Submitted:
26 April 2023
Posted:
27 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Leishmania Isolates
2.1. Parasite Culture and DNA Extraction
2.2. Molecular Identification of Leishmania panamensis Isolates
2.2.1. PCR Amplification
2.2.2. Sanger Sequencing and Species Discrimination by Phylogenetic Analysis
2.3. PCR Amplification and Sequencing of Gene Loci used in The MLST Approach
2.4. Sequence Assembling, Haplotype Construction, and Determination of Diversity Indexes
2.5. Determination of L. panamensis DSTs and Their Geographical Distribution
2.6. Phylogenetic Analyzes of L. panamensis DSTs
2.7. Clonal Complex Determination
2.8. Global Ancestry of Local Leishmania panamensis Isolates
3. Results
3.1. Identification of Leishmania panamensis Isolates
3.2. Genetic Diversity of L. panamensis Haplotypes
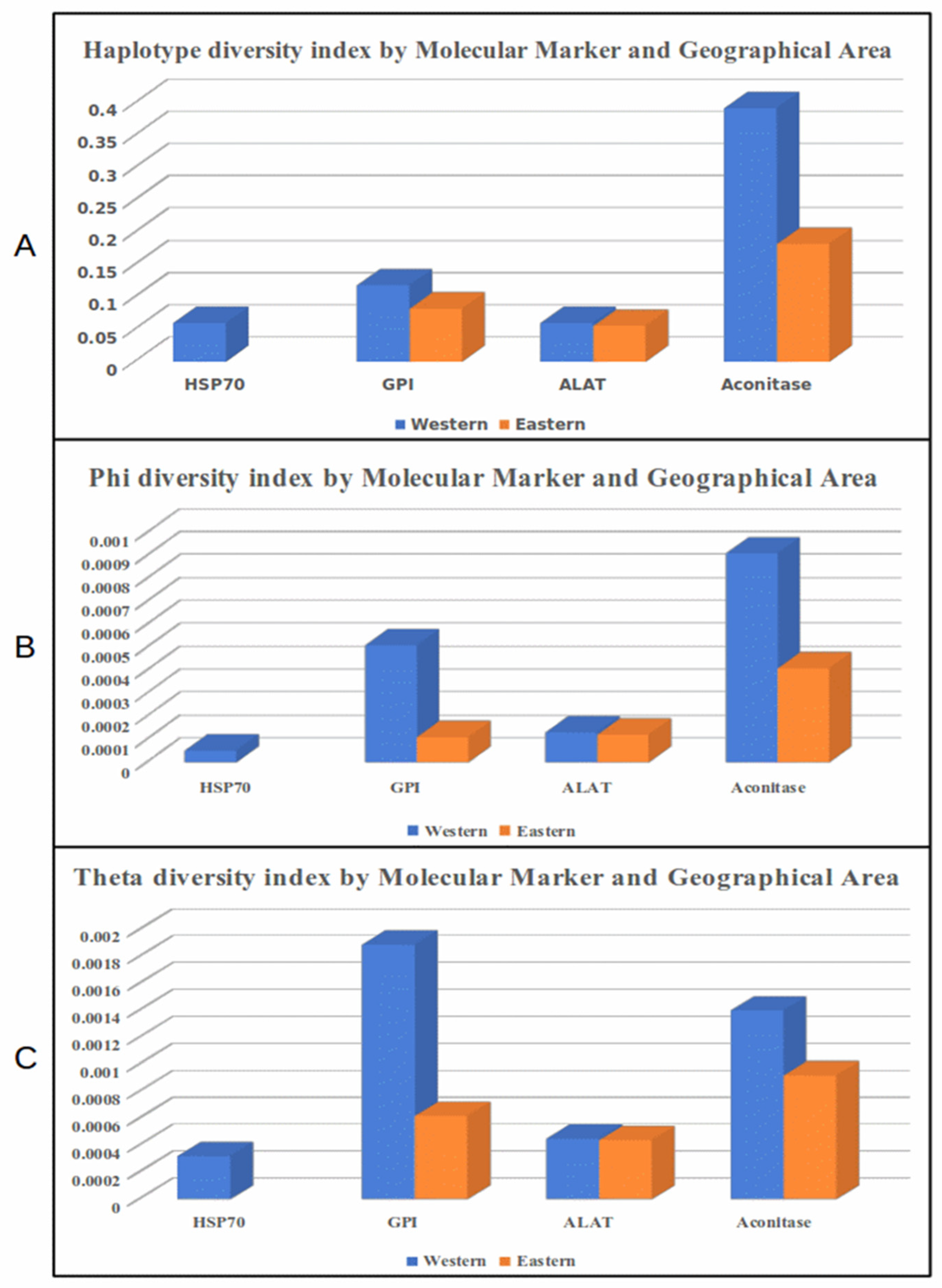
3.3. Diversity Indexes by Locus and Geographical Region
3.4. Genotyping Approach by MLST
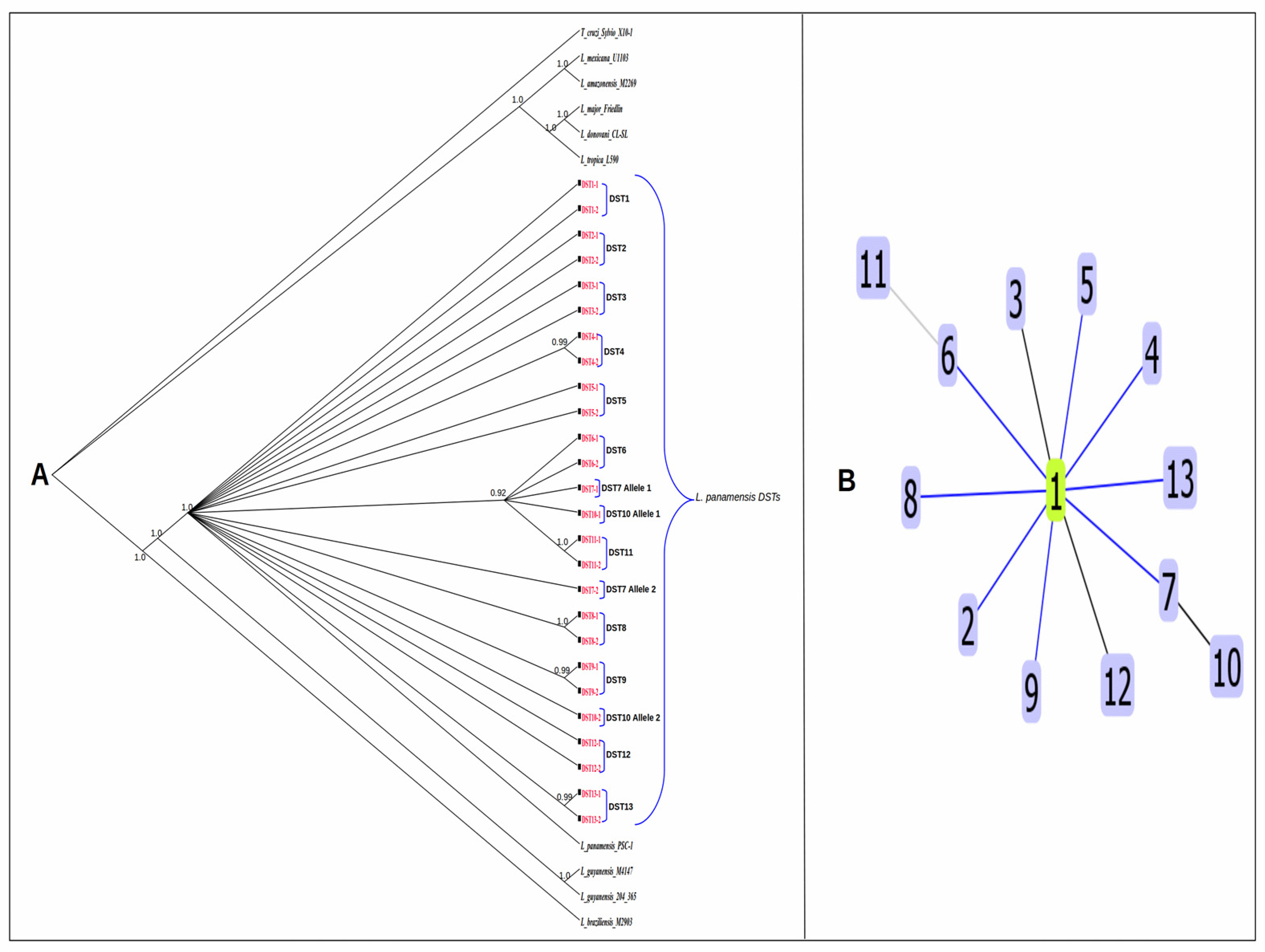
3.5. Geographic Distribution of DSTs
3.6. Haplotype Resolution of local L. panamensis Isolates
3.7. Phylogenetic Analysis and Clonal Complex Determination of L. panamensis DSTs
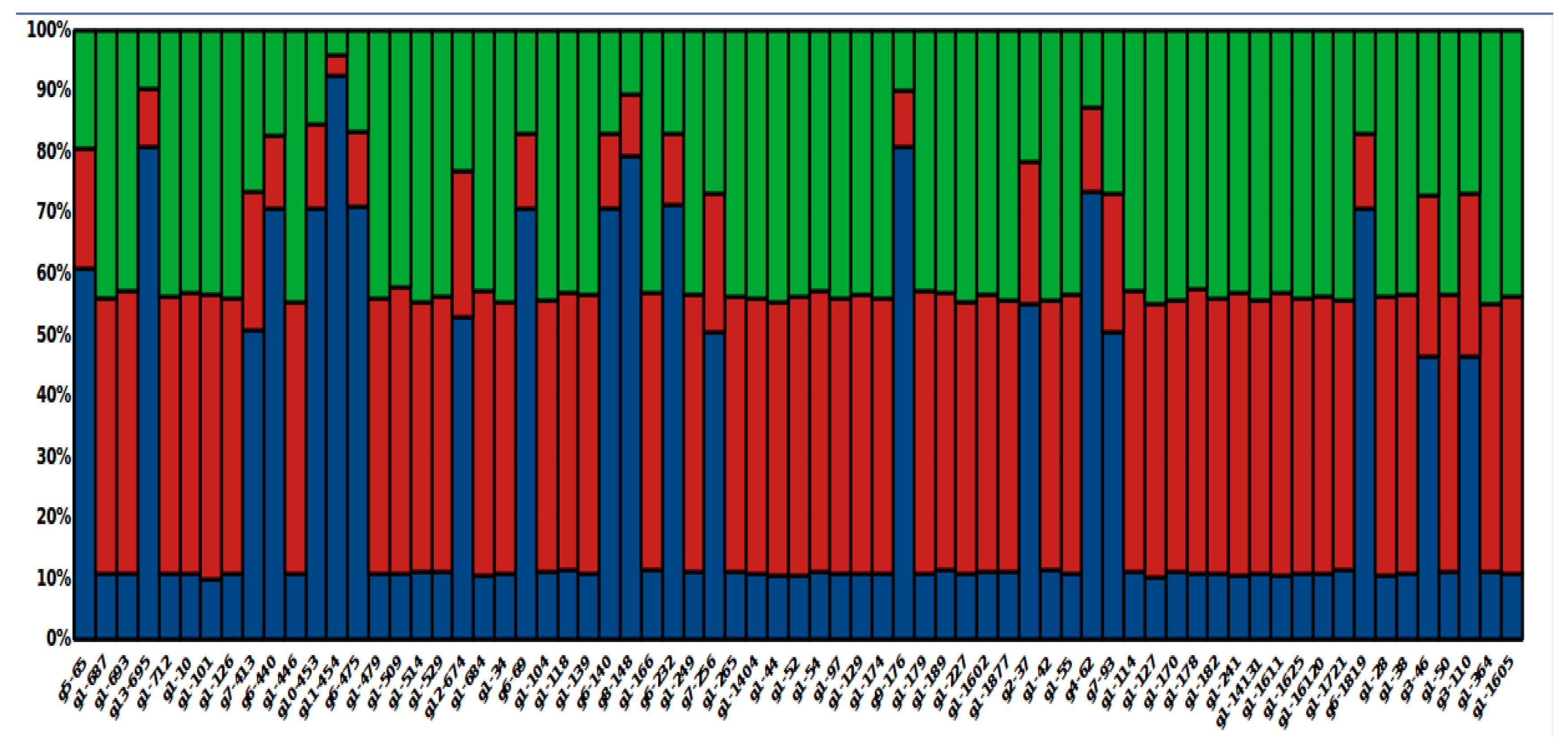
3.8. Global Ancestry of Local L. panamensis Isolates
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Moura, T.R.; Oliveira, F.; Novais, F.O.; Miranda, J.C.; Clarêncio, J.; Follador, I.; Carvalho, E.M.; Valenzuela, J.G.; Barral-Netto, M.; Barral, A.; et al. Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl. Trop. Dis. 2007, 1, 1–10. [CrossRef]
- Vanloubbeeck, Y.; Jones, D.E. The immunology of Leishmania infection and the implications for vaccine development. In Proceedings of the Annals of the New York Academy of Sciences; New York Academy of Sciences, 2004; Vol. 1026, pp. 267–272. [CrossRef]
- Pan American Health Organization. Leishmaniasis: Epidemiological Report in the Americas.; Washington, D.C, 2019; pp 2-3.
