1. Introduction
Rice false smut is a disease on rice spikes that occurs from the flowering to the milking stage[
1,
2]. It’s typical and visible symptom is the replacement of rice grains with false smut balls[
3,
4]. It occurs mainly in Asian countries such as China, Japan, Korea, the Philippines and India, and is one of the most devastating diseases in the world’s major rice producing regions[
5,
6,
7,
8]. In recent years, due to the promotion of short-stalked compact and high-yielding rice varieties, indica-japonica interspecific hybrid rice combinations, changes in cultivation patterns, and the excessive use of nitrogen fertilizer during the tillering and gestation periods, the occurrence of rice false smut has become increasingly serious and has gradually risen from a previously minor or sporadic disease to become one of the three new major diseases of rice in China[
9,
10].
The damages caused not only result in decrease of rice quality and yields, but also the generation of mycotoxin ustiloxins on infected rice spikelets[
11,
12]. As antimitotic cyclopeptide mycotoxins, the ustiloxins produced within false smut ball can inhibit microtubule assembly and cell skeleton formation, which poses a serious threat to farmland preservation, ecosystems, as well as the safety of humans and animals[
13]. Strategies are therefore urgently needed to management of this devastating disease.
It is widely accepted that
Ustilaginoidea virens (telemorph
Villosiclava virens) is the causing agent of rice false smut[
14,
15]. As a typical airborne disease, virulent pathogen spores land on the surface of a rice spikelet, germinate hyphae as well as false smut ball on the spikelet[
12,
16,
17,
18,
19]. Thus, the epidemic of rice false smut is closely related to the amount of
U. virens spores in the field, and rice false smut diagnosis combining of accurate detection and spore quantification is of significantly important for its prevention and management[
5,
20]. Traditionally, the microscopic counting of spores after capture is widely used in rice false smut diagnosis, however this method requires specialist taxonomic technicians[
21]. Given that the complexity of environmental samples and human subjectivity, microscopic analysis is difficult to provide reliable data with high efficiency. Recently, quantitative real-time PCR (q-PCR) technique has been applied for the identification and quantification of pathogen in airborne disease early warning[
22]. However, this technique is susceptible to interference from environmental dust and other pathogens, making it difficult to quantify the low concentrations of spores captured[
21].
Loop-mediated isothermal amplification (LAMP) is a non-PCR-based nucleic acid amplification technique that can be used for the molecular detection of various bacteria, viruses, fungi in disease diagnosis[
23,
24,
25]. The LAMP reaction is carried out at a constant temperature (between 60 and 65°C) in less than an hour by use of two pairs of primers, an inner primer (FIP/BIP) and an outer primer (F3/B3), to recognize specific nucleic acid sequences of monitored targets[
26,
27,
28]. The quantitative-LAMP (q-LAMP) assay (DiaSorin S.p.A., Saluggia, Italy) is a technical improvement over the classical LAMP, which combine LAMP technology with the real-time fluorescence quantitative PCR technique[
29]. It is based on the addition of nucleic acid fluorescent dyes (SYBR Green, etc.) or the synthesis of fluorescent probes (TaqMan probes, hybridization probes, etc.) resulting in a more sophisticated method suitable for the needs of field diagnosis[
30]. In this study, we aimed to design and develop a specific and sensitive q-LAMP assay for detection and quantification
U. virens, which can be applied in the early diagnosis of rice false smut for preventing the widespread spreading of this devastating airborne disease. And this study is the also first report describing a quantitative diagnostic test for detection of
U. virens using q-LAMP.
2. Results
2.1. Design of primers for U. virens detection
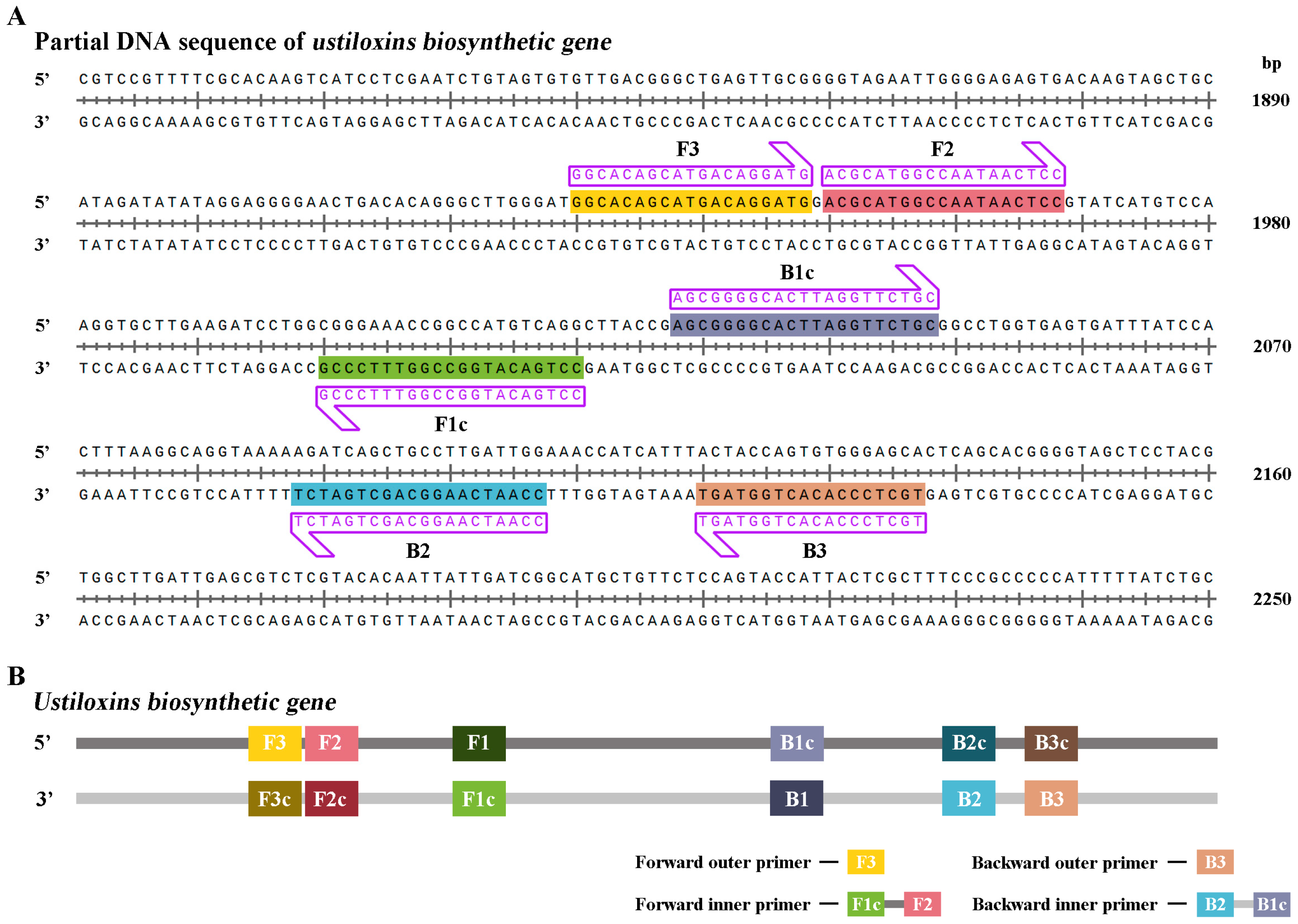
The best LAMP primers UV-2 were designed based on the ustiloxins biosynthetic gene sequence of
U. virens (NCBI accession number: BR001221.1) that did not show any similarities to other sequences available in the National Center for Biotechnology Information (NCBI) GenBank database to allow specific amplification of
U. virens (
Figure 1,
Table 1)[
31,
32,
33]. Additionally, the primer sets UV-2 met the requirement that ΔG values of less than or equal to -4 Kcal/mol at the 3’end of F3/B3 and F2/B2 and 5’ends of F1c and B1c.
Table 1.
The sequences of species-specific primers used in the quantitative loop-mediated isothermal amplification (q-LAMP) assay and quantitative real-time PCR (q-PCR) assay.
