1. Introduction
Globally, the estimated area of harvested beans in 2021 was 35.92 mil. hectares, with another 2.72 mil. hectares dedicated to broad (horse) bean cultivation. Total production of beans was 33.68 mil. tons [
1]. The genus
Phaseolus includes approximately 80 cultivated and wild species, among which the most widespread and important species is
Phaseolus vulgaris L. [
2]. The common bean is susceptible to many diseases and pests, depending on its geographic location, growing conditions, nutrition, and protection. Diseases and pests may cause significant yield losses, a reduction in germination, and a reduction in bean quality. The most important pathogens of common beans also include viruses, among them the Bean common mosaic virus (BCMV) and Bean golden yellow mosaic virus (BGYMV) [
3], both from the genus
Potyvirus. These viruses belong to the three groups of viruses that cause devastating diseases and the highest losses in the yield and quality of a broad range of cultivated crops [
4]. BCMV is transmitted to host plants within the legume families through seeds and pollen, by aphid species, and by mechanical friction. It has a wide host range, including over 100 plants in 44 genera, and is one of the most widespread plant viruses infecting legumes under natural conditions [
5].
According to symptoms on beans, isolates of BCMV have been classified into seven [
6], and later eight [
7] pathotype groups. Meanwhile, based on their serological reactivity, BCMVs were divided into two serogroups, A and B [
8]. Later, based on peptide profile and nucleotide sequence analyses of coat protein (CP) and the 3'-end noncoding region (3'-UTR), the Potyviridae Study Group of the Plant Virus Subcommittee of ICTV [
9] accepted that the common bean mosaic necrosis virus (BCMNV) and BCMV were differentiated from serogroups A and B, respectively. The serological relationships among BCMV strains and other potyviruses tend to be ambiguous and complex. Rapid evolution in adapting to the environment, different hosts, and overcoming multiple resistance genes in host plants is typical for viruses, including the BCMV. Another source of emerging genetic variation are recombinations, which benefit viruses in overcoming host resistance and successful infection [
10,
11]. Many new strains or pathotypes do not behave like traditional BCMV strains and cannot be distinguished, even by the partial sequence of the CP and 3'-UTR of the virus genome [
12,
13]. Therefore, molecular characterization using whole-genome sequence analysis is currently used as a precise and reliable criterion for the differentiation of BCMV isolates and strains [