- Alvar, J.; Vélez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; de Boer, M. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [CrossRef]
- Schönian, G.; Mauricio, I.; Cupolillo, E. Is it time to revise the nomenclature of Leishmania? Trends Parasitol. 2010, 26, 466–469. [CrossRef]
- Christensen, H.A.; De Vasquez, A.M.; Petersen, J.L. Short report: Epidemiologic studies on cutaneous leishmaniasis in eastern Panama. Am. J. Trop. Med. Hyg. 1999, 60, 54–57. [CrossRef]
- Miranda, A.; Carrasco, R.; Paz, H.; Pascale, J.M.; Samudio, F.; Saldaña, A.; Santamaría, G.; Mendoza, Y.; Calzada, J.E. Molecular epidemiology of American tegumentary leishmaniasis in Panama. Am. J. Trop. Med. Hyg. 2009, 81, 565–571. [CrossRef]
- Del Miranda, A.C.; González, K.A.; Samudio, F.; Pineda, V.J.; Calzada, J.E.; Capitan-Barrios, Z.; Jiménez, A.; Castillo, J.; Mendoza, Y.; Suárez, J.A.; et al. Molecular identification of parasites causing cutaneous leishmaniasis in Panama. Am. J. Trop. Med. Hyg. 2021, 104, 1326–1334. [CrossRef]
- Vasquez, A.N.A.M.D.E.; Saenz, R.E.; Petersen, J.L.; Johnson, C.M. Leishmania Mexicana complex: Human infections in the Republic of Panama. Am. J. Trop. Med. Hyg. 1990, 43, 619–622. [CrossRef]
- Miranda, A.; Samudio, F.; Saldaña, A.; Castillo, J.; Brandão, A.; Calzada, J.E. The calmodulin intergenic spacer as molecular target for characterization of Leishmania species. Parasites and Vectors 2014, 7, 1–9. [CrossRef]
- Miranda, A.; Samudio, F.; González, K.; Saldaña, A.; Brandão, A.; Calzada, J.E. Calmodulin polymerase chain reaction-restriction fragment length polymorphism for leishmania identification and typing. Am. J. Trop. Med. Hyg. 2016, 95, 383–387. [CrossRef]
- Davila, M.; Pineda, V.; Calzada, J.E.; Saldaña, A.; Samudio, F. Evaluation of cytochrome b sequence to identify Leishmania species and variants: The case of Panama. Mem. Inst. Oswaldo Cruz 2021, 116, 1–10. [CrossRef]
- Miranda, A.; Carrasco, R.; Paz, H.; Pascale, J.M.; Samudio, F.; Saldaña, A.; Santamaría, G.; Mendoza, Y.; Calzada, J.E. Molecular epidemiology of American tegumentary leishmaniasis in Panama. Am. J. Trop. Med. Hyg. 2009, 81, 565–571. [CrossRef]
- Hashiguchi, Y.; Gomez, E.L.; Kato, H.; Martini, L.R.; Velez, L.N.; Uezato, H. Diffuse and disseminated cutaneous leishmaniasis: Clinical cases experienced in Ecuador and a brief review. Trop. Med. Health 2016, 44, 1–9. [CrossRef]
- Osorio, L.E.; Castillo, C.M.; Ochoa, M.T. Mucosal Leishmaniasis due to Leishmania (Viannia) panamensis in Colombia: Clinical characteristics. Am. J. Trop. Med. Hyg. 1998, 59, 49–52. [CrossRef]
- Freites, C.O.; Gundacker, N.D.; Pascale, J.M.; Saldaña, A.; Diaz-Suarez, R.; Jimenez, G.; Sosa, N.; García, E.; Jimenez, A.; Suarez, J.A. First case of diffuse leishmaniasis associated with leishmania panamensis. Open Forum Infect. Dis. 2018, 5, 1–3. [CrossRef]
- Grimaldi, G.; Tesh, R.B.; McMahon-Pratt, D. A review of the geographic distribution and epidemiology of leishmaniasis in the New World. Am. J. Trop. Med. Hyg. 1989, 41, 687–725. [CrossRef]
- Mauricio, I.L.; Yeo, M.; Baghaei, M.; Doto, D.; Pratlong, F.; Zemanova, E.; Dedet, J.P.; Lukes, J.; Miles, M.A. Towards multilocus sequence typing of the Leishmania donovani complex: Resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int. J. Parasitol. 2006, 36, 757–769. [CrossRef]