Table 1.
The sequences of species-specific primers used in the quantitative loop-mediated isothermal amplification (q-LAMP) assay and quantitative real-time PCR (q-PCR) assay.
| Serial number |
Sequence (5′-3′) |
| UV-2 |
F3 |
GGCACAGCATGACAGGATG |
| B3 |
TGCTCCCACACTGGTAGT |
| FIP(F1c-F2) |
CCTGACATGGCCGGTTTCCCGACGCATGGCCAATAACTCC |
| BIP(B1c-B2) |
AGCGGGGCACTTAGGTTCTGCCAATCAAGGCAGCTGATCT |
Figure 1.
The species-specific primers for detecting Ustilaginoidea virens in the quantitative loop-mediated isothermal amplification (q-LAMP) and quantitative real-time PCR (q-PCR). (A) The species-specific primers designed based on the sequence of the ustiloxins biosynthetic gene segments for identification and quantification of U. virens in q-LAMP assay and q-PCR assay. The forward and reverse primer sequences were highlighted with shade and arrow for orientation. (B) Model for q-LAMP and q-PCR primers design. Primers for q-LAMP assays consist of F3, B3, FIP, and BIP. FIP is a hybrid primer consisting of the F1c and F2 sequences; BIP is a hybrid primer consisting of the B1c and B2 sequences. Primers for q-PCR assays consist of F3 and B3.
Figure 1.
The species-specific primers for detecting Ustilaginoidea virens in the quantitative loop-mediated isothermal amplification (q-LAMP) and quantitative real-time PCR (q-PCR). (A) The species-specific primers designed based on the sequence of the ustiloxins biosynthetic gene segments for identification and quantification of U. virens in q-LAMP assay and q-PCR assay. The forward and reverse primer sequences were highlighted with shade and arrow for orientation. (B) Model for q-LAMP and q-PCR primers design. Primers for q-LAMP assays consist of F3, B3, FIP, and BIP. FIP is a hybrid primer consisting of the F1c and F2 sequences; BIP is a hybrid primer consisting of the B1c and B2 sequences. Primers for q-PCR assays consist of F3 and B3.
2.2. Optimization of the q-LAMP assay
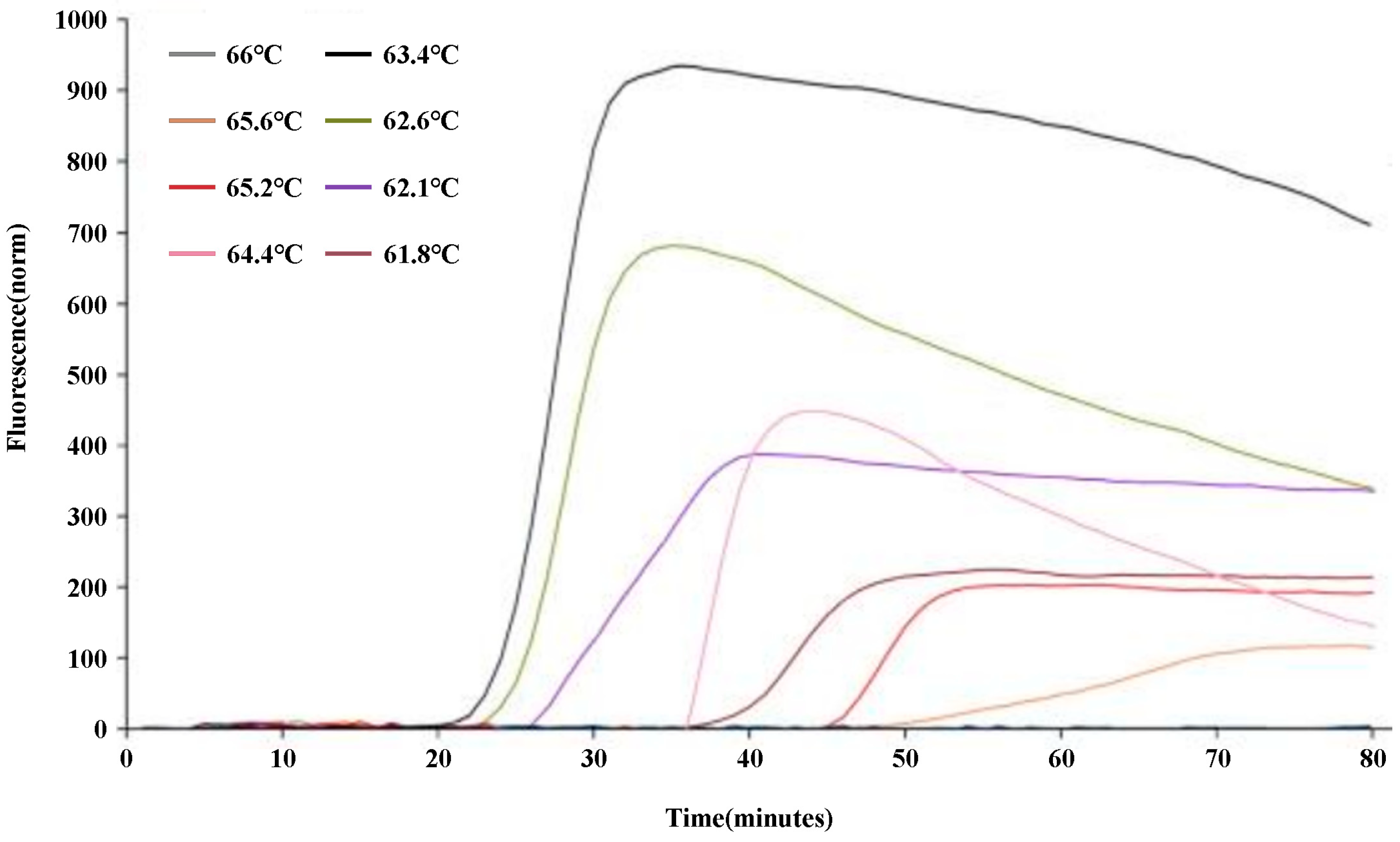
To optimize the q-LAMP assay system, the q-LAMP assay was carried out using the primer sets UV-2 at temperatures ranged from 61.8℃ to 66℃. As shown in
Figure 2, the fluorescence quantitative results found that the strongest fluorescence intensity and the shortest reaction time were obtained when the reaction temperature was 63.4℃ (which reaching the amplification peak at 30 min). Thus, 63.4℃ was chosen as the reaction temperature to carry out the optimal q-LAMP assays.
Figure 2.
Optimization of the q-LAMP assay via reaction temperature screening. The influence of temperature ranged from 61.8℃ to 66℃ in the q-LAMP detection system showed that the strongest fluorescence intensity and the shortest reaction time were obtained at the 63.4℃ (black line).
Figure 2.
Optimization of the q-LAMP assay via reaction temperature screening. The influence of temperature ranged from 61.8℃ to 66℃ in the q-LAMP detection system showed that the strongest fluorescence intensity and the shortest reaction time were obtained at the 63.4℃ (black line).
