I. Introduction
In the realm of scientific research, the vast amount of information present in academic literature, reports, and studies can pose a difficulty for researchers in locating pertinent data and drawing valuable conclusions. As a solution to this problem, there is an increasing demand for tools that enable researchers to efficiently navigate and scrutinize the vast quantity of data at their disposal.
Several influential articles have explored the use of artificial intelligence (AI) to help researchers in this regard. For example, [
1] discussed the application of deep learning to computational biology and the potential for using neural networks to predict RNA binding sites, [
2] provided an overview of the history and current applications of AI in healthcare, while [
3] examined the history and state of the art of machine learning in medical diagnosis. In addition, [
4] provided a survey of recent research using machine learning in natural sciences, including molecular and materials science, single-cell genomics, and Earth system science. In line with these studies, [
5] investigated the effect of the topological shape and orientation of oxide fillers on the properties of polymer/ceramic nanocomposites using high-throughput phase-field simulations.
One such tool that addresses this need is DataDiscoveryLab, a software system that utilizes computer vision algorithms and NLP techniques to parse academic articles into text and figures. This creates two separate databases that can be used to find similarities between users’ research questions, experimental setups, and existing literature. The system then utilizes this data to provide researchers with the most relevant pathways to research and articles to read, ensuring that they can make informed decisions about their work. Also, here we would like to add that it can be enlarged by using every laboratory’s Standard operating procedures (SOPs) to analyze different methods’ roles.
Auto-GPT, a recent example of an AI-powered task management system, is an innovative application that utilizes AI to perform autonomous tasks [
6]. Several influential projects and articles have explored the use of AI in this regard [
7]. For instance, [
8] introduced the Transformer architecture for sequence-to-sequence tasks, which has since become a cornerstone of modern natural language processing and highly known tools like ChatGPT by OpenAI. [
9] introduced the BERT architecture, currently one of the most widely used pre-trained language models for NLP tasks. For instance on the image recognition side, [
10] introduced the AlexNet architecture, which marked the beginning of the deep learning revolution in computer vision. [
11] introduced the YOLO object detection algorithm, known for its fast inference speed and high accuracy. [
12] introduced the GAN architecture, which has since become one of the most popular and versatile frameworks for generative modeling in computer vision. These articles are important building blocks for Auto-GPT and other AI systems that aim to automate tasks and improve efficiency by utilizing text and image-based deep learning models.
By using some of these tools from these two domains, DataDiscoveryLab can be a powerful tool that can help researchers save time and resources while gaining valuable insights from existing literature. With the ability to parse large volumes of data and find relevant information quickly, this system can significantly enhance the research process, ultimately leading to more impactful research outcomes.
According to [
13], every year 7 million articles are published and from them, 1.8 million of them have more than 5 citations. This means two things, one, publishing is increasing day by day, and two, we cannot keep up with every published article, and we may be missing the critical articles that can help specifically with our research problems. Here our purpose is to create a bridge with nearly every published article, standard operating procedure, and documentation of scientific devices and software to find patterns for near Artificial General Intelligence Generative academic Artificial Intelligence. We are doing it by integrating already existing features of different deep learning models, such as DeepSearch by IBM to parse and retrieve the relevant information for us, Look, Read, and Enrich by[
14] to create an improved deep learning figure and caption fusing model by their using, text-embedding-ada-002 by [
15] to create embeddings to train, in latter stages, LLMs which will be one of the paramount parts of this project. So far, we have created a pipeline to retrieve titles, introductions, methods, conclusions, captions, and references of articles with DeepSearch, a Docker image and a container for Look, Read, and Enrich, and vectorized articles with text-embedding-ada-002 for more than 8000 articles. In short, our plan is to increase the number by collaborating with journals and start to integrate the platform by collaborating with companies about scientific devices and software to start a new era of scientific discoveries.
II. Article Database and Sub Databases Based on Article Database
Article mining is the process of extracting valuable information and insights from scientific articles using automated techniques. This involves collecting a corpus of articles, often from online databases or other sources, and pre-processing the text data to extract relevant features, such as keywords, abstracts, or citation networks. Machine learning algorithms can then be applied to these features to uncover patterns, relationships, or other meaningful information.
For example, in the study, [
16], article mining was used to extract sentence-level linguistic features from a corpus of scientific abstracts, which were then used to build models that predict the readability of the abstracts. This type of article mining can be valuable for understanding the structure and content of scientific writing, as well as for developing tools to improve scientific communication and accessibility.