- Zemanová, E.; Jirků, M.; Mauricio, I.L.; Horák, A.; Miles, M.A.; Lukeš, J. The Leishmania donovani complex: Genotypes of five metabolic enzymes (ICD, ME, MPI, G6PDH, and FH), new targets for multilocus sequence typing. Int. J. Parasitol. 2007, 37, 149–160. [CrossRef]
- Boité, M.C.; Mauricio, I.L.; Miles, M.A.; Cupolillo, E. New Insights on Taxonomy, Phylogeny and Population Genetics of Leishmania (Viannia) Parasites Based on Multilocus Sequence Analysis. PLoS Negl. Trop. Dis. 2012, 6, 1–14. [CrossRef]
- Marco, J.D.; Barroso, P.A.; Locatelli, F.M.; Cajal, S.P.; Hoyos, C.L.; Nevot, M.C.; Lauthier, J.J.; Tomasini, N.; Juarez, M.; Estévez, J.O.; et al. Multilocus sequence typing approach for a broader range of species of Leishmania genus: Describing parasite diversity in Argentina. Infect. Genet. Evol. 2015, 30, 308–317. [CrossRef]
- Lauthier, J.J.; Ruybal, P.; Barroso, P.A.; Hashiguchi, Y.; Marco, J.D.; Korenaga, M. Development of a Multilocus sequence typing (MLST) scheme for Pan-Leishmania. Acta Trop. 2020, 201, 1–13. [CrossRef]
- Restrepo, C.M.; Llanes, A.; De La Guardia, C.; Lleonart, R. Genome-wide discovery and development of polymorphic microsatellites from Leishmania panamensis parasites circulating in central Panama. Parasit. Vectors 2015, 8, 527. [CrossRef]
- Yamada, K.; Valderrama, A.; Gottdenker, N.; Cerezo, L.; Minakawa, N.; Saldaña, A.; Calzada, J.E.; Chaves, L.F. Macroecological patterns of American Cutaneous Leishmaniasis transmission across the health areas of Panamá (1980–2012). Parasite Epidemiol. Control 2016, 1, 42–55. [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [CrossRef]
- Herrera, G.; Hernández, C.; Ayala, M.S.; Flórez, C.; Teherán, A.A.; Ramírez, J.D. Evaluation of a Multilocus Sequence Typing (MLST) scheme for Leishmania (Viannia) braziliensis and Leishmania (Viannia) panamensis in Colombia. Parasit. Vectors 2017, 10, 236. [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Varlamov, A.; Vaskin, Y.; Efremov, I.; German Grehov, O.G.; Kandrov, D.; Rasputin, K.; Syabro, M.; et al. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [CrossRef]
- Rozas, J.; Rozas, R. DnaSP, DNA sequence polymorphism: An interactive program for estimating population genetics parameters from DNA sequence data. Comput. Appl. Biosci. 1995, 11, 621–625. [CrossRef]
- Tomasini, N.; Lauthier, J.J.; Llewellyn, M.S.; Diosque, P. MLSTest: Novel software for multi-locus sequence data analysis in eukaryotic organisms. Infect. Genet. Evol. 2013, 20, 188–196. [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. Sea view version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [CrossRef]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [CrossRef]
- Veland, N.; Boggild, A.K.; Valencia, C.; Valencia, B.M.; Llanos-Cuentas, A.; Van Der Auwera, G.; Dujardin, J.C.; Arevalo, J. Leishmania (Viannia) species identification on clinical samples from cutaneous leishmaniasis patients in Peru: Assessment of a molecular stepwise approach. J. Clin. Microbiol. 2012, 50, 495–498. [CrossRef]
- Santamaría, A.M.; Vásquez, V.; Rigg, C.; Samudio, F.; Moreno, D.; Romero, L.; Saldaña, A.; Chaves, L.F.; Calzada, J.E. Plasmodium vivax genetic diversity in Panama: Challenges for malaria elimination in Mesoamerica. Pathogens 2021, 10, 1–18. [CrossRef]
- Instituto Geográfico Nacional “Tommy Guardia” Atlas Nacional de la República de Panamá; 5th edition.; Instituto Tommy Guardia: Panama, 2016; ISBN 978-9962-11-048-4.