14]. Such detailed characterization allows the discovery of new BCMV pathotypes, their comparison with previously known ones, and the detection of mutations throughout the entire genome of the virus. In addition to the discovery of new virus pathotypes, new virus hosts are also being discovered. Reports about the first occurrences of BCMV appear continuously, either describing the virus occurrence in a specific country and region, or the discovery of infections caused by the virus in a previously undescribed host species. BCMV is also hosted by wild legumes [
15,
16,
17], growing in agro-ecological interfaces, which represent a potential reservoir of viral infection for cultivated crops in adjacent fields.
The objectives of this study were to: i) isolate and identify the virus from a crownvetch plant with symptoms of its presence; ii) confirm the presence of BCMV and perform genomic and proteomic analyses of the isolate; and iii) confirm a new, previously unreported BCMV host, the crownvetch.
2. Results
2.1. BCMV in Crownvetch
The plant of crownvetch (
Securigera varia L. Lassen, synonym
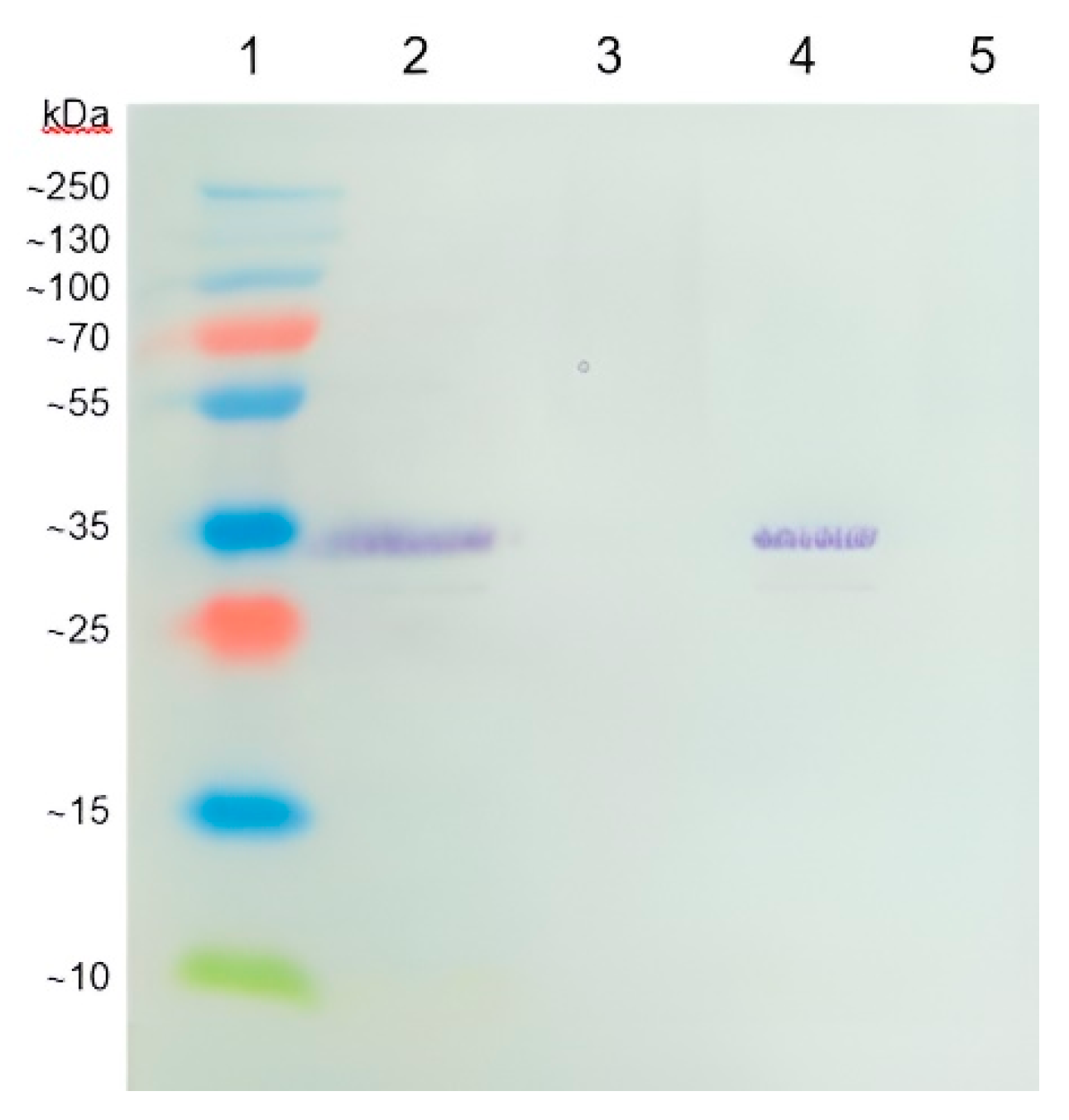
Coronilla varia L.) collected in the locality Čachtice (Slovakia) manifested symptoms of disease caused by virus. Plant was immunochemically analyzed and Western blotting analysis confirmed the presence of a specific BCMV-related protein from a positive BCMV control sample (
Figure 1).
The manufacturer of the polyclonal antibody does not declare the method of obtaining it. However, probably the oligonucleotide sequences from the capsid protein with antigenic potential could have been used for immunization of animals. The expected size of the BCMV capsid protein was calculated to be 32.12 kDa, corresponding to the size of the protein fragment in the gel. However, more important is the fact that the protein fragment from the BCMV positive control isolate BCMV PV 0915 was identical to the fragment present in the symptomatic crownvetch plant. The BCMV isolate from crownvetch was designated by the code BCMV SVK.
2.2. LC-MS/MS Detection of Viral Proteins
Proteins were extracted from young leaves of tobacco plants artificially infected by BCMV SVK, and the viral proteins were detected by a medium-resolution nano-Liquid Chromatography-Electrospray Ionization-Quadrupole-Time of Flight (nanoLC-ESI-Q-TOF). The total covered sequences were 251 and 199 amino acids (17 and 13 peptides), respectively. Peptides detected were from 3 out of 10 viral proteins: HC-Pro (up to 14.2% coverage i.e., 65 out of 457 amino acids), CI (up to 16.9% coverage i.e., 107 out of 634 amino acids), and CP (up to 27.5% coverage i.e., 79 out of 287 amino acids). LC-MS/MS analysis of the compared peptide fragments from the viral CP protein confirmed the identity between BCMV SVK and BCMV PV0915 as well as differences in two amino acids (
Figure 2). This directly confirmed the results from the proteomic in silico analysis, which are shown in
Table 1.