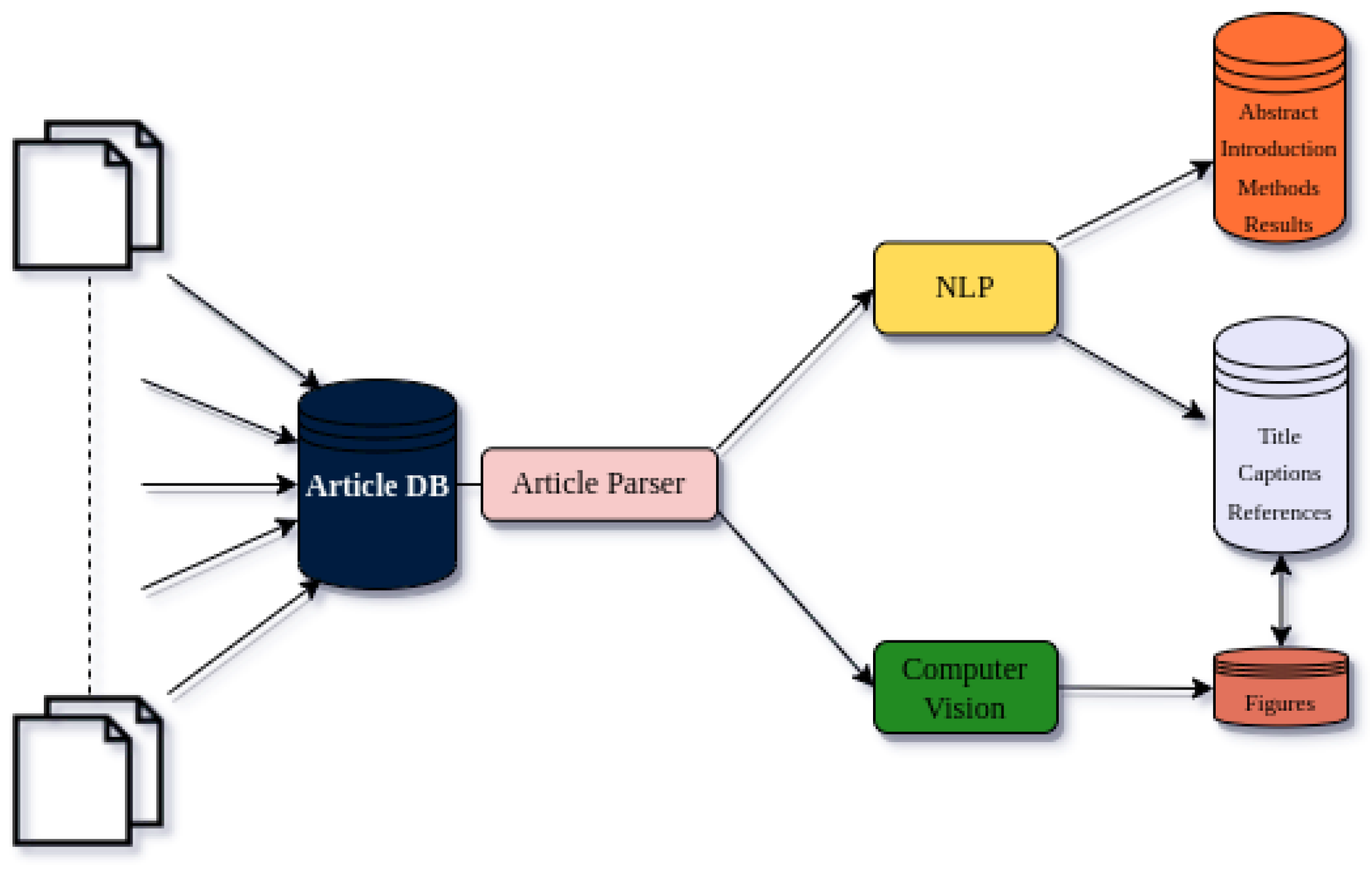
The creation of the article database is a crucial part of the DataDiscoveryLab software system, which utilizes article mining to provide researchers with a comprehensive approach to analyzing academic articles. This software system can collect articles from different scientific journals; yet for the current construction of the demo, we will be solely using arxiv.org, and parsing them into text and figures, which are then analyzed using NLP and computer vision algorithms to create three separate sub-databases.
The first sub-database includes titles, captions, and references from the articles, allowing researchers to quickly identify articles relevant to their research question. The second sub-database includes abstracts, introductions, methods, and results from the articles, providing researchers with more detailed information about the articles and a deeper understanding of the research presented. This would enable us to lessen the computational power by first finding the category of researchers’ questions and setups by finding connections with the first sub-database. After that, we would be able to connect to the specific studies to analyze each of the questions in specific research areas.
Another important part of this is to consider scientific figures due to their ability to deliver some of the most significant information in themselves. Therefore analyzing them is more than crucial. Scientific mining, also known as figure analysis or image analysis, is the process of automatically extracting information from scientific figures, such as graphs, plots, and diagrams, in order to gain insights into the underlying data. This approach has become increasingly important in recent years as the volume of scientific literature has grown exponentially, making it difficult for researchers to manually analyze and interpret the vast amount of visual data presented in scientific articles.
The third database is an image-based database that contains the figures from the articles, created by using computer vision algorithms to detect and extract individual figures. This database can be particularly useful for researchers working in fields that rely heavily on visual data. This would be enhanced as our database of articles will increase, yet we are aware that it should be strengthened by using different deep-learning models and their combinations.
The combination of these three sub-databases with the use of NLP and computer vision algorithms will allow researchers to gain a more complete understanding of the research presented in the articles. Additionally, the quick and efficient analysis of large volumes of data makes it easier for researchers to find relevant information and make informed decisions about their research. On the whole, the integration of article mining in the DataDiscoveryLab software system can provide researchers with a powerful tool for navigating the vast amount of scientific literature available today.
Figure 1.
Article Database and sub-databases nucleation.
Figure 1.
Article Database and sub-databases nucleation.
III. Prompt-Based Search
Recent articles have explored the potential benefits and drawbacks of using conversational AI technology with prompt-based search in scientific research [
17,
18]. While there are concerns about transparency, accountability, and bias, there are also potential advantages of using this technology.
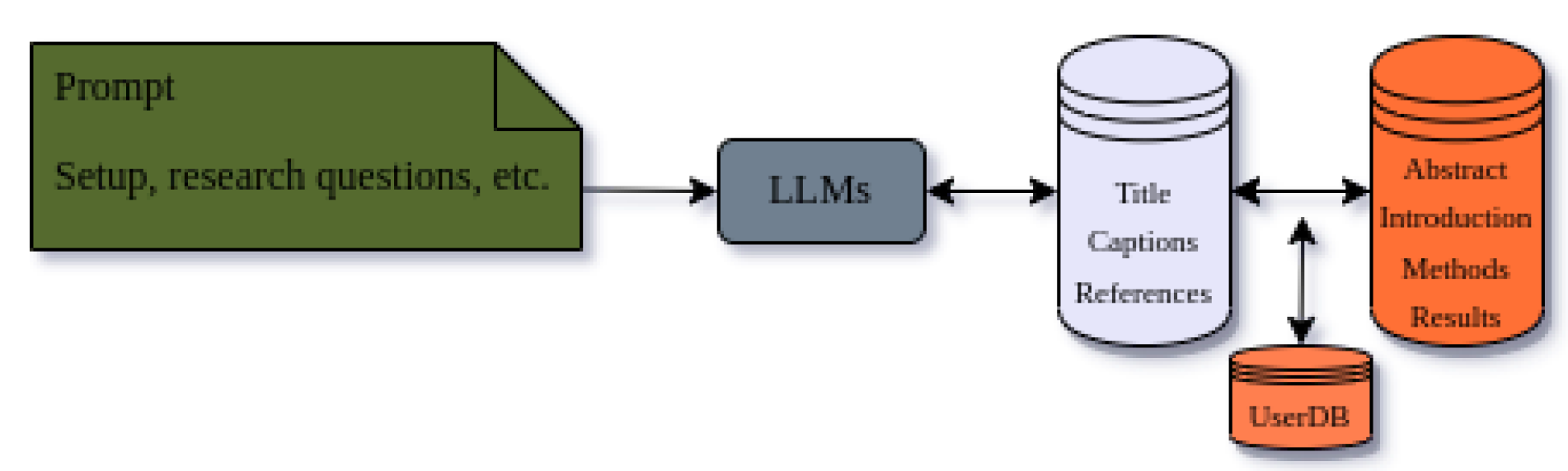
One way researchers can utilize this technology is through systems like DataDiscoveryLab. By using Large Language Models (LLMs) to analyze and understand research questions, DataDiscoveryLab can find connections with relevant articles, extract the most important information, and provide researchers with a comprehensive and efficient method for analyzing academic articles and gaining insights from existing literature.
The use of LLMs in scientific research is promising, and the study by [
19] demonstrates the potential benefits of using prompt-based LLMs in medicine, where accurate and reliable information is crucial for patient care. By using prompt-based strategies, the authors were able to develop an LLM model that could accurately and reliably answer medical questions. This has important implications for the medical field, as it could potentially improve the speed and accuracy of diagnoses, reduce the workload of healthcare professionals, and provide patients with more accessible and reliable information.
In this context just showing the capabilities of OpenAI’s GPT-4 [
20] is enough to comprehend how these kinds of systems can benefit our software in the picture.
Moreover, LLMs are continuing to develop and become more sophisticated, with the potential to revolutionize scientific research in fields where large amounts of data need to be analyzed and interpreted. Systems like DataDiscoveryLab can play a crucial role in this evolution, by providing researchers with the tools they need to efficiently analyze vast amounts of articles.
In conclusion, the potential benefits of using prompt-based search in scientific research are clear. By generating personalized and relevant search prompts, these systems can help researchers and medical professionals access and interpret large volumes of complex information, ultimately leading to improved outcomes and discoveries. DataDiscoveryLab is a powerful tool that harnesses the potential of LLMs in scientific research and provides researchers with the ability to gain insights from existing literature in a comprehensive and efficient manner.
Figure 2.
Analysis of Researchers’ Questions among text-based databases.
Figure 2.
Analysis of Researchers’ Questions among text-based databases.
IV. Data-Based Search
Nowadays, most scientists are about or already embraced deep learning algorithms at their cores due to large datasets of scientific data. For instance, as the field of Earth system science has seen a dramatic increase in data availability, recent advances in statistical modeling and machine learning have provided exciting new avenues for extracting knowledge from these vast amounts of data. Machine learning has become an essential part of geoscientific processing schemes, and it has co-evolved with data availability over the past decade, leading to early landmarks in the classification of land cover and clouds. The combination of unprecedented data sources increased computational power, and recent advances in statistical modeling and machine learning,[
21] offer promising new opportunities for expanding our understanding of the Earth system. Deep learning, in particular, holds great potential for building new data-driven models of Earth system components, providing new insights into complex interactions within the Earth system.
Machine learning has also shown significant potential in scientific research in molecular and materials science[
22]. Despite the challenges associated with its use, researchers can overcome these limitations by making their data accessible in a computer-readable format, exploring meta-learning, and developing efficient chemical representations.