- Chaves, L.F.; Calzada, J.E.; Valderrama, A.; Saldaña, A. Cutaneous Leishmaniasis and Sand Fly Fluctuations Are Associated with El Niño in Panamá. PLoS Negl. Trop. Dis. 2014, 8, 1–11. [CrossRef]
- Ready, P.D. Leishmaniasis emergence and climate change. OIE Rev. Sci. Tech. 2008, 27, 399–412. [CrossRef]
- Hlavacova, J.; Votypka, J.; Volf, P. The effect of temperature on Leishmania (Kinetoplastida: Trypanosomatidae) development in sand flies. J. Med. Entomol. 2013, 50, 955–958. [CrossRef]
- Llanes, A.; Cruz, G.; Morán, M.; Vega, C.; Pineda, V.J.; Ríos, M.; Penagos, H.; Suárez, J.A.; Saldaña, A.; Lleonart, R.; et al. Genomic diversity and genetic variation of Leishmania panamensis within its endemic range. Infect. Genet. Evol. 2022, 103, 1–11. [CrossRef]
- Valderrama, A.; Tavares, M.G.; Filho, J.D.A. Phylogeography of the Lutzomyia gomezi (Diptera: Phlebotominae) on the Panama Isthmus. Parasites and Vectors 2014, 7, 1–11. [CrossRef]
- Rougeron, V.; Bañuls, A.L.; Carme, B.; Simon, S.; Couppié, P.; Nacher, M.; Hide, M.; De Meeûs, T. Reproductive strategies and population structure in Leishmania: Substantial amount of sex in Leishmania Viannia guyanensis. Mol. Ecol. 2011, 20, 3116–3127. [CrossRef]
- Rougeron, V.; De Meeûs, T.; Hide, M.; Waleckx, E.; Bermudez, H.; Arevalo, J.; Llanos-Cuentas, A.; Dujardin, J.C.; De Doncker, S.; Le Ray, D.; et al. Extreme inbreeding in Leishmania braziliensis. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 10224–10229. [CrossRef]
- Herrer, A.; Christensen, H.A. Epidemiological patterns of cutaneous leishmaniasis in Panama. III. Endemic persistence of the disease. Am. J. Trop. Med. Hyg. 1976, 25, 54–58. [CrossRef]
- González, K.; Calzada, J.E.; Saldaña, A.; Rigg, C.A.; Alvarado, G.; Rodríguez-Herrera, B.; Kitron, U.D.; Adler, G.H.; Gottdenker, N.L.; Chaves, L.F.; et al. Survey of wild mammal hosts of cutaneous leishmaniasis parasites in Panamá and Costa Rica. Trop. Med. Health 2015, 43, 75–8. [CrossRef]
- Dutari, L.C.; Loaiza, J.R. American Cutaneous Leishmaniasis in Panama: A historical review of entomological studies on anthropophilic Lutzomyia sand fly species. Parasit. Vectors 2014, 7, 218. [CrossRef]
- Christensen, H.A.; De Vasquez, A.M. The tree-buttress biotope: A pathobiocenose of Leishmania braziliensis. Am. J. Trop. Med. Hyg. 1982, 31, 243–251. [CrossRef]
- Herrer, A.; Christensen, H.A.; Beumer, R.J. Reservoir hosts of cutaneous leishmaniasis among Panamanian forest mammals. Am. J. Trop. Med. Hyg. 1973, 22, 585–591. [CrossRef]
- Valderrama, A.; Tavares, M.G.; Filho, J.D.A. Anthropogenic influence on the distribution, abundance, and diversity of sandfly species (Diptera: Phlebotominae: Psychodidae), vectors of cutaneous leishmaniasis in Panama. Mem. Inst. Oswaldo Cruz 2011, 106, 1024–1031. [CrossRef]
- PAHO. Manual de Procedimientos para Vigilancia y Control de Las Leishmaniasis en las Américas; Organización Panamericana de la Salud: Washington, D.C, 2019; ISBN 978-92-75-32063-1. [CrossRef]
- Myers, N.; R. A. Mittermeier; C. G. Mittermeier; G. A. B. da Fonseca and J. Kent. Biodiversity hotspots for conservation priorities. Nature. 2000, 403:853-858. [CrossRef]
- García-Moreno, J., A. G. Navarro-Sigüenza, A. T. Peterson, and L. A. Sánchez-González. Genetic variation coincides with geographic structure in the common bush-tanager (Chlorospingus opthalmicus) complex from Mexico. Mol. Phylogenet. Evol 2004, 206, 33, 186-196. [CrossRef]
- Patino L.H.; Muñoz M.; Cruz-Saavedra L.; Muskus, C.; Ramírez J.D. Genomic Diversification, Structural Plasticity, and Hybridization in Leishmania (Viannia) braziliensis. Front. Cell. Infect. Microbiol. 2020, 10, 582196. [CrossRef]
- Patiño L.H.; Muñoz M.; Pavia P.; Muskus C.; Shaban M.; Paniz-Mondolfi A.; Ramírez J.D. Filling the gaps in Leishmania naiffi and Leishmania guyanensis genome plasticity. G3, 2022, 12, 1, jkab377. [CrossRef]

| Target gene | Enzyme entry | Gene length (bp) | Chromosomal location | Amplicon size (bp) | Primers sequences | Reference |
|---|---|---|---|---|---|---|
| Heat Shock Protein 70 | AAG01344.1 | 2,566 | 28 | 1364 | PLeishF: GATGGTGCTGCTGAAGATGA PLeishR: GGTCATGATCGGGTTGCATR |
This study |
| Glucose-6-phosphate isomerase | EC 5.3.1.9 | 2,084 | 12 | 1,745 | GPIextF: AAT GTT CTT CAT ACC CCT CT GPIextR: TTC CGT CCG TCT CCT G GPIintF: TGG GAT TGG CGG CAG CGA CCTT GPIintR: CGC CAC AGG TAC TGG TCG T |
[37] |
| Alanine aminotransferase | EC 2.6.1.21 | 1,493 | 12 | 589 | ALAT.F: GTGTGCATCAACCCMGGGAA ALAT.R: CGTTCAGCTCCTCGTTCCGC |
[21] |
| Aconitase | EC 4.2.1.3 | 2,690 | 18 | 579 | ACO.F: CAAGTTCCTGRCGTCTCTGC ACO.R: GAGTCCGGGTATAGCAKCCC |
[21] |
| Loci | s | h | hd | π | ϴ |
|---|---|---|---|---|---|
| HSP70 | 2 | 3 | 0.029 | 0.00002 | 0.00028 |
| GPI | 12 | 6 | 0.099 | 0.00030 | 0.00217 |
| ALAT | 2 | 3 | 0.057 | 0.00012 | 0.00077 |
| Aconitase | 4 | 5 | 0.290 | 0.00067 | 0.00162 |
| Western N=34 |
Eastern N=35 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | s | h | hd | π | ϴ | s | h | hd | π | ϴ |
| HSP70 | 2 | 3 | 0.060 | 0.00005 | 0.00032 | 0 | 1 | 0 | 0 | 0 |
| GPI | 9 | 4 | 0.118 | 0.00051 | 0.00189 | 3 | 3 | 0.082 | 0.00011 | 0.00062 |
| ALAT | 1 | 2 | 0.060 | 0.00013 | 0.00045 | 1 | 2 | 0.055 | 0.00012 | 0.00044 |
| Aconitase | 3 | 4 | 0.392 | 0.00091 | 0.00140 | 2 | 3 | 0.182 | 0.00041 | 0.00092 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