2.3. Genomic Analysis
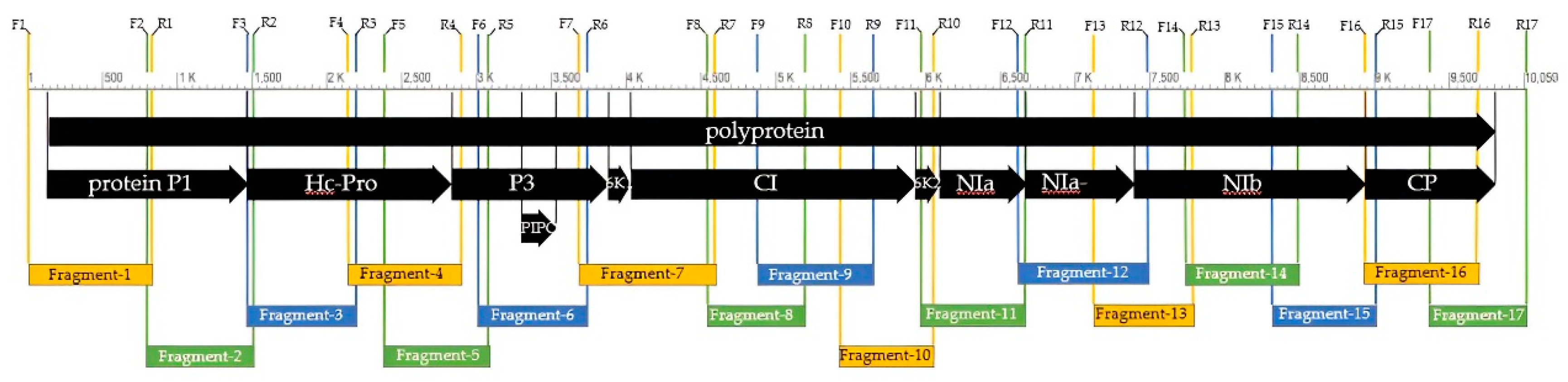
PCR products amplified by designed primer pairs covered the full length of the BCMV genome. Adjacent fragments partially overlapped to completely cover the entire genome (
Figure 3). The whole genome sequence of the BCMV SVK isolate has been deposited in the GenBank DNA sequence database (
https://www.ncbi.nlm.nih.gov/genbank/) under the accession code OQ448156. Its genome has 10,050 nucleotides and encodes a single polyprotein of 3,222 amino acids.
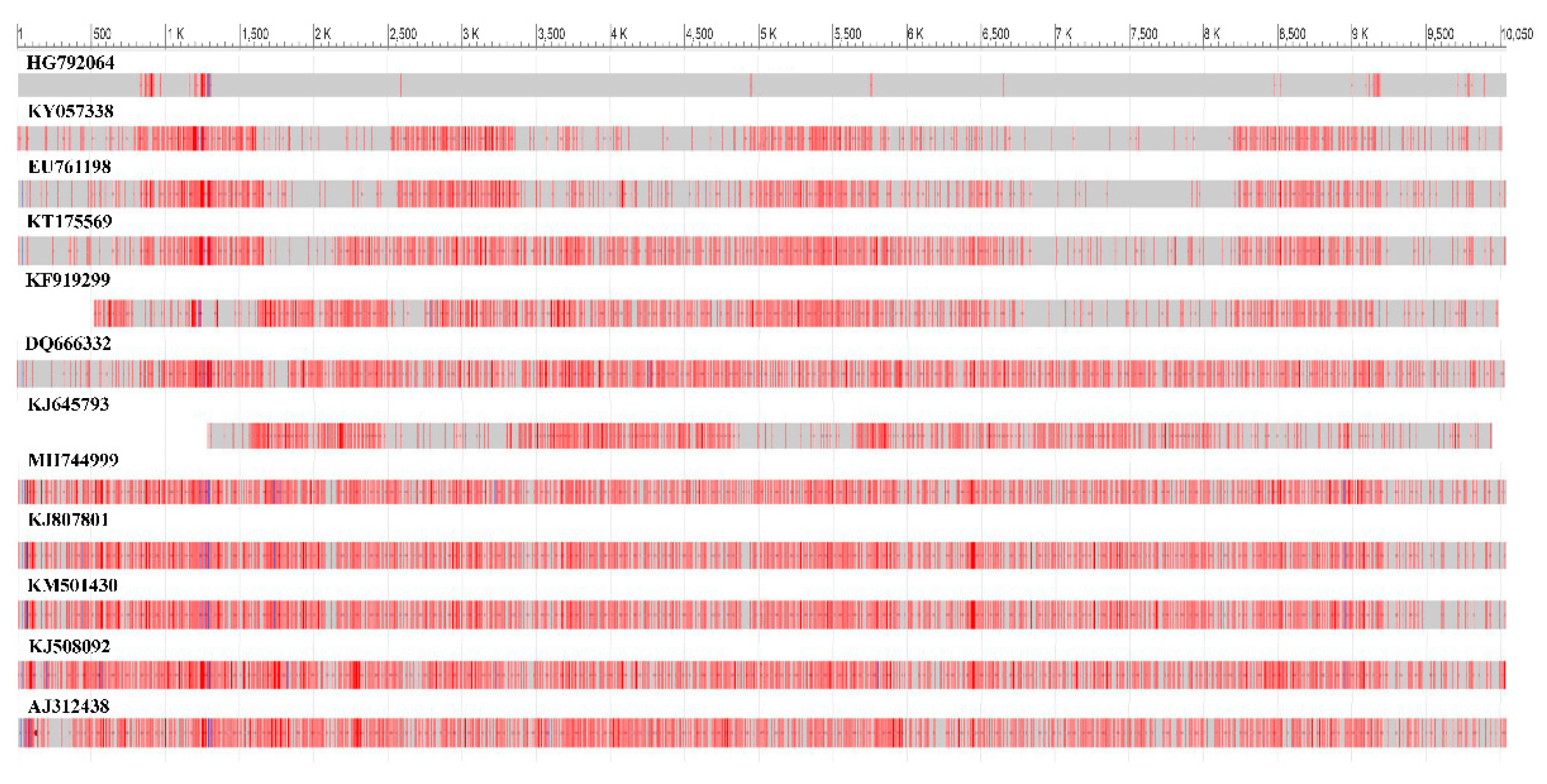
Isolate of the BCMV SVK obtained from symptomatic crownvetch plant was compared in the full length of the genome sequence with twelve BCMVs selected from the GenBank database to access their genetic patterns using the BLAST tool. Widespread nucleotide differences compared to selected BCMVs were detected throughout the genome of BCMV SVK. Identity in nucleotide sequences between them varied in range 99.09%–82.92% (
Figure 4). The most similar genome was the BCMV isolate PV 0915 (GenBank accession HG792064) isolated from common bean. Thus, it differed from BCMV SVK in 0.91% of nucleotides.