2.3. Specificity validation of the q-LAMP assay system
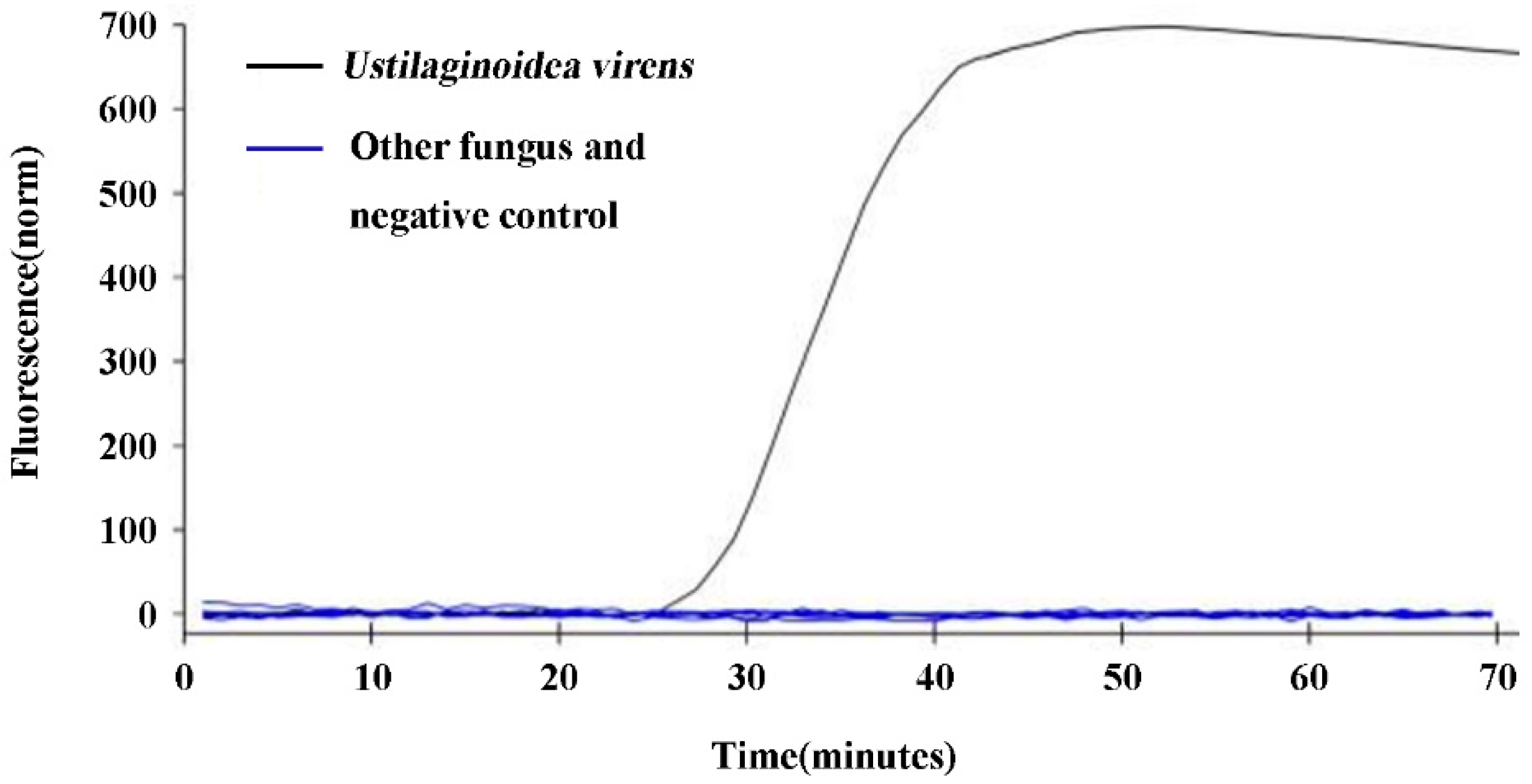
The specificity validation of design q-LAMP assay system (using primer sets UV-2 and 63.4℃as reaction temperature) was tested using
U. virens stain and the other nine fungi. The results showed that fluorescence signals were detected in the samples with DNA template of
U. virens, while the samples with DNA template of the other 9 fungi or ddH
2O (negative control) did not show any fluorescence signal (
Figure 3), indicating that design q-LAMP assay system was highly specific for the detection of
U. virens.
Figure 3.
Specificity validation of the q-LAMP assay system. The q-LAMP assay system (using primer sets UV-2 and 63.4℃ as reaction temperature) was highly specific for the detection of U. virens. The q-LAMP assay system showed that fluorescence signals were only detected in the samples with DNA template of U. virens (black line), while the samples with DNA template of the other 9 fungi (including Fusarium fujikuroi, F. oxysporum, F. proliferatum, F. solani, F. graminearum, Penicillium sp, Pyricularia oryzae, Alternaria alternata and Rhizoctonia solani) or negative control (nucleic acid-free water) did not show any fluorescence signal.
Figure 3.
Specificity validation of the q-LAMP assay system. The q-LAMP assay system (using primer sets UV-2 and 63.4℃ as reaction temperature) was highly specific for the detection of U. virens. The q-LAMP assay system showed that fluorescence signals were only detected in the samples with DNA template of U. virens (black line), while the samples with DNA template of the other 9 fungi (including Fusarium fujikuroi, F. oxysporum, F. proliferatum, F. solani, F. graminearum, Penicillium sp, Pyricularia oryzae, Alternaria alternata and Rhizoctonia solani) or negative control (nucleic acid-free water) did not show any fluorescence signal.
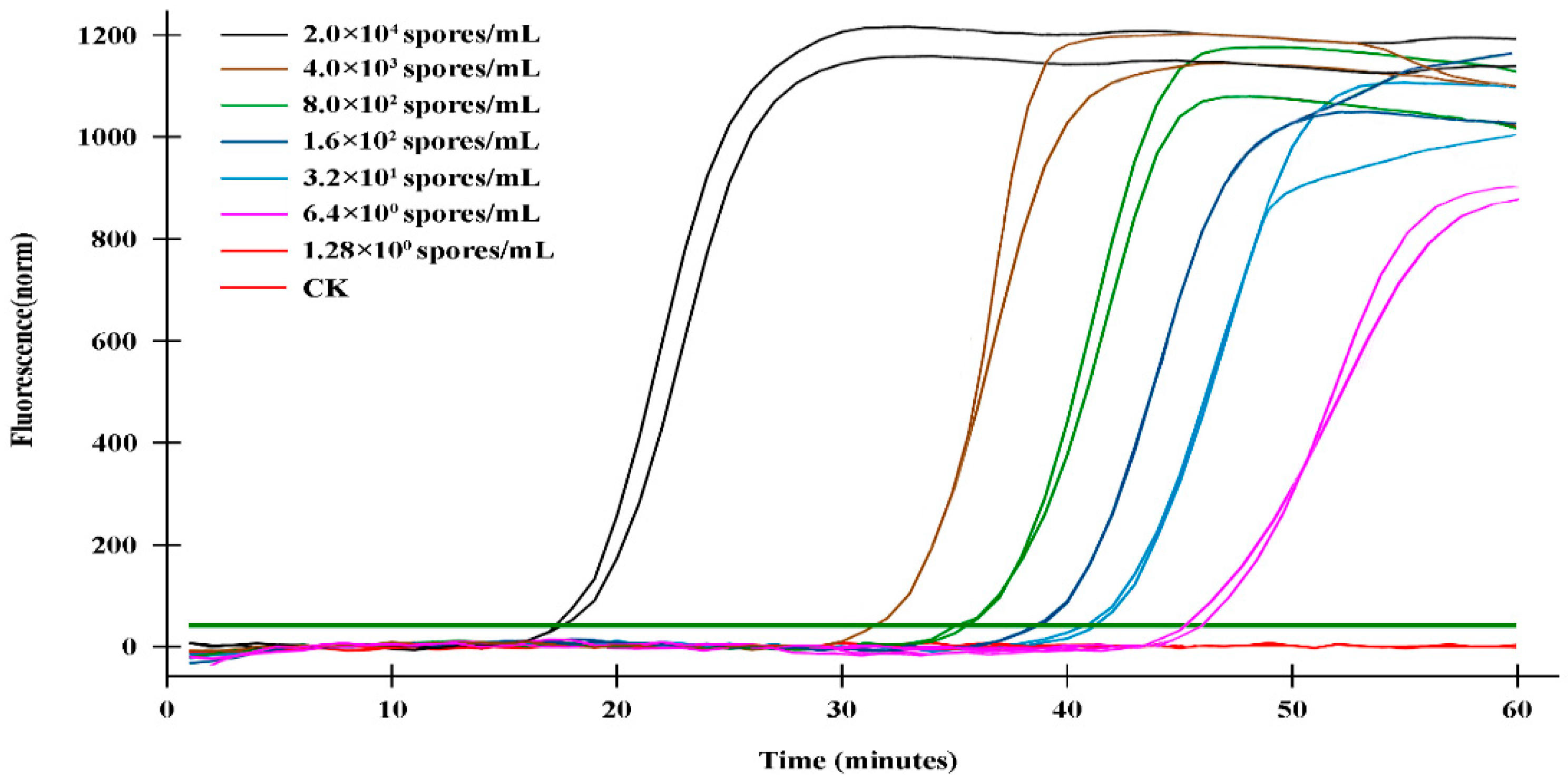
2.4. Sensitivity validation of the q-LAMP assay system
The sensitivity validation of the q-LAMP assay was determined using the genomic DNA of gradient dilution of
U. virens spore as templates under optimal condition (using primer sets UV-2 and 63.4℃ as reaction temperature). As shown in
Table 2 and
Figure 4, the fluorescence signals were detected in the samples with DNA template of 2×10
4 spores/mL, 4×10
3 spores/mL, 8×10
2 spores/mL, 1.6×10
2 spores/mL, 32 spores/mL and 6.4 spores/mL within 60 min, while no signals were detected in sample with DNA template of 1.28 spores/mL. Thus, theoretically q-LAMP assay was able to detect sample with concentration of 6.4 spores/mL. We also compared the sensitivity of q-LAMP assay system with quantitative real-time PCR (q-PCR) for
U. virens detection. The q-PCR assay was carried out using the primer set F3/B3 and the effective amplification reactions were detected at samples with spore concentrations of 2×10
4, 4×10
3, 8×10
2, 1.6×10
2 spores/mL, but not 32 spores/mL (
Supplementary Figure S1), indicating the theoretically q-PCR assay was ineffective in detecting samples with spore concentration lower than 32 spores/mL. Thus, the q-LAMP assay system is more sensitive and efficient compared to q-PCR system.
Table 2.
The time for reaching fluorescence signal peak and fluorescence signal detection in the q-LAMP assays for testing samples with known spore concentration.
Table 2.
The time for reaching fluorescence signal peak and fluorescence signal detection in the q-LAMP assays for testing samples with known spore concentration.
| Spore concentration (spores/mL) |
Time a (min) |
Fluorescence signals b
|
The Log10 value of spore number |
| 2×104
|
17.90 |
+ |
4.30 |
| 4×103
|
31.44 |
+ |
3.60 |
| 8×102
|
34.68 |
+ |
2.90 |
| 1.6×102
|
37.92 |
+ |
2.20 |
| 3.2×101
|
41.16 |
+ |
1.51 |
| 6.4 |
44.40 |
+ |
0.81 |
| 1.28 |
/ |
- |
0.20 |
| CKc
|
/ |
- |
/ |
Figure 4.
Sensitivity validation of q-LAMP system. The fluorescence signals in q-LAMP assays were detected in the samples with DNA template of 2×104 spores/mL, 4×103 spores/mL, 8×102 spores/mL, 1.6×102 spores/mL, 32 spores/mL and 6.4 spores/mL within 60 min, while no signals were detected in sample with DNA template of 1.28 spores/mL and CK. The bolded dark green line (horizontal) indicates fluorescence threshold. Fluorescence signals above this threshold marked as a successful detection for U. virens in q-LAMP assays.
Figure 4.
Sensitivity validation of q-LAMP system. The fluorescence signals in q-LAMP assays were detected in the samples with DNA template of 2×104 spores/mL, 4×103 spores/mL, 8×102 spores/mL, 1.6×102 spores/mL, 32 spores/mL and 6.4 spores/mL within 60 min, while no signals were detected in sample with DNA template of 1.28 spores/mL and CK. The bolded dark green line (horizontal) indicates fluorescence threshold. Fluorescence signals above this threshold marked as a successful detection for U. virens in q-LAMP assays.
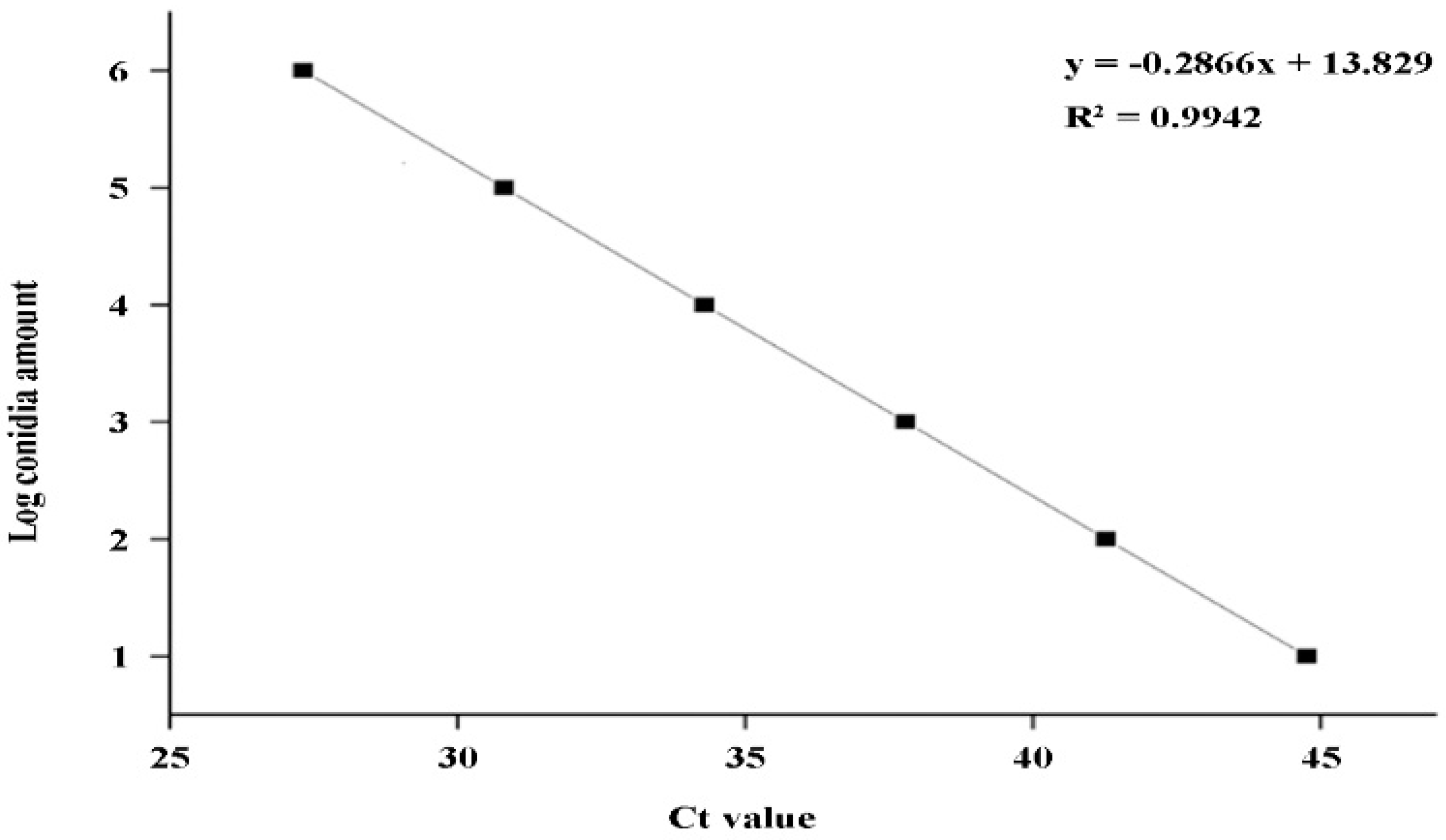
2.5. Establishment of a DNA standard curve for q-LAMP detection of U. virens
A standard curve between the amplification time (x) and the Log
10 value of spore number (y) was constructed based on the q-LAMP assay results listed in
Table 2: y = -0.2866x+13.829 (
Figure 5) , and the formula used for calculating spore number is: 10
0.65y , and the correlation coefficient R²= 0.9942 showing a good linear relationship.
Figure 5.
Standard curve of q-LAMP detection system. A standard curve between logarithmic values of the spore number (y) and the amplification time quantitated using the cycle threshold (Ct) values (x): y = -0.2866x+13.829. The correlation coefficient (R²) is 0.9942 showing a good linear relationship.
Figure 5.
Standard curve of q-LAMP detection system. A standard curve between logarithmic values of the spore number (y) and the amplification time quantitated using the cycle threshold (Ct) values (x): y = -0.2866x+13.829. The correlation coefficient (R²) is 0.9942 showing a good linear relationship.
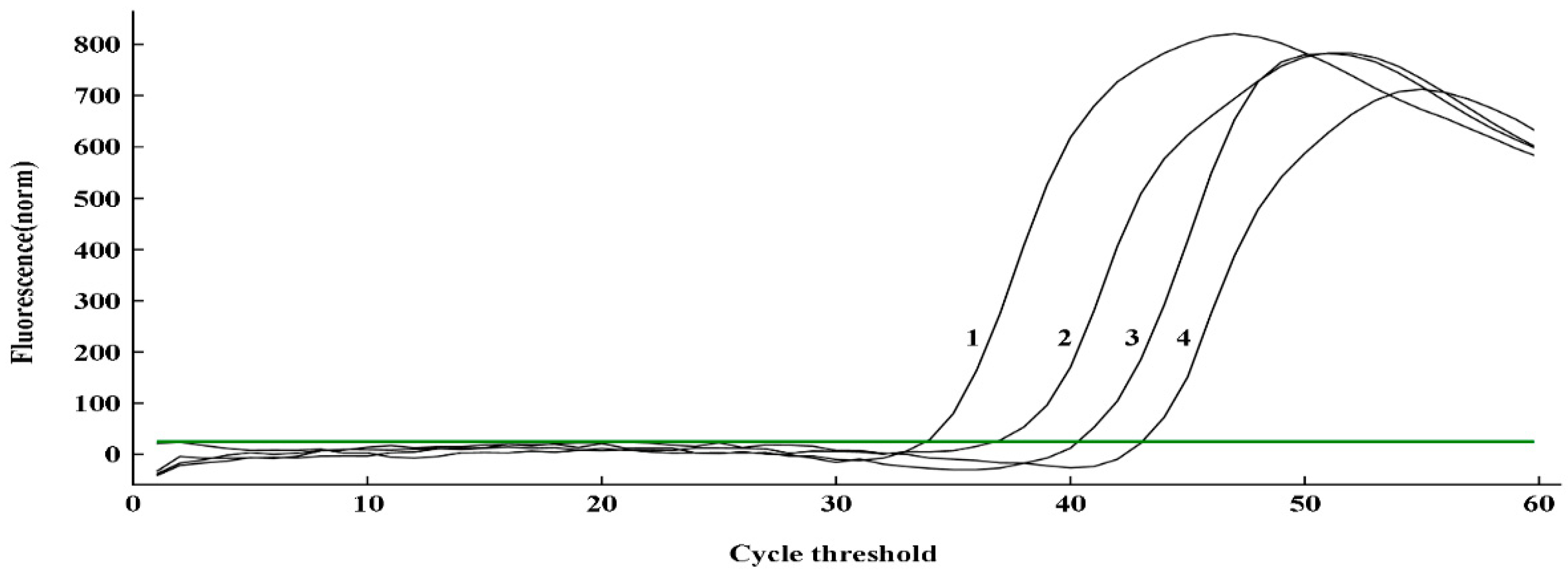
2.6. Application of q-LAMP assay for U. virens spore calculating
The standard curve of q-LAMP was applied to calculate
U. virens spore number on tapes and each tape sample contained 450, 116, 29 and 9 manually added spores, respectively. As showed in
Table 3 and
Figure 6, the amplification times for the tested samples were 34.03, 37.12, 40.46 and 43.17 mins, corresponding to 446.07, 118.51, 28.29 and 8.85 predicted spores per tape, respectively, which is super close to the actual spore number on each Melinex tape. Thus, this q-LAMP system can efficiently quantitate
U. virens spore number with high accuracy.
Table 3.
Quantitative detection of U. virens spores using q-LAMP system.
Table 3.
Quantitative detection of U. virens spores using q-LAMP system.
| Manually added spores (spores/mL) |
Time a (min) |
Predictive spores (spores/mL) |
| 450 |
34.03 |
446.07 |
| 116 |
37.12 |
118.51 |
| 29 |
40.46 |
28.29 |
| 9 |
43.17 |
8.85 |
Figure 6.
Quantitative detection of U. virens spores on Melinex tape using q-LAMP system. Serial numbers 1, 2, 3, and 4 represent 450, 116, 29, and 9 spores, respectively. The green line (horizontal) indicates fluorescence threshold. Fluorescence signals above this threshold marked as a successful detection for U. virens in q-LAMP assays.
Figure 6.
Quantitative detection of U. virens spores on Melinex tape using q-LAMP system. Serial numbers 1, 2, 3, and 4 represent 450, 116, 29, and 9 spores, respectively. The green line (horizontal) indicates fluorescence threshold. Fluorescence signals above this threshold marked as a successful detection for U. virens in q-LAMP assays.
2.7. Field application of q-LAMP assay system
The q-LAMP system results showed that spores of
U. virens were first observed on August 27, 2018, while the results obtained by microscope showed that spores were observed for the first time on September 2. Then, the number of spores began to rise rapidly and reached its peak on September 20, and obvious symptoms of rice false trot were found in the field on September 25. In the next year (2019), the q-LAMP system and microscope manual observation were used to monitor the spores of
U. virens in the field again. The results showed that the q-LAMP detection detected the spores for the first time on August 31, while the microscope observation found only a handful of spores on September 6, and then the concentration of spores reached its peak on September 30. The symptoms of rice false smut were found in the field on October 5. Through monitoring the dynamic change of spore number of
U. virens in the field for two consecutive years(
Figure 7), it was obviously found that the q-LAMP system was faster and more timely than the traditional microscope observation method.
Figure 7.
Field application of U. virens spores using q-LAMP system. (A) Flow chart of field U. virens spore sample detection, q-LAMP assay system and microscope observation were used for the collected samples respectively. (B) a and b are the results of U. virens spore concentration measured by different methods in rice fields in 2018 and 2019 respectively (q-LAMP assay system is the gray line, microscope observation method is the blue line), in which the green circle is the first detection of spores by q-LAMP assay system, the orange triangle is the first observation of spores by microscope observation, and the red arrow is the occurrence time of rice false smut in the field.
Figure 7.
Field application of U. virens spores using q-LAMP system. (A) Flow chart of field U. virens spore sample detection, q-LAMP assay system and microscope observation were used for the collected samples respectively. (B) a and b are the results of U. virens spore concentration measured by different methods in rice fields in 2018 and 2019 respectively (q-LAMP assay system is the gray line, microscope observation method is the blue line), in which the green circle is the first detection of spores by q-LAMP assay system, the orange triangle is the first observation of spores by microscope observation, and the red arrow is the occurrence time of rice false smut in the field.
3. Discussion
Currently, rice false smut disease caused by
U. virens is one of the most devastating rice diseases in China, as well as many other countries[
34]. The occurrence of rice false smut disease not only results in decrease of rice quality and serious loss of rice yield, but also threatens food safety due to its production of toxic mycotoxins within false smut balls [
10,
35]. However, it has been found that the rice false smut disease is difficult to control. As a typical airborne disease, the epidemic of rice false smut is closely related to the amount of
U. virens spores in the fields, thus early detection and warning are critical for preventing and mitigating rice false smut. In this study, a q-LAMP assay system was developed. The result showed that the species-specific primer sets UV-2 in q-LAMP assay system could correctly distinguish
U. virens from the other 9 air-dispersal fungi including
Fusarium fujikuroi,
F. oxysporum,
F. proliferatum,
F. solani,
F. graminearum,
Penicillium sp,
Pyricularia oryzae,
Alternaria alternata and Rhizoctonia solani (
Figure 3). Additionally, sensitivity validation found that the q-LAMP assay was able to detect a concentration of 6.4
U. virens spores/mL at an optimal reaction temperature of 63.4℃ within 60 min (
Figure 4), and the q-LAMP assay can even achieve accurate quantitative detection when there were only 9
U. virens spores on the Melinex tape (
Figure 6). Moreover, there is a good linear relationship between the spore amount (y) and the amplification time (x) (
Figure 5), which enables the accurate quantification of
U. virens and provides early diagnosis of
U. virens infection via q-LAMP assay.
For
U. virens diagnosis, besides traditional disease diagnosis that includes identification of symptoms, isolation of pathogens and microscopic techniques, a conventional nested-PCR assay has been developed for detection
U. virens in rice [
36]. However, the nested-PCR has less sensitivity and cannot be used in accurate quantification of
U. virens[
37]. Recently, the q-PCR technique and q-LAMP assay have been applied for the identification and quantification of pathogen in disease diagnosis. In this study, we have established these two systems for
U. virens quantification. The q-PCR assay was carried out using the primer set F3/B3 and the effective amplification reactions were detected at samples with spore concentrations of 2×10
4, 4×10
3, 8×10
2, 1.6×10
2 spores/mL, but not 32 spores/mL (
Supplementary Figure S1), indicating a lower sensitivity of q-PCR for
U. virens detection compared to q-LAMP assay system.
Rice false smut has no symptoms in the early stage and can only be identified in the late stage when the smut ball appears. Chemical control is the main means of rice false smut prevention and control[
38]. The previous study showed that the first 4~15 d of ear bud breakage was the main period of control, and the first 4~7 d of control was the best[
39]. If the key window stage for infection of
U. virens on rice is not been grasped, the efficacy of management will be disappointment[
16,
39]. Therefore, for rice false smut that rely on airborne transmission, early detection and early warning can provide appropriate guidance for disease prevention and control. In this study, we collected spore samples of
U. virens from the field for two consecutive years for q-LAMP assay system and microscope observation. Compared with manual observation, q-LAMP assay system could detect spores in the air more accurately and quickly, providing theoretical basis for precise fungicide application(
Figure 7). Therefore q-LAMP assay with higher efficiency and sensitivity is a better choice for early diagnosis of rice false smut.
In conclusion, this is the first assay developed for the detection of U. virens using q-LAMP assays. Compared with other U. virens detection methods, the newly developed LAMP assay has stronger operability, specificity as well as sensitivity, and is more suitable for quantitative detection of U. virens in the early disease diagnosis.
4. Materials and Methods
4.1. Fungal isolates
Isolates of Ustilaginoidea virens and the 9 other fungal pathogens used in this study were isolated and identified in our lab, and the detail information of each fungus was listed in
Table 4. Isolates were maintained on potato dextrose agar (PDA, prepared by 200 g potato, 20 g glucose and 20 g agar per 1 L pure water) slants at 4°C.
Table 4.
The information of strains used in the specificity validation of the q-LAMP assay system.
Table 4.
The information of strains used in the specificity validation of the q-LAMP assay system.
| Species |
Isolate NO. |
Host |
Origin |
| Fusarium fujikuroi |
/ |
Rice |
Zhejiang, China |
| F. oxysporum |
ACCC30927a
|
Rice |
Hainan, China |
| F. proliferatum |
CICC2489b
|
Rice |
Anhui, China |
| F. solani |
ACCC37119 |
Rice |
Hebei, China |
| F. graminearum |
ACCC37680 |
Wheat |
Jiangxi, China |
|
Penicillium sp. |
ACCC31507 |
Soil |
Shandong, China |
| Ustilaginoidea virens |
ACCC2711 |
Rice |
Hunan, China |
| Pyricularia oryzae |
ACCC37631 |
Rice |
Fujian, China |
| Alternaria alternata |
ACCC36843 |
Rice |
Hainan, China |
| Rhizoctonia solani |
ACCC36246 |
Rice |
Beijing, China |
4.2. DNA templet preparation from mycelium and spores for q-PCR and q-LAMP analysis
For mycelial DNA templet preparation, after mycelia grew covering two-thirds of the PDA plate surfaces, the hyphae were then transferred to a mortar and ground with liquid nitrogen. The resultant powder was then placed into a 2-mL centrifuge tube and the mycelial DNA of each fungus was extracted using a Genomic DNA Kit (Sangon Biotech Co., Ltd, Shanghai) according to the manufacturer’s instructions. The extracted DNA was used as DNA template in q-LAMP analyses and stored at -20℃. For spores DNA templet preparation, after growing on PDA medium at 25℃ in darkness for 20 days, 5-mm diameter mycelial plugs taken from colony margin were placed into the potato sucrose (PS, prepared by 200 g potato and 20 g per 1 L pure water) medium at 25℃ 150 rpm, in darkness for 7 days. Spores separated from medium with filtration through four layers of lens tissue and washed twice with distilled water. Then spores were diluted with 10% sodium dodecylsulfate (SDS) solution into a series of concentration gradients. 1-mL spore suspension sample with known concentration mixed with 200-μL 10% Chelex-100 solution[
21], 50-μL 10% SDS solution and 0.4-g acid-washed glass beads was placed into a 2.0-mL centrifuge tube. The sample was lysed by Fast Prep Apparatus (JXFSTPRP-24L, Jingxin Technology, Shanghai, China) for 40s at speed of 6 m/s, placed in boiling water bath for 5 minutes. The grinding and heating septs were repeated for three times, after which the sample was placed on ice for 2 min. The cooled lysate was used directly as DNA template in q-PCR and q-LAMP analyses and stored at -20℃.
4.3. Design of q-LAMP primers for U. virens detection
Ustiloxin A and Ustiloxin B of
U. virens are synthesized by ustiloxins biosynthetic gene that was found to be species-specific to
U. virens [
13,
40]. Thus, the sequence of ustiloxins biosynthetic gene (NCBI accession number: BR001221.1) was chosen for q-LAMP primers design using Primer Explore V5 (online web service,
http://primerexplorer.jp/e/) ensuring the specificity and accuracy of q-LAMP assay system for
U. virens detection. The q-LAMP primers contain forward inner primer (FIP), backward inner primer (BIP), and two outer (F3 and B3) primers (
Figure 1B). The primers were design according to the following rules: ΔG values of less than or equal to -4 Kcal/mol at the 3’end of F3/B3 and F2/B2 and 5’ends of F1c and B1c.
4.4. Determination of optimum condition of the q-LAMP assay
To better facilitate the efficiency of q-LAMP reaction, the LAMP reaction system was improved via screening for the optimal reaction temperature based on a reference from Notomi[
41]. The LAMP reaction was carried out in the following reaction mixtures containing 0.25 μM·L
-1 of the primers FIP and BIP, 0.2 μM·L
-1 of the primers F3 and B3, 1.0 mM·L
-1 betaine, 2.0 mM·L
-1 dNTPs (Takara Bio Inc., 108 USA), 25 mM·L
-1 Tris-HCl (pH 8.8), 12.5 mM·L
-1 KCl, 12.5 mM·L
-1 (NH
4)
2SO
4, 10 mM·L
-1 MgCl
2, 0.125% (v/v) Triton X-100, 0.2 U·L
-1 of Bst DNA polymerase (New England Biolabs, 110 Beijing), 0.5 μL 1×SYBR Green I and 1 μL of DNA template extracted as described above, and the volume was adjusted to 25 μL with nucleic acid-free water. The screened reaction temperature gradients were 61.8°C, 62.1°C, 62.6°C, 63.4°C, 64.4°C, 65.2°C, 65.6°C and 66°C. LAMP reactions were performed using a Bio-Rad quantitative fluorescent PCR instrument (Bio-Rad CFX96, Hercules, California, USA) for 80 cycles each, each cycle for 60 s, and the reaction was terminated at 80°C for 10 min. Optimal reaction temperature screening experiments were repeated three times.
4.5. Validation of the specificity for q-LAMP assay systems
The specificity of the reaction system was tested by performing q-LAMP reactions at the optimal reaction temperature with UV-2 primers in above 25-μL reaction mixtures for 70 min. The assay results were compared with the DNA of
U. virens and the 9 other fungi listed in
Table 4. The nucleic acid-free water was set as negative control. The results were rigorously validated with the assessment that the detectable peak of fluorescence signals detected by Bio-Rad CFX96 as positive; no fluorescence signal as negative. The specificity testing experiment was repeated three times.
4.6. Sensitivity validation of q-LAMP and q-PCR assay systems
The sensitivity validation of q-LAMP reactions was performed at the optimal reaction temperature with UV-2 primers in above 25-μL reaction mixtures for 60 min. 1 μL of DNA lysate from U. virens spores with known concentration was used as a DNA template in the LAMP reaction system. The nucleic acid-free water was used as a DNA template in the negative control (CK). The detectable peak of fluorescence signals detected by Bio-Rad CFX96 was regarded as positive, while no fluorescence signal was regarded as negative. Sensitivity assay experiments were repeated three times. The sensitivity of the q-PCR reaction system was assayed by performing q-PCR amplification using primers UV-2 F3/B3. The q-PCR reaction system was 12.5 μL SYBR® Premix Ex Taq II (Tli RNaseH Plus, 2×), 1.0 μL of forward primer F3 (10 μM), 1.0 μL of reverse primer B3 (10.0 μM), 1.0 μL DNA template (in CK, nucleic acid-free water was used as DNA template), and the volume was adjusted to 25 μL with nucleic acid-free water. The reaction conditions were: pre-denaturation at 95℃for 2 min, denaturation at 95℃ for 5 s, annealing at 60℃ for 30 s, extension at 72℃for 6 s. The fluorescence signal was collected during the extension for a total of 40 cycles, and finally the lysis curve was plotted. The detectable peak of fluorescence signals detected by Bio-Rad CFX96 was regarded as positive, while no fluorescence signal was regarded as negative. Sensitivity assay experiments were repeated three times.
4.7. Establishment of standard curves for q-LAMP assay systems
A standard curve was constructed using software SPSS 13.0 by analyzing the association of logarithmic values of the spore number (y) and the amplification time quantitated using the cycle threshold (Ct) values (x). The correlation coefficient R2 was used for assessing the linear relationship between the spore number in sample (y) and amplification time (x).
4.8. Calculating of U. virens spore using q-LAMP system
Spores of U. virens were artificially added to each of the four Melinex tape (1 cm×2 cm) in the ultra-clean bench, with 450, 116, 29 and 9 spores in each tape. The collected spore-adsorbed Melinex tape was cut and placed into 2-mL centrifuge tubes, and the genomic DNA of the spores on the Melinex tape was then extracted according to the method mentioned above. 1 μL of the cooled lysate was used directly as DNA template. q-LAMP was performed with the optimal reaction conditions in above 25-μL reaction mixtures for 60 min, and the cycle threshold (Ct) values detected by Bio-Rad CFX96 was recorded as the amplification time (x). The linearized equation for the standard curve was used for converting the amplification time to the corresponding spore number. Then the calculated spore number was compared to the amount of actual added (listed above) to test the accuracy efficiency of this q-LAMP system.
4.9. Field application of q-LAMP assay by U. virens
An air borne spore catcher (DIANJIANG, DJ-0723) with Melinex tape was established in Yongyou 1540 cultivation area, Jiangtang Village, Jinhua City, Zhejiang Province for the collection of spores of U. virens. and samples were collected at six-day intervals for 11 consecutive times starting on August 9, 2018. Similarly, starting on August 13 of the following year (2019), 11 consecutive samples were collected every six days. The spores of U. virens were adsorbed to the Melinex tape and the tapes (1 cm×1 cm) with spores were cut and placed in a 2-mL centrifuge tube, and the conidial DNA was extracted according to the methods mentioned above. 1 μL of the cooled lysate was used directly as DNA template. q-LAMP was performed with the optimal reaction conditions in above 25-μL reaction mixtures for 60 min, and the amplification time was recorded. According to the established standard curve, the number of spores was calculated. The spore population of U. virens in the glass slide was recorded by q-LAMP assay at six-day intervals. Meanwhile, the spores of U. virens (1 cm×1 cm) adsorbed on the slide were suspended in 1-mL ddH2O, and the spore suspension was counted using a hemocytometer under the microscope to determine the spore concentration.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1. Sensitivity validation of q-PCR detection system.
Author Contributions
Conceptualization, T.M. and C.Z.; methodology, C.Z., Y.Z., and X.L.; software, T.M. and Y.Z; validation, T.M. and C.Z.; formal analysis, Y.Z., S.Z.; investigation, S.Z. and X.L.; writing—original draft preparation, Y.Z. and T.M.; writing—review and editing, T.M. and C.Z.; visualization, J.W. and X.L.; supervision, T.M. and C.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key Research and Development Project of Zhejiang Province, China (2015C02019), Science &Technology Program of Agriculture and Country in Zhenhai District.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wei, S.; Wang, Y.; Zhou, J.; Xiang, S.; Sun, W.; Peng, X.; Li, J.; Hai, Y.; Wang, Y.; Li, S. The conserved effector UvHrip1 interacts with OsHGW, and infection of Ustilaginoidea virens regulates defense- and heading date-related signaling pathway. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, J. Arabidopsis thaliana: A model host plant to study plant–pathogen interaction using rice false smut isolates of Ustilaginoidea virens. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Song, J.H.; Wei, W.; Lv, B.; Lin, Y.; Yin, W.X.; Peng, Y.L.; Schnabel, G.; Huang, J.B.; Jiang, D.H.; Luo, C.X. Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ. Microbiol. 2016, 18, 3840–3849. [Google Scholar] [CrossRef]
- Meng, S.; Xiong, M.; Jagernath, J.S.; Wang, C.; Qiu, J.; Shi, H.; Kou, Y. UvAtg8-Mediated autophagy regulates fungal growth, stress responses, conidiation, and pathogenesis in Ustilaginoidea virens. Rice 2020, 13, 56. [Google Scholar] [CrossRef]
- Devi, T.K.; Singh, N.I. Aerobiology and epidemiology of false smut disease of rice by Ustilagnoidea virens (Syn. Claviceps oryzae sativae) in Thoubal District. J. Mycopatholog. Res. 2007. [Google Scholar]
- Rush, M.C.; Shahjahan, A.; Jones, J.P.; Groth, D.E. Outbreak of false smut of rice in Louisiana. Plant Dis. 2000, 84, 100. [Google Scholar] [CrossRef]
- Ashizawa, T.; Takahashi, M.; Moriwaki, J.; Hirayae, K. Quantification of the rice false smut pathogen Ustilaginoidea virens from soil in Japan using real-time PCR. Eur. J. Plant Pathol. 2010, 128, 221–232. [Google Scholar] [CrossRef]
- Tanaka, E.; Ashizawa, T.; Sonoda, R.; Tanaka, C. Villosiclava virens gen. nov., comb. nov., teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon 2008, 106, 491–501. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, K.; Fang, A.; Han, Y.; Yang, J.; Xue, M.; Bao, J.; Hu, D.; Zhou, B.; Sun, X.; et al. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat. Commun. 2014, 5, 3849. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.L.; Xie, X.W.; Zhang, F.; Wang, S.; Liu, X.Z.; Zhu, L.H.; Xu, J.L.; Gao, Y.M.; Li, Z.K. Detection of quantitative resistance loci associated with resistance to rice false smut (Ustilaginoidea virens) using introgression lines. Plant Pathol. 2014, 63, 365–372. [Google Scholar] [CrossRef]
- Sun, X.; Kang, S.; Zhang, Y.; Tan, X.; Yu, Y.; He, H.; Zhang, X.; Liu, Y.; Wang, S.; Sun, W.; et al. Genetic diversity and population structure of rice pathogen Ustilaginoidea virens in China. PLoS One 2013, 8, e76879. [Google Scholar] [CrossRef]
- Zhou, L.; Lu, S.; Shan, T.; Wang, P.; Wang, S. Chemistry and biology of mycotoxins from rice false smut pathogen; Mycotoxins: Properties, Applications and Hazards, 2012, 109-130.
- Fu, X.; Wang, A.; Wang, X.; Lin, F.; He, L.; Lai, D.; Liu, Y.; Li, Q.X.; Zhou, L.; Wang, B. Development of a monoclonal antibody-based icELISA for the detection of Ustiloxin B in rice false smut balls and rice grains. Toxins 2015, 7, 3481–3496. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Yu, J.; Cao, H.; Song, T.; Pan, X.; Qi, Z.; Du Y; Zhang, R. ; Huang, S.; Liu, W. et al. SUN-Family protein UvSUN1 regulates the development and virulence of Ustilaginoidea virens. Front. Microbiol. 2021, 12, 739453. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, M.; Song, T.; Cao, H.; Pan, X.; Yong, M.; Qi, Z.; Du Y; Zhang, R.; Yin, X. et al. A homeobox transcription factor UvHOX2 regulates chlamydospore formation, conidiogenesis, and pathogenicity in Ustilaginoidea virens. Front. Microbiol. 2019, 10, 1071. [CrossRef] [PubMed]
- Tang, Y.; Jin, J.; Hu, D.; Yong, M.; Xu, Y.; He, L. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 2013, 62, 1–8. [Google Scholar] [CrossRef]
- Hu, Y. Infection processes of Ustilaginoidea virens during artificial inoculation of rice panicles. Eur. J. Plant Pathol. 2014, 139. [Google Scholar] [CrossRef]
- Song, J.; Wei, W.; Lv, B.; Yang, L.; Yin, W.; Peng, Y.; Schnabel, G.; Huang, J.; Jiang, D.; Luo, C. Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ. Microbiol. 2016, 18, 3840–3849. [Google Scholar] [CrossRef]
- Yong, M.; Deng, Q.; Fan, L.; Miao, J.; Lai, C.; Chen, H.; Yang, X.; Wang, S.; Chen, F.; Jin, L. The role of Ustilaginoidea virens sclerotia in increasing incidence of rice false smut disease in the subtropical zone in China. Eur. J. Plant Pathol. 2018, 150, 669–677. [Google Scholar] [CrossRef]
- Tsukui, T.; Nagano, N.; Umemura, M.; Kumagai, T.; Terai, G.; Machida, M.; Asai, K. Ustiloxins, fungal cyclic peptides, are ribosomally synthesized in Ustilaginoidea virens. Bioinformatics 2015, 31, 981–985. [Google Scholar] [CrossRef]
- Wang, Q.W.; Zhang, C.Q. Q-LAMP Assays for the detection of Botryosphaeria dothidea causing Chinese hickory canker in trunk, water, and air samples. Plant DIS. 2019, 103, 3142–3149. [Google Scholar] [CrossRef]
- Harrison, N.A.; Womack, M.; Carpio, M.L. Detection and characterization of a lethal yellowing (16SrIV) group Phytoplasma in canary island date palms affected by lethal decline in Texas. Plant Dis. 2002, 86, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Aryan, E.; Makvandi, M.; Farajzadeh, A.; Huygen, K.; Bifani, P.; Mousavi, S.L.; Fateh, A.; Jelodar, A.; Gouya, M.M.; Romano, M. A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of Mycobacterium tuberculosis complex. Microbiol. Res. 2010, 165, 211–220. [Google Scholar] [CrossRef]
- McKenna, J.P.; Fairley, D.J.; Shields, M.D.; Cosby, S.L.; Wyatt, D.E.; McCaughey, C.; Coyle, P.V. Development and clinical validation of a loop-mediated isothermal amplification method for the rapid detection of Neisseria meningitidis. Dia. Microbiol. Infect. Dis. 2011, 69, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Xie, Z.; Zhao, G.; Liu, J.; Pang, Y.; Deng, X.; Xie, Z.; Fan, Q.; Luo, S. A loop-mediated isothermal amplification assay for the visual detection of duck circovirus. Virol. J. 2014, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, M.; Shams, S.; Majidzadeh-A, K. Developing a real-time quantitative loop-mediated isothermal amplification assay as a rapid and accurate method for detection of Brucellosis. J. Appl. Microbiol. 2013, 115, 828–834. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef]
- Stella, S.; Gottardi, E.M.; Favout, V.; Barragan, G.E.; Errichiello, S.; Vitale, S.R.; Fava, C.; Luciano, L.; Stagno, F.; Grimaldi, F.; et al. The q-LAMP method represents a valid and rapid alternative for the detection of the BCR-ABL1 rearrangement in Philadelphia-positive Leukemias. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Wang, Y.; Li, K.; Xu, G.; Chen, C.; Song, G.; Dong, Z.; Lin, L.; Wang, Y.; Xu, Z.; Yu, M.; et al. Low-cost and scalable platform with multiplexed microwell array biochip for rapid diagnosis of COVID-19. Research 2021, 2021, 2813643. [Google Scholar] [CrossRef]
- Huang, Y.; Tang, X.; Zheng, L.; Huang, J.; Zhang, Q.; Liu, H. Development of generic immuno-magnetic bead-based enzyme-linked immunoassay for Ustiloxins in rice coupled with enrichment. Toxins 2021, 13. [Google Scholar] [CrossRef]
- Wang, X.; Fu, X.; Lin, F.; Sun, W.; Yang, L. The contents of ustiloxins A and B along with their distribution in rice false smut balls. Toxins 2016, 8, 262. [Google Scholar] [CrossRef]
- Fu, R.; Chen, C.; Wang, J.; Liu, Y.; Zhao, L.; Lu, D. Transcription profiling of rice panicle in response to crude toxin extract of Ustilaginoidea virens. Front. Microbiol. 2022, 13, 701489. [Google Scholar] [CrossRef]
- Abbas, H.K.; Shier, W.T.; Cartwright, R.D.; Sciumbato, G.L. Ustilaginoidea virens infection of rice in Arkansas: toxicity of false smut galls, their extracts and the ustiloxin fraction. American Journal of Plant Sciences 2014, 5, 3166–3176. [Google Scholar] [CrossRef]
- Sun, X.; Shu, K.; Zhang, Y.; Tan, X.; Yu, Y.; He, H.; Zhang, X.; Liu, Y.; Wang, S.; Sun, W. Genetic diversity and population structure of rice pathogen Ustilaginoidea virens in China. PLoS One 2013, 8. [Google Scholar] [CrossRef]
- L. , Y.; Zhou; K.; Izumitsu; R.; Sonoda; T.; Nakazaki; E.; Tanaka PCR-based specific detection of Ustilaginoidea virens and Ephelis japonica. J. Phytopathol. 2003, 9, 513–518. [Google Scholar] [CrossRef]
- Li, H.; Ni, D.H.; Duan, Y.B.; Chen, Y.; Li, J.; Song, F.S.; Li, L.; Wei, P.C.; Yang, J.B. Quantitative detection of the rice false smut pathogen Ustilaginoidea virens by real-time PCR. Genet. Mol. Res. 2013, 12, 6433–6441. [Google Scholar] [CrossRef]
- WANG, Z.; YANG, X.; LYU, L.; YUAN, B.; CHANG, X.; ZHANG, S. Progress and prospective of Villosiclava virens infection mechanism. Hubei Agric. Sci. 2019, 58, 5. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Q.; Luo, H. Test pesticides against rice false smut and choose optimum application period. J. Huazhong Agric. Univ. 2007, 26, 178. [Google Scholar]
- Fu, X.; Xie, R.; Jian, W.; Chen, X.; Wang, X.; Sun, W.; Meng, J.; Lai, D.; Zhou, L.; Wang, B. Development of colloidal gold-based lateral flow immunoassay for rapid qualitative and semiquantitative analysis of ustiloxins A and B in rice samples. Toxins 2017, 9, 79. [Google Scholar] [CrossRef]
- Tomlinson, J.; Boonham, N. Real-Time LAMP for Chalara fraxinea diagnosis. Methods Mol Biol 2015, 1302, 75–83. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).