Another example can be the partnership between the Scientific Computing Department at RAL and the Alan Turing Institute is a promising development in the field of machine learning and AI. The examples discussed in the article demonstrate the potential of machine learning to enable breakthroughs in a wide range of scientific fields. With the continued growth of Big Scientific Data generated by UK national facilities, the use of machine learning and other AI technologies will likely become increasingly prevalent in scientific research, enabling researchers to gain valuable insights from vast amounts of data.[
23]
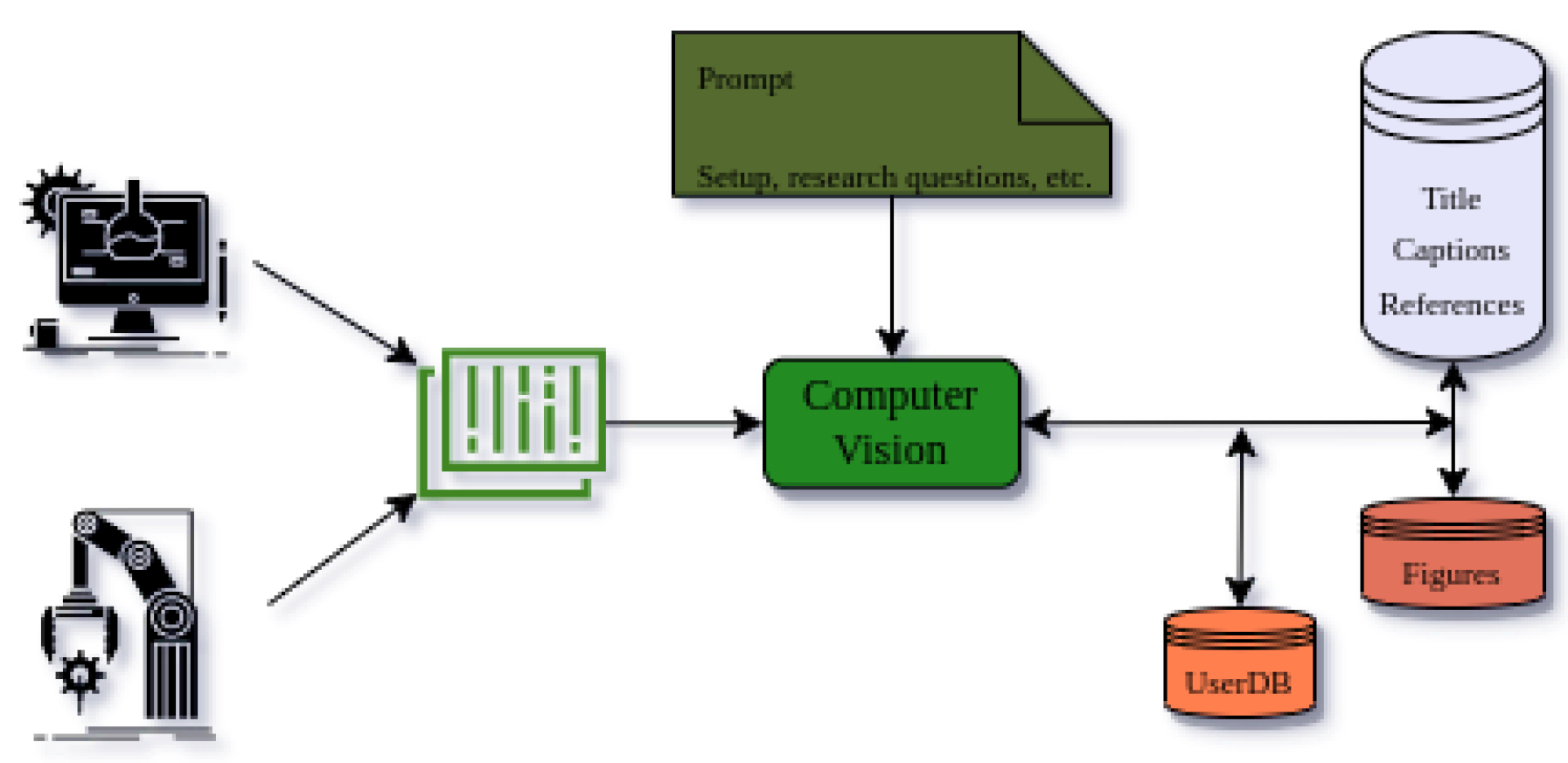
The DataDiscoveryLab can be at the forefront of using artificial intelligence (AI) to revolutionize the scientific research community’s understanding and analysis of scientific data. With its ability to quickly and accurately process vast amounts of information, AI has become an essential tool for identifying patterns and trends in scientific experimental and simulation data. All of them can be done by connecting scientific devices and software to this platform to easily retrieve scientists’ data from their studies to analyze the potential. Also, we would like to add that these data will be interpreted by the figures of articles, and the provided data by researchers, from their devices and software, will be transformed into figures and with their prompts about their methods, setups, and research questions the understanding of their figures will increase. This feature is particularly valuable for researchers working in fields that rely heavily on visual data, such as biology, chemistry, and physics. By analyzing figures and associated text data, researchers can gain valuable insights into the data and conclusions presented in academic articles.
The result of this analysis could be added to the user database, which contains all relevant information and insights gained from the analysis. By combining computer vision algorithms with natural language processing-based analysis, the DataDiscoveryLab system can provide researchers with a powerful tool for gaining insights from academic articles. This technology has the potential to revolutionize the way researchers approach their work, enabling them to make informed decisions about their research and gain a deeper understanding of their field.
Figure 3.
Analysis of Researchers’ Data with Questions among text and figure-based databases.
Figure 3.
Analysis of Researchers’ Data with Questions among text and figure-based databases.
V. Loop of Excellence
Re-evaluating input data with the outputs of AI is a powerful technique that can significantly enhance the accuracy and reliability of AI systems. By utilizing this technique, researchers can refine and improve their AI algorithms by continuously analyzing the outcomes of the algorithms and using them to improve the input data.
Image refinement is a powerful technique used in various fields to improve the quality of images. In cryo-electron microscopy, image refinement has enabled the determination of high-resolution structures of challenging macromolecules. The maximum-likelihood approach proposed by [
24] has become an essential tool for analyzing single-particle images, revolutionizing the field. Similarly, in computer graphics and artificial intelligence, image refinement has led to the development of a breakthrough approach for synthesizing photorealistic images from semantic layouts. [
25]’s approach uses cascaded refinement networks to produce photographic images that conform to a given semantic layout, without relying on computationally expensive light transport simulation. In social media platforms, image refinement plays a crucial role in improving the quality of social media annotations. Image refinement involves improving the quality of human-provided tags by organizing data for manual labeling, improving the quality of human-provided tags, or recommending tags for manual selection, instead of relying solely on automatic tagging. The importance of image refinement in social media platforms is emphasized by [
26]’s survey, which highlights the need for a more thorough empirical comparison of different approaches. To sum up, image refinement is an essential tool for improving the quality of images and has applications in various fields, including cryo-electron microscopy, computer graphics, and social media platforms.
In recent years, researchers have proposed various methods for text refinement, including manual and automatic approaches. Automatic methods are gaining popularity due to their ability to process large-scale data collections. One method [
27] involves mining anchor text for generating query refinements automatically. Anchor text provides valuable information for query refinement and can produce high-quality suggestions. Semantic analysis, multilingual text processing, and domain knowledge integration are some challenges that need to be addressed in text refinement.
In the realm of text-to-image synthesis, a new approach [
28] called DM-GAN has been proposed, which utilizes a dynamic memory module to refine the image contents accurately. The model outperforms existing text-to-image synthesis methods and addresses issues related to image accuracy and diversity.
Additionally, an article highlights the benefits of text-based refinement tools in improving the accuracy of decision-making and knowledge-intensive tasks. The article introduces Reflexion [
29] and presents two experiments, AlfWorld and HotPotQA, to demonstrate its effectiveness. The experiments show that Reflexion can improve the performance of agents in complex tasks and can enable discovery in previously challenging environments.
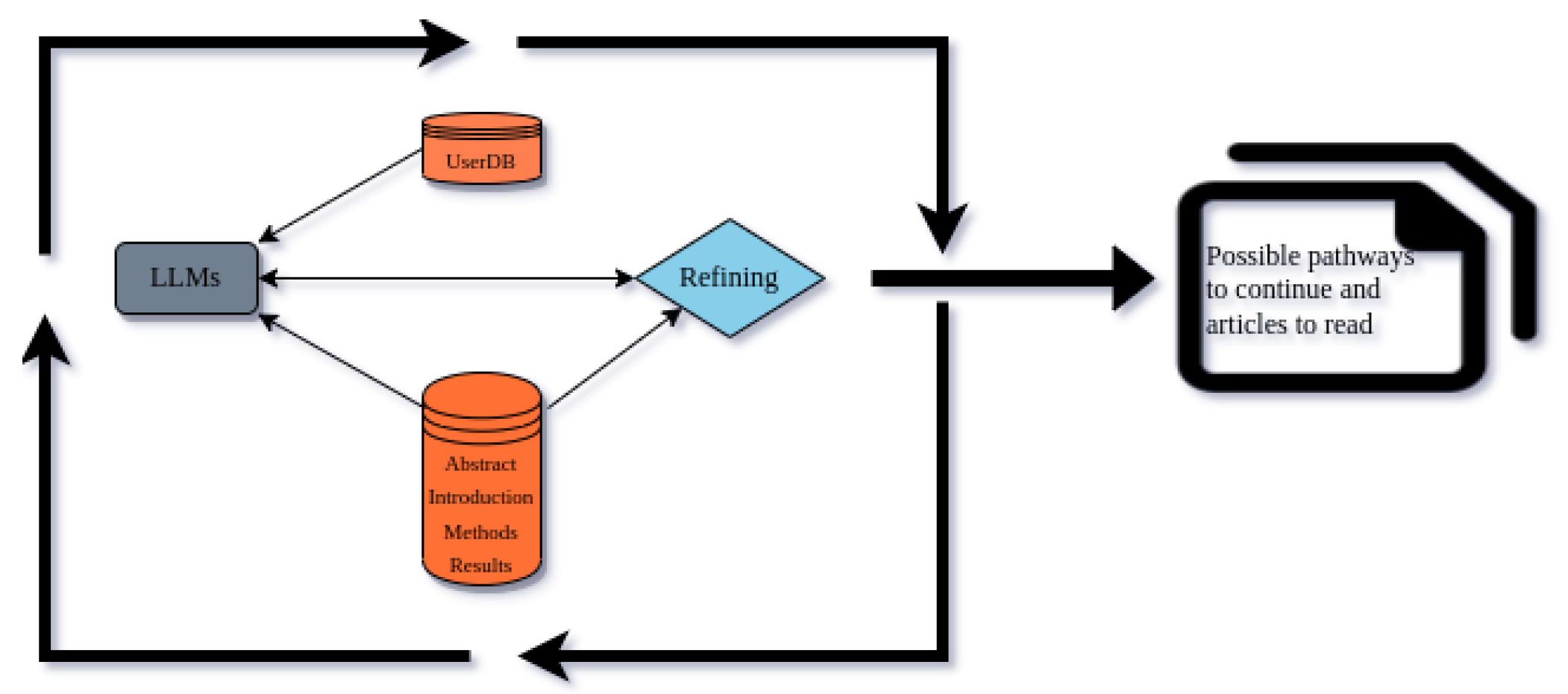
The DataDiscoveryLab system can be an example of a tool that uses this approach to continuously refine and optimize its analysis, like those mentioned above. The system analyzes the database that contains abstracts, introductions, methods, and results of articles, along with the user database that has been fed with the results of researchers’ prompts and data, using LLMs to identify patterns and connections between the different sources of information. The analysis is then refined according to the database, allowing the system to gain a deeper understanding of the research presented in the articles and the insights gained from the user database.
This iterative process is known as the Loop of Excellence, representing a continuous cycle of refinement and optimization that provides researchers with the best possible insights and outcomes. By leveraging advanced data analysis techniques and machine learning algorithms, the DataDiscoveryLab system provides researchers with a powerful tool for gaining insights from academic articles and advancing their research.
Figure 4.
Usage of Users’ Databases for nucleating recommendations.
Figure 4.
Usage of Users’ Databases for nucleating recommendations.
VI. Conclusion
The abundance of information available in the form of academic articles, reports, and studies can make it challenging for researchers to find relevant data and gain insights from the existing literature. To address this need, the DataDiscoveryLab software system has been developed to provide researchers with a comprehensive approach to analyzing academic articles.
Utilizing advanced computer vision algorithms and NLP techniques, the system can quickly parse articles into text and figures, creating three separate databases. These databases can be used to find similarities between users’ research questions and existing literature and can integrate users’ data from scientific software and devices. This creates a loop that can lead to the best answer and pathways for research, ultimately providing researchers with the most relevant articles to read.
The creation of these databases, along with the use of advanced data analysis techniques, provides researchers with a clear path forward for their research and helps them make informed decisions about their work. By enabling researchers to parse large volumes of data and find relevant information quickly, DataDiscoveryLab significantly enhances the research process and can lead to more impactful research outcomes.
In general, the integration of article mining in the DataDiscoveryLab software system provides researchers with a powerful tool for navigating the vast amount of scientific literature available today. With the ability to efficiently analyze academic articles, this system has the potential to revolutionize the way researchers approach their work and ultimately contribute to more meaningful scientific advancements.
Appendix A. Methods
Appendix A.1. Creating Text-Based Article Database
To create a comprehensive article database, the DataDiscoveryLab software system utilizes IBM’s DeepSearch to retrieve text-based information such as references, abstracts, introductions, methods, and results. This information is then parsed and stored in the appropriate sub-databases. Additionally, web scraping techniques are used to retrieve titles and affiliations.
DeepSearch has been previously used in various publications such as [
30,
31,
32], and [
33]. These publications demonstrate the effectiveness of DeepSearch in extracting and analyzing text-based data.
The article database and sub-databases allow researchers to quickly identify relevant articles and gain a deeper understanding of the research presented in those articles. By utilizing DeepSearch and other techniques for data retrieval, the DataDiscoveryLab software system provides a comprehensive approach to analyzing academic articles.
After analyzing articles to parse them into text-based versions, the system uses natural language processing (NLP) methods to clean and then embed them with tools like OpenAI text-embedding-ada-002[
15]. This ensures that the text-based information is accurate and easily searchable.
Appendix A.2. Creating Figure-Based Article Database
To create a comprehensive article database, the DataDiscoveryLab software system utilizes a range of tools and techniques for extracting and analyzing scientific figures. One such tool is Deepfigures-open, which is based on the article [
34]. Deepfigures-open has been widely used in other research projects, including [
35,
36,
37,
38,
39], and [
40].
The DataDiscoveryLab software system also leverages other resources to enrich the figure-based article database. For instance, the software uses EXSCLAIM, based on the article [
41], to extract labeled images from articles. Additionally, the system employs EXACT, based on the article [
42], to facilitate collaborative annotation of images. Other tools, such as ArtPop, based on the article [
43], and TEMExtraction, based on the article [
44], are also can be used to enhance the figure-based database. As well as the MedICaT dataset, based on the article [
44] which includes medical images, captions, and textual references, can be used in conjunction with other resources to enhance medical image analysis and natural language processing tasks. These tools will be to increase the quality of classification process for creating Knowledge-based Graphs, which is one of our plans for the future to increase this system’s capabilities.
The DataDiscoveryLab software system integrates these various tools and techniques into a single framework called Look, Read, and Enrich, which is based on the article [
14]. This framework allows researchers to quickly search, browse, and explore the figure-based database to gain insights into the research presented in the articles by using graph-based knowledge which would be helpful for us to get to the Generative Artificial Intelligence level. In this context, constructing the data and prompts retrievals from researchers and their systems using sequence-to-graph models like [
45,
46], and many others for processing data and transformers models like [
20,
47], and [
7] for specialized pseudo-knowledge-based-graph sentiment analysis can increase this system’s abilities on generating new research possibilities.
Overall, the creation of the figure-based article database, along with the text-based and sub-databases, will provide researchers with a powerful tool for exploring and analyzing academic articles.
References
- C. Angermueller, T. Pärnamaa, L. Parts, and O. Stegle, “Deep learning for computational biology,” Molecular Systems Biology, vol. 12, no. 7, p. 878, July 2016. [Online]. Available: https://doi.org/10.15252/msb.20156651. [CrossRef]
- F. Jiang, Y. Jiang, H. Zhi, Y. Dong, H. Li, S. Ma, Y. Wang, Q. Dong, H. Shen, and Y. Wang, “Artificial intelligence in healthcare: past, present and future,” Stroke and Vascular Neurology, vol. 2, no. 4, pp. 230–243, June 2017. [Online]. Available: https://doi.org/10.1136/svn-2017-000101. [CrossRef]
- I. Kononenko, “Machine learning for medical diagnosis: history, state of the art and perspective,” Artificial Intelligence in Medicine, vol. 23, no. 1, pp. 89–109, 2001. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S093336570100077X. [CrossRef]
- R. Roscher, B. Bohn, M. F. Duarte, and J. Garcke, “Explainable machine learning for scientific insights and discoveries,” IEEE Access, vol. 8, pp. 42 200–42 216, 2020. [Online]. Available: https://doi.org/10.1109/access.2020.2976199. [CrossRef]
- W. Li, T. Yang, C. Liu, Y. Huang, C. Chen, H. Pan, G. Xie, H. Tai, Y. Jiang, Y. Wu, Z. Kang, L.-Q. Chen, Y. Su, and Z. Hong, “Optimizing piezoelectric nanocomposites by high-throughput phase-field simulation and machine learning,” Advanced Science, vol. 9, no. 13, p. 2105550, Mar. 2022. [Online]. Available: https://doi.org/10.1002/advs.202105550. [CrossRef]
- Y. N. Yohei Nakajima, “Yoheinakajima/babyagi.” [Online]. Available: https://github.com/yoheinakajima/babyagi.
- Significant-Gravitas, “Significant-gravitas/auto-gpt: An experimental open-source attempt to make gpt-4 fully autonomous.” [Online]. Available: https://github.com/Significant-Gravitas/Auto-GPT.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. Kaiser, and I. Polosukhin, “Attention Is All You Need,” arXiv e-prints, p. arXiv:1706.03762, June 2017. [CrossRef]
- J. Devlin, M.-W. Chang, K. Lee, and K. Toutanova, “BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding,” arXiv e-prints, p. arXiv:1810.04805, Oct. 2018. [CrossRef]
- A. Krizhevsky, I. Sutskever, and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems, F. Pereira, C. Burges, L. Bottou, and K. Weinberger, Eds., vol. 25. Curran Associates, Inc., 2012. [Online]. Available: https://proceedings.neurips.cc/paper_files/paper/2012/file/c399862d3b9d6b76c8436e924a68c45b-Paper.pdf.
- J. Redmon and A. Farhadi, “Yolov3: An incremental improvement,” arXiv preprint arXiv:1804.02767, 2018. [CrossRef]
- I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, and Y. Bengio, “Generative Adversarial Networks,” arXiv e-prints, p. arXiv:1406.2661, June 2014.
- M. Fire and C. Guestrin, “Over-optimization of academic publishing metrics: observing goodhart’s law in action,” GigaScience, vol. 8, no. 6, May 2019. [Online]. Available: https://doi.org/10.1093/gigascience/giz053. [CrossRef]
- J. M. Gomez-Perez and R. Ortega, “Look, read and enrich. learning from scientific figures and their captions,” arXiv e-prints, vol. arXiv:1909.09070, 2019. [Online]. Available: https://ui.adsabs.harvard.edu/abs/2019arXiv190909070G. [CrossRef]
- R. Greene, T. Sanders, L. Weng, and A. Neelakantan, “New and improved embedding model,” Retrieved from https://openai.com/blog/new-and-improved-embedding-model, 2022.
- D. Kozlowski, J. Dusdal, J. Pang, and A. Zilian, “Semantic and relational spaces in science of science: Deep learning models for article vectorisation,” Scientometrics, vol. 126, no. 7, p. 5881–5910, 2021. [CrossRef]
- E. A. van Dis, J. Bollen, W. Zuidema, R. van Rooij, and C. L. Bockting, “Chatgpt: five priorities for research,” Nature, vol. 614, no. 7947, pp. 224–226, 2023. [CrossRef]
- B. Gordijn and H. t. Have, “Chatgpt: evolution or revolution?” Medicine, Health Care and Philosophy, pp. 1–2, 2023. [CrossRef]
- K. Singhal, S. Azizi, T. Tu, S. S. Mahdavi, J. Wei, H. W. Chung, N. Scales, A. Tanwani, H. Cole-Lewis, S. Pfohl, P. Payne, M. Seneviratne, P. Gamble, C. Kelly, N. Scharli, A. Chowdhery, P. Mansfield, B. Aguera y Arcas, D. Webster, G. S. Corrado, Y. Matias, K. Chou, J. Gottweis, N. Tomasev, Y. Liu, A. Rajkomar, J. Barral, C. Semturs, A. Karthikesalingam, and V. Natarajan, “Large Language Models Encode Clinical Knowledge,” arXiv e-prints, p. arXiv:2212.13138, Dec. 2022. [CrossRef]
- OpenAI, “Gpt-4 technical report,” 2023. [CrossRef]
- M. Reichstein, G. Camps-Valls, B. Stevens, M. Jung, J. Denzler, and N. Carvalhais, “Deep learning and process understanding for data-driven earth system science,” Nature, vol. 566, no. 7743, pp. 195–204, 2019. [CrossRef]
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev, and A. Walsh, “Machine learning for molecular and materials science,” Nature, vol. 559, no. 7715, pp. 547–555, 2018. [CrossRef]
- T. Hey, K. Butler, S. Jackson, and J. Thiyagalingam, “Machine learning and big scientific data,” Philosophical Transactions of the Royal Society A, vol. 378, no. 2166, p. 20190054, 2020. [CrossRef]
- F. J. Sigworth, “A maximum-likelihood approach to single-particle image refinement,” Journal of structural biology, vol. 122, no. 3, pp. 328–339, 1998. [CrossRef]
- Q. Chen and V. Koltun, “Photographic Image Synthesis with Cascaded Refinement Networks,” arXiv e-prints, p. arXiv:1707.09405, July 2017. [CrossRef]
- C. Wang, F. Jing, L. Zhang, and H.-J. Zhang, “Content-based image annotation refinement,” in 2007 IEEE Conference on Computer Vision and Pattern Recognition, 2007, pp. 1–8. [CrossRef]
- R. Kraft and J. Zien, “Mining anchor text for query refinement,” in Proceedings of the 13th international conference on World Wide Web, 2004, pp. 666–674. [CrossRef]
- M. Zhu, P. Pan, W. Chen, and Y. Yang, “Dm-gan: Dynamic memory generative adversarial networks for text-to-image synthesis,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), June 2019. [CrossRef]
- N. Shinn, B. Labash, and A. Gopinath, “Reflexion: an autonomous agent with dynamic memory and self-reflection,” arXiv preprint arXiv:2303.11366, 2023. [CrossRef]
- C. Lin, P.-H. Wang, Y. Hsiao, Y.-T. Chan, A. C. Engler, J. W. Pitera, D. P. Sanders, J. Cheng, and Y. J. Tseng, “Essential step toward mining big polymer data: Polyname2structure, mapping polymer names to structures,” ACS Applied Polymer Materials, vol. 2, no. 8, pp. 3107–3113, 2020. [CrossRef]
- M. Manica, C. Auer, V. Weber, F. Zipoli, M. Dolfi, P. Staar, T. Laino, C. Bekas, A. Fujita, H. Toda, S. Hirose, and Y. Orii, “An information extraction and knowledge graph platform for accelerating biochemical discoveries,” arXiv e-prints, p. arXiv:1907.08400, 2019. [CrossRef]
- P. L. Dognin, I. Melnyk, I. Padhi, C. Nogueira dos Santos, and P. Das, “DualTKB: A dual learning bridge between text and knowledge base,” arXiv e-prints, p. arXiv:2010.14660, 2020. [CrossRef]
- P. W. Staar, M. Dolfi, C. Auer, and C. Bekas, “Corpus conversion service,” in Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 2018. [CrossRef]
- N. Siegel, N. Lourie, R. Power, and W. Ammar, “Extracting scientific figures with distantly supervised neural networks,” arXiv e-prints, p. arXiv:1804.02445, 2018. [CrossRef]
- W. Ammar, D. Groeneveld, C. Bhagavatula, I. Beltagy, M. Crawford, D. Downey, J. Dunkelberger, A. Elgohary, S. Feldman, V. Ha, R. Kinney, S. Kohlmeier, K. Lo, T. Murray, H.-H. Ooi, M. Peters, J. Power, S. Skjonsberg, L. L. Wang, C. Wilhelm, Z. Yuan, M. van Zuylen, and O. Etzioni, “Construction of the literature graph in semantic scholar,” arXiv e-prints, May 2018. [CrossRef]
- J. Bhatt, K. A. A. Hashmi, M. Z. Afzal, and D. Stricker, “A survey of graphical page object detection with deep neural networks,” Applied Sciences, vol. 11, no. 12, p. 5344, 2021. [CrossRef]
- Q. Wang, M. Li, X. Wang, N. Parulian, G. Han, J. Ma, J. Tu, Y. Lin, H. Zhang, W. Liu, A. Chauhan, Y. Guan, B. Li, R. Li, X. Song, Y. R. Fung, H. Ji, J. Han, S.-F. Chang, J. Pustejovsky, J. Rah, D. Liem, A. Elsayed, M. Palmer, C. Voss, C. Schneider, and B. Onyshkevych, “Covid-19 literature knowledge graph construction and drug repurposing report generation,” arXiv e-prints, Jul 2020. [CrossRef]
- M. Li, L. Cui, S. Huang, F. Wei, M. Zhou, and Z. Li, “Tablebank: A benchmark dataset for table detection and recognition,” arXiv e-prints, 2019.
- X. Zhong, E. ShafieiBavani, and A. J. Yepes, “Image-based table recognition: data, model, and evaluation,” arXiv e-prints, 2019.
- M. Li, Y. Xu, L. Cui, S. Huang, F. Wei, Z. Li, and M. Zhou, “Docbank: A benchmark dataset for document layout analysis,” arXiv e-prints, 2020. [Online]. Available: https://ui.adsabs.harvard.edu/abs/2020arXiv200601038L. [CrossRef]
- E. Schwenker, W. Jiang, T. Spreadbury, N. Ferrier, O. Cossairt, and M. K. Y. Chan, “Exsclaim!–an automated pipeline for the construction of labeled materials imaging datasets from literature,” arXiv e-prints, vol. arXiv:2103.10631, 2021. [CrossRef]
- C. Marzahl, M. Aubreville, C. A. Bertram, J. Maier, C. Bergler, C. Kröger, J. Voigt, K. Breininger, R. Klopfleisch, and A. Maier, “Exact: a collaboration toolset for algorithm-aided annotation of images with annotation version control,” Scientific Reports, vol. 11, p. 4343, 2021. [CrossRef]
- J. P. Greco and S. Danieli, “Artpop: A stellar population and image simulation python package,” The Astrophysical Journal, vol. 941, no. 1, p. 26, 2022. [CrossRef]
- S. Subramanian, L. L. Wang, S. Mehta, B. Bogin, M. van Zuylen, S. Parasa, S. Singh, M. Gardner, and H. Hajishirzi, “Medicat: A dataset of medical images, captions, and textual references,” arXiv e-prints, vol. arXiv:2010.06000, 2020. [Online]. Available: https://doi.org/10.48550/arXiv.2010.06000. [CrossRef]
- W. Hu, Y. Yang, Z. Cheng, C. Yang, and X. Ren, “Time-series event prediction with evolutionary state graph,” 2020. [CrossRef]
- X. Zhang, M. Zeman, T. Tsiligkaridis, and M. Zitnik, “Graph-guided network for irregularly sampled multivariate time series,” 2022. [CrossRef]
- Y. Anand, Z. Nussbaum, B. Duderstadt, B. Schmidt, and A. Mulyar, “Gpt4all: Training an assistant-style chatbot with large scale data distillation from gpt-3.5-turbo,” https://github.com/nomic-ai/gpt4all, 2023.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).