2.4. Proteomic Analysis
Comparative analysis of the amino acids revealed a difference of 1.55% in the amino acids between the polyproteins of BCMV PV 0915 and BCMV SVK. The highest number of differences was in the P1 protein (39 different amino acids), followed by coat protein (CP) with 7 different amino acids. Two substitutions were in the Hc-pro protein, and one substitution each in the cylindrical inclusion protein CI and the nuclear inclusion b protein Nib, respectively (
Table 1).
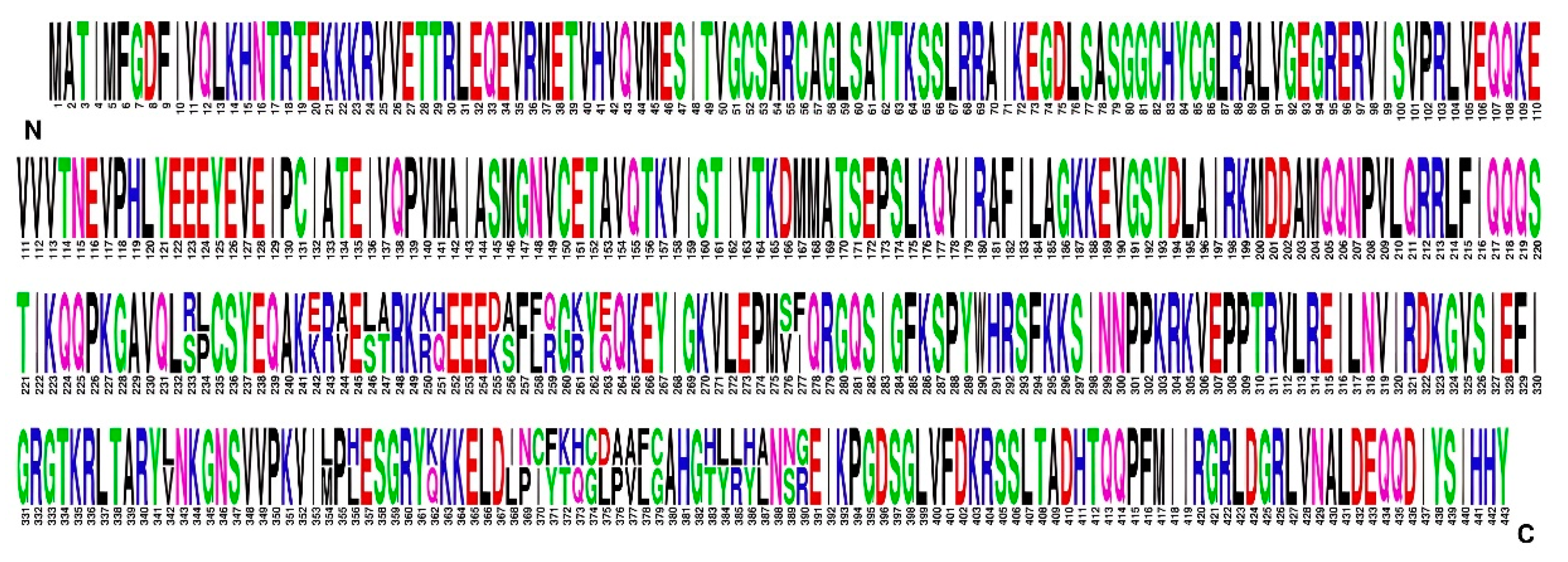
Substitutions in the most variable P1 protein, which contains 443 amino acids, were in two regions. One was between amino acids 233 and 277 and contained differences in 16 amino acids. The second region was between amino acids 342 and 390 and contained differences in 23 amino acids (
Figure 5). BCMV SVK and BCMV PV0915 differed in 2.44% of amino acids. The amino acid composition of the complete P1 protein of the BCMV SVK isolate differed by 8.8% amino acids from BMCV PV 0915.
Within the CP protein, there were two substitutions at amino acids 22
nd and 285
th and five amino acid substitutions in the region between 68
th and 85
th amino acids (
Figure 6).
The conserved motifs in BCMV SVK proteins were compared with the conservative motifs of 62 BCMVs [
18]. Complete identity was found in four conserved motifs of the P1 protein. Another conserved motif, common for the P1 protein of BCMVs (HX
10DX
29SGX
21RG), is in the C-terminal region of this protein. Its composition was HX
75DX
29SGX
21RG. However, this motif in the BCMV SVK isolate was significantly different from all others in BCMVs. The match was at the end of the motif (toward the C-terminus). The difference is at the beginning of the motif (in the opposite direction). Within the HC-Pro (many activities and functions), P3 (possible protease activity), CI (potential helicase activity), NIa-Pro (with the proteolytic activity), NIb (RNA-dependent polymerase activity), and CP (coat) protein regions, all conserved motifs published in BCMVs [
18] were present in BCMV SVK.
2.5. Phylogenetic Comparison of BCMV SVK
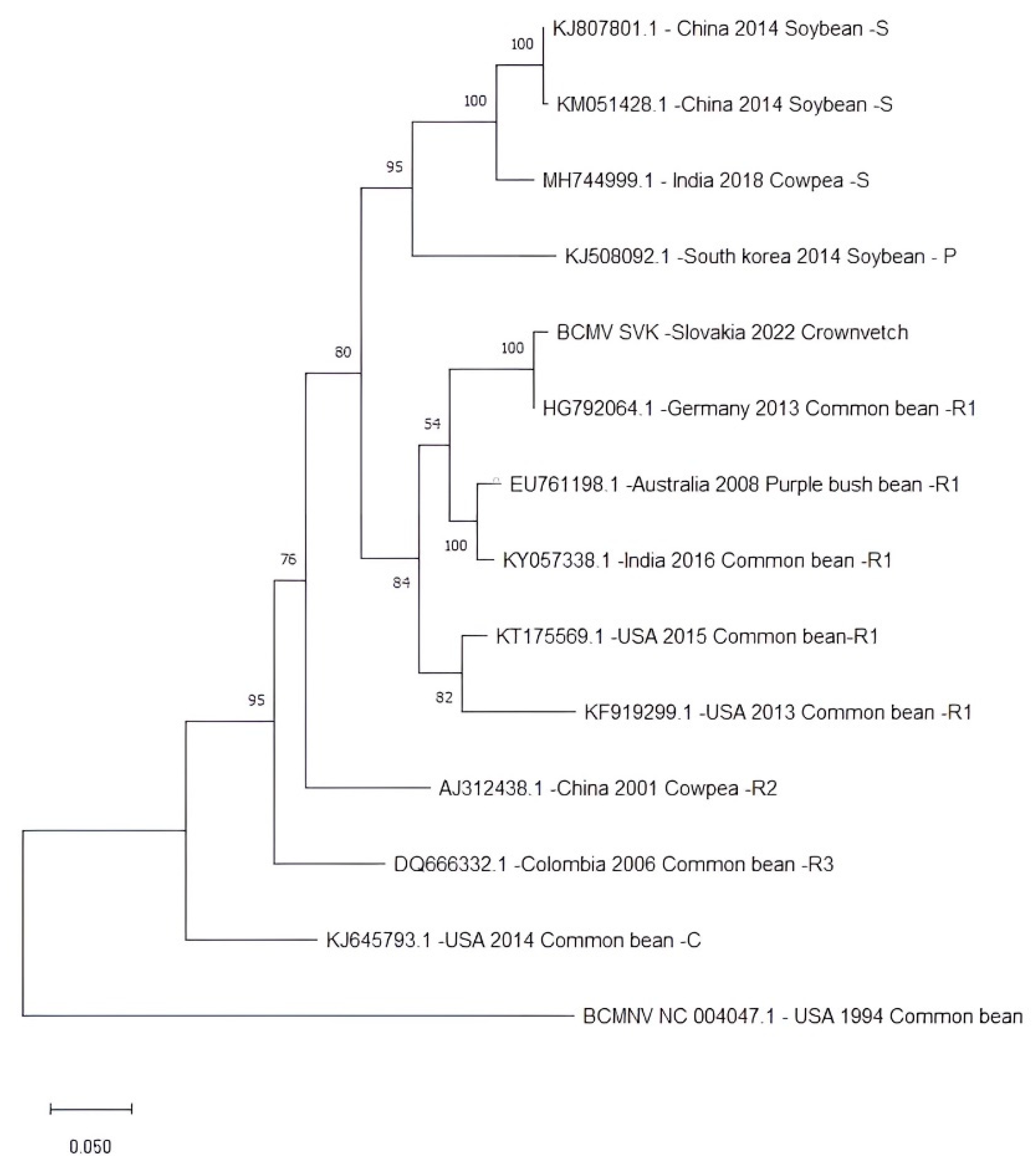
The nucleotide sequence of isolate BCMV SVK was compared with 13 complete BCMV genomes representing non-recombinant isolates from the three main phylogroups (S, P, and C) and recombinant isolates from the phylogroups R1, R2, and R3. This phylogenetic classification relates to the tendency to sort isolates according to their original host plant [
19]. The isolate BCMV SVK was classified into the R1 recombinant phylogroup together with the most similar BCMV PV 0915 (
Figure 7).
The analysis revealed a clear differentiation of isolates into the six phylogroups mentioned above. Phylogroup S (soybean) consisted of isolates that originated in China and India and were isolated from soybean. Phylogroup P (peanut) consisted of only one isolate from South Korea obtained from soybean. One isolate, originating from the USA and isolated from common bean, formed a separate branch representing phylogroup C (common bean). Others have created three separate phylogroups of isolates from recombinant isolates R1, R2, and R3 from Australia, India, the USA, China, and Colombia.
3. Discussion
The BCMV occurs in natural conditions mainly in
Phaseolus species, especially in
Phaseolus vulgaris (order Fabales, family Fabaceae). BCMV naturally infects wild and cultivated legumes, but the experimental host range is much wider [
20,
21,
22]. The plant from which the BCMV SVK isolate was obtained and characterized in this work was crownvetch growing wild on the edge of vegetable gardens. This species from the family Fabaceae is usually assigned to the group of non-crop plants but is commonly found at the agro-ecological interface where virus interactions between cultivated and wild species take place. Somewhere, crownvetch is cultivated, harvested, and used as fresh feed or hay for ruminants, or as a substrate to produce biomethane and renewable energy [
23]. Its breeding programs are also being considered [
24]. The crownvetch did not appear in a broad list of cultivated and wild BCMV hosts from six susceptible plant families [
25]. To date, there has been no published report on the occurrence of BCMV on the crownvetch (
Securigera varia L. Lassen, synonym
Coronilla varia L.).
The complexity of the population genetic structure of BCMV is based on differences between isolates originating from different locations and different hosts. Thus, adaptation of BCMV populations is driven to a large extent by both the host species and geographic location. Virus populations have a high potential for genetic variation through mutations and recombinations. On the other side, a factor that limits this variation is natural selection, an important evolutionary force acting on the proteins responsible for different virus functions [
26]. Combinations of host and virus are different. Selection contributes to the emergence of new hosts or novel strains of viruses [
27].
Nucleotide and amino acid substitutions across the potyviral genome are not randomly distributed [
28]. Substitutions preferentially accumulate in hypervariable areas at a higher frequency than in the rest of the genome. A comparison of the complete genomic or polyprotein sequences for 95 potyviruses showed that amino acid substitutions occurred mainly in the N-terminal part of P1, the N- and C-terminal parts of HC-Pro, the C-terminal part of P3, VPg, the C-terminal part of NIb, and the N-terminal part of CP [
28]. This is true for potyviruses in general, but it may be different for individual potyviruses. The BCMVs have the most substitutions within the P1 protein. Nucleotide and amino acid substitutions are widespread throughout the P1 protein, although there is a tendency towards their higher frequency in the N-terminal part of the protein [
28]. This characteristic was not fully confirmed for the P1 protein of the BMCV SVK isolated from crownvetch, because substitutions were concentrated closer to the C-terminal parts of the protein. Four of the five conserved amino acid motifs in the P1 protein of the BCMV SVK isolate were identical to those of BCMVs. Although there was a difference in the fifth (HX
75DX
29SGX
21RG) of the conserved motifs in the BCMV SVK isolate, the effect of this change on its self-hydrolytic ability was not evident. The P1 protein of potyviruses is associated with their pathogenicity and host specificity [
29,
30,
31]. Variations at many codons of the P1 protein should be responsible for the molecular adaptation of certain genotypes to increase the diversification of the virus population [
19]. These differences could be largely responsible for the ability of BCMV to infect a new host, the crownvetch plants. If the P1 protein were not functional, it would not sufficiently separate P1 and HC-Pro, and other important functions of the virus would likely be impaired. Despite the change, or perhaps because of the change in one conserved motif in P1 protein, the proteolytic competence of P1 protein was maintained also in the crownvetch host and the P1 protein was functional.
The presented work reveals the potential of crownvetch to be a host of BCMV and to participate in the emergence of novel genetic variants of the virus. This could have negative impacts on the cultivation of economically important legumes, especially in areas with agroecological interfaces where common beans and related BCMV host species, including crownvetch, are in contact.
4. Materials and Methods
4.1. Plant Material
Plants of crownvetch (Securigera varia L. Lassen, synonym Coronilla varia L.) were searched and collected during a collecting expedition in western Slovakia (Čachtice and Plavecký Štvrtok) and the BCMV was isolated from crownvetch plants manifesting symptoms of disease caused by virus. Leaves from symptomatic plant were collected and stored at -80 °C. Seeds of tobacco (Nicotiana benthamiana Domin) were obtained from the collections of genetic resources of the Slovak Genbank (Research Institute of Plant Production, Piešt’any, Slovakia).
4.2. Artificial Infections of Plants
Tobacco plants were inoculated at the stage of two true leaves using homogenized leaf tissue from crownvetch plant that was positively tested for the presence of BCMV. The inoculum was resuspended in Norit-buffer (0.05 M sodium/potassium phosphate buffer, pH 7.0, 1 mM EDTA, 5 mM diethyldithiocarbamic acid, and 5 mM thioglycolic acid, Merck KGaA, Darmstadt, Germany). Plants were inoculated by mechanical wounding using sterile silicon carbide powder (Merck KGaA, Darmstadt, Germany) and cultivated in growing chambers at 24-25 °C with a photoperiod of 16 hours of light and 8 hours of darkness at 152 µmol m-2 s-1 intensity. Uninfected control plants were grown in isolation under the same conditions. Symptoms of virus disease occurred during the cultivation of tobacco plants.
4.3. Immunochemical Detection
Proteins were extracted from plants using the Pierce Plant Total Protein Extraction Kit (Thermo Fisher Scientific, Waltham, MA, USA). Equivalent volumes of protein extract (50 μg per lane) were loaded into polyacrylamide gels containing the denaturing agent, sodium dodecyl sulfate. The presence of BCMV in samples was detected using polyclonal antibodies against Bean common mosaic virus (BCMV) (Leibniz Institute, DSMZ-German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany) diluted in a ratio of 1:500. The secondary antibody used was the anti-Rabbit IgG, peroxidase-linked species-specific whole antibody (from donkey) (Thermo Fisher Scientific, Waltham, MA, USA), diluted in a ratio of 1:4000. Positive control used in immunochemical detection was the BCMV isolate PV 0915 (Leibniz Institute, DSMZ-German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany), isolated from common bean, and its geographic origin is in Türkiye. The size of detected proteins was determined by the molecular weight markers (PageRulerTM Prestained Protein Ladder 10 to 180 kDa and PageRulerTM Plus Prestained Protein Ladder 10 to 250 kDa, Thermo Fisher Scientific, Waltham, MA, USA). Immunocomplexes were visualized using 1-StepTM Ultra TMB-Blotting Substrate Solution (Thermo Fisher Scientific, Waltham, MA, USA) and recorded using the photo documentation system Azure 400 (Azure Biosystems Inc., Dublin, CA, USA).
4.4. LC-MS/MS Analysis
Tobacco plants (
Nicotiana benthamiana Domin) were inoculated with a tested BCMV isolate and cultivated in growing chambers. Two young leaves were cut from each plant for the extraction of proteins. This procedure was repeated twice for each plant, producing two protein samples per plant. In all samples, BCMV viral proteins were detected by a medium resolution nano-Liquid Chromatography-Electrospray Ionization-Quadrupole-Time of Flight (nanoLC-ESI-Q-TOF), as described before [
32], confirming the pathogenicity of the isolate. Proteins were extracted from young leaves using the double crushing step extraction protocol [
32] including the RuBisCO depletion step by phytate and Ca
2+ ions. Mass spectrometry measurements were carried out using the UHPLC Dionex Ultimate 3000 RSLCnano (Dionex, Sunnyvale, CA, USA) connected to the mass spectrometer ESI-Q-TOF Maxis Impact (Bruker, Billerica, MA, USA), and the spectra were acquired as before [
32]. The peptides in raw spectra were identified and quantified by MaxQuant 1.6.17.0 [
33] for Windows using the same parameters as before [
32]. The spectra were searched against all known
Nicotiana proteins downloaded from the UniProt protein knowledgebase [
34] together with all known BCMV protein sequences, all known BCMV protein sequences downloaded from the NCBI Protein database [
35], and BCMV SVK isolate protein sequence. The same scripts as previously [
32] were used for detailed sequence coverage.
4.5. Sequencing Analyses
RNA was isolated using the NucleoSpin RNA Plant, Mini Kit for RNA from Plant (Macherey-Nagel, Düren, Germany) and TRIzolTM Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The concentration of isolated RNA was determined spectrophotometrically (NanoDrop 1000 Spectrophotometer, Thermo Fisher Scientific, Waltham, MA, USA), and its quality has been verified by electrophoresis in 1.5% agarose-formaldehyde gel stained with ethidium bromide. The RevertAid First Strand cDNA Synthesis Kit (Fermentas GmbH, St. Leon-Rot, Germany) was used for the first-strand cDNA synthesis.
Based on the assumption of homology between our isolate of BCMV (BCMV_SVK) and other previously sequenced pathotypes, the primers (
Table 2) were designed by the Primer3 software [
36] using the complete sequence of BCMV isolate Viva2 (GenBank accession MH024839, [
37]) using the SnapGene software (GSL Biotech LLC, San Diego, CA, USA) so that PCR products cover the entire genome and neighboring fragments would partially overlap to get full length genome information during sequencing.
PCR fragments (2, 4, 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17) (
Table 2) were amplified under the same reaction conditions in a 25 µL reaction mixture containing 1×PCR buffer, 1.5 mM MgCl
2, 10 pM of both primers, 0.2 mM each of dNTPs, 0.5 U Platinum
TM Taq DNA polymerase (Invitrogen Corp., Carlsbad, CA, USA), and 100 ng of reverse transcript template from a BCMV-positive plant. PCR reactions were performed in the Mastercycler
® ep (Eppendorf, Hamburg, Germany) thermal cycler using the following parameters: 94 °C for 3 min, followed by 30 cycles, each consisting of 94 °C for 1 min, 56 °C for 60 s, 72 °C for 1 min, and the final extension at 72 °C for 10 min. Fragments (1, 3, 5, 6, and 7) (
Table 2) were obtained under the same reaction conditions, but with the addition of Q-solution reagent and the annealing temperature reduced to 54°C. PCR products were verified by electrophoresis in 1.0% agarose–formaldehyde gel stained with ethidium bromide.
After electrophoretic separation in 1.0 % (w/v) agarose gels pre-stained with ethidium bromide, amplicons were extracted from the agarose gel using the Agarose Gel DNA Extraction Kit (Roche Diagnostics GmbH, Mannheim, Germany). Concentrations of isolated DNA fragments were determined spectrophotometrically (Nanodrop 1000 Spectrophotometer, Thermo Fisher Scientific, Waltham, MA, USA). Sanger sequencing of both DNA strands was performed with a sequencing service (Eurofins Genomics Sequencing GmbH, Ebersberg/Köln, Germany).
Nucleotide and amino acid sequences were analyzed using the software CLC Bio Main Workbench (Qiagen N.V., Venlo, The Netherlands) and SnapGene (GSL Biotech LLC, Boston, MA, USA). The maximum likelihood phylogenetic tree of selected BCMV isolates was constructed by the bootstrapping method with 1000 replications using the software MEGA11 (Molecular Evolutionary Genetics Analysis Version 11, [
38] with BCMNV as an outgroup. The comparison of nucleotide sequences between BCMVs was performed using the Basic Local Alignment Search Tool (BLAST) [
39].
5. Conclusions
Crownvetch (Securigera varia L. Lassen) was confirmed as a new host species for BCMV. At the same time, a novel virus isolate was isolated from it, whose genome has 10,050 nucleotides and encodes one polyprotein with 3,222 amino acids. Amino acid comparison revealed substitutions in 50 amino acids against the most similar BCMV isolate PV 0915. BCMV isolates with complete genomic sequences were obtained mainly from beans, soybeans, cowpeas, and peanuts. The BCMV SVK isolate from the crownvetch joined the group of only four BCMV isolates obtained from other plant species with completely sequenced genomes (Nandina domestica Thunb., Sesamum indicum L., Cudrania tricuspidata (Carr.) Bur., and Macroptilium atropurpureum (DC.) Urb.). Therefore, crownvetch has been included among the risk hosts and sources of BCMV spreading, especially in agroecological interfaces where Phaseolus spp. are cultivated on the other side.
Author Contributions
Conceptualization, D.M.; methodology, D.M.; software, R.H. and J.K.; validation, D.M. and J.K.; formal analysis, D.M., S.G., R.H., P.C. and M.H.; investigation, D.M., S.G., R.H., P.C., and M.H.; resources, D.M.; data curation, D.M., R.H., P.C. and J.K.; writing—original draft preparation, J.K.; writing—review and editing, J.K.; visualization, R.H., P.C. and J.K.; supervision, D.M.; project administration, D.M.; funding acquisition, D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Slovak Research and Development Agency, projects numbers APVV-20-0015 and APVV-21-0289, and by the Operational Programme Integrated Infrastructure funded by the European Regional Development Found, projects number ITMS: 313011AVA9 (“PandemicFood”) and ITMS: 313011ASN4 (“Addressing the societal threats posed by the COVID-19 pandemic”).
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#home (accessed on 7 March 2023).
- OECD (2016), “Common bean (Phaseolus vulgaris)”, in Safety Assessment of Transgenic Organisms in the Environment, Volume 6: OECD Consensus Documents, OECD Publishing, Paris. [CrossRef]
- Broughton, W.J.; Hernandez, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.) – model food legumes. Plant Soil 2003, 252, 55–128. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Naidu, R.A. Global dimensions of plant virus diseases: Current status and future perspectives. Annu. Rev. Virol. 2019, 6, 387–409. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Feng, X. Bean common mosaic disease: Etiology, resistance resource, and future prospects. Agronomy 2023, 13, 58. [Google Scholar] [CrossRef]
- Drijfhout, E.; Silbernagel, M.J.; Burke, D.W. Differentiation of strains of Bean common mosaic virus. Neth. J. Plant Pathol. 1978, 84, 13–26. [Google Scholar] [CrossRef]
- Feng, X.; Myers, J.R.; Karasev, A.V. Bean common mosaic virus isolate exhibits a novel pathogenicity profile in common bean, overcoming the bc-3 resistance allele coding for the mutated eIF4E translation initiation factor. Phytopathology 2015, 105, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Vetten, H.J.; Lesemann, D.-E.; Maiss, E. Serotype A and B strains of bean common mosaic virus are two distinct potyviruses. In Potyvirus Taxonomy, Archives of Virology; Barnett, O.W., Ed.; Springer: Vienna, Austria, 1992; Volume 5, pp. 415–431. [Google Scholar] [CrossRef]
- Taxonomy and classification of legume-infecting potyviruses. Arch. Virol. 1994, 139, 231–235. [CrossRef]
- Revers, F.; Le Gall, O.; Candresse, T.; Le Romancer, M.; Dunez, J. Frequent occurrence of recombinant potyvