin Transl Allergy 2019, 9, 31. [Google Scholar] [CrossRef]
| Allergic Rhinitis | Allergic Asthma | Atopic Dermatitis | |
|---|---|---|---|
| n=150 | 50 | 50 | 50 |
| Age (y.o.) median (range) | 27.5 (9-70) | 32 (8-67) | 26 (9-62) |
| <20 y.o. (n= 54) | 22 (40.7%) | 16 (29.6%) | 16 (29.6%) |
| ≥20 y.o. (n=96) | 28 (29,1%) | 34 (35.4%) | 34 (35.4%) |
| Sex (F/M) | 29/21 | 32/18 | 22/28 |
| Food Allergy (n=23) | 8 (34.7%) | 6 (20.0%) | 9 (39.1%) |
| Drug Allergy (n=11) | 2 (18.1%) | 6 (54.5%) | 3 (27.27%) |
| SPT+ to any aeroallergen | 50 (100%) | 50 (100%) | 50 (100%) |
| Total IgE (IU/ml) median (range) | 56 (23.88-1083) | 674 (56.5-19,993) | 898 (81.81-17,420) |
| Blood Eosinophils/mm3 median (range) | 340 (140-530) | 365 (20-1,290) | 375 (25-1,880) |
| Family History of Atopy | 36 (72%) | 38 (76%) | 39 (78%) |
| Positive SPT | N (%) |
|---|---|
| HDM and/or SM | 146 (97.33) |
| Cat and/or dog dander | 46 (30.66) |
| Pollen | 14 (9.33) |
| Cockroach | 9 (6) |
| Molds | 8 (5.33) |
| Mite allergen |
Mean sIgE (M±SD) | No. of sensitized patients (All) | Mean sIgE in AR (M±SD) |
No. of sensitized patients (AR) |
Mean sIgE in asthma (M±SD) | No. of sensitized patients (A) | Mean sIgE in atopic dermatitis (M±SD) | No. of sensitized patients (AD) |
|---|---|---|---|---|---|---|---|---|
| Der f 2 | 25.6±14.2 | 126 (84) | 17.17±0.2 | 39 (78) | 26.2±11.41 | 43 (86) | 32.7±0.31 | 46 (92) |
| Der p 2 | 24.1±15.3 | 125 (83) | 16.19±0.1 | 39 (78) | 24.79±12.3 | 43 (86) | 30.27±2.96 | 46 (92) |
| Der p 23 | 17.15±0.3 | 123 (82) | 13.44±1.2 | 38 (76) | 13.15±6.25 | 42 (84) | 23.88±12.7 | 47 (94) |
| Lep d 2 | 9.68± 1.5 | 114 (76) | 9.92±0.2 | 36 (72) | 8.16±5.23 | 40 (80) | 9.53±7.47 | 42 (84) |
| Der p 1 | 14.9±3.34 | 109 (72.6) | 11.7±4.3 | 34 (68) | 13±9.3 | 37 (74) | 19.19±14.5 | 43 (86) |
| Der f 1 | 9.79±2.51 | 104 (69.3) | 3.63±0.89 | 26 (52) | 10.36±8.32 | 38 (76) | 15.2±4.1 | 43 (86) |
| Gly d 2 | 7.72±1.53 | 101 (67.3) | 7.34±0.34 | 37 (74) | 5.03±3.9 | 32 (64) | 9.5±3.81 | 36 (72) |
| Der p 21 | 13.71±3.6 | 100 (66.6) | 7.38±0.36 | 24 (48) | 11.85±4.17 | 36 (72) | 21.65±0.22 | 42 (84) |
| Der p 5 | 14.07±1.0 | 96 (64) | 8.14±0.72 | 22 (44) | 11.82±1.66 | 36 (72) | 20.9±2.3 | 40 (80) |
| Der p 7 | 11.01±1.9 | 82 (54.6) | 5.08±0.39 | 16 (32) | 9.14±4.21 | 29 (58) | 18.37±1.9 | 37 (74) |
| Blo t 5 | 8.96±2.84 | 76 (50.6) | 7.85±0.53 | 23 (46) | 5.97±0.2 | 28 (56) | 12.65±2.75 | 28 (56) |
| Tyr p 2 | 4.18±0.2 | 68 (45.3) | 4.61±1,02 | 23 (46) | 2.15±2 | 23 (46) | 5.5±0.22 | 25 (50) |
| Blo t 21 | 8.64±0.31 | 66 (44) | 10.46±0.8 | 27 (54) | 7.1±5.3 | 23 (46) | 8.42±0.1 | 20 (40) |
| Der p 20 | 1.94±0.5 | 22 (14.6) | 1.72±0.08 | 5 (10) | 1.53±0.09 | 8 (16) | 2.62±0.5 | 9 (18) |
| Der p 10 | 0.86±0.23 | 16 (10.6) | 1.67±0.06 | 7 (14) | 0.67±2.3 | 8 (16) | <0.35 | 2 (4) |
| Blo t 10 | 0.83±0.41 | 14 (9.3) | 1.78±0.09 | 6 (12) | 0.62±0.2 | 8 (16) | <0.35 | 1 (2) |
| Der p 11 | 0.75±0.3 | 3 (2.0) | <0.35 | 0 (0) | <0.35 | 1 (2) | <0.35 | 2 (4) |
| Number of identified mite allergens | Allergic Rhinitis | Asthma | Atopic Dermatitis |
|---|---|---|---|
| 0 | 0 | 3 | 1 |
| 1 | 1 | 1 | 2 |
| 2 | 0 | 1 | 0 |
| 3 | 2 | 1 | 1 |
| 4 | 5 | 1 | 0 |
| 5 | 3 | 0 | 0 |
| 6 | 5 | 4 | 1 |
| 7 | 6 | 1 | 0 |
| 8 | 3 | 4 | 5 |
| 9 | 7 | 5 | 5 |
| 10 | 4 | 3 | 5 |
| 11 | 6 | 11 | 11 |
| 12 | 5 | 5 | 10 |
| 13 | 1 | 7 | 7 |
| 14 | 1 | 2 | 0 |
| 15 | 1 | 0 | 1 |
| 16 | 0 | 1 | 1 |
| 17 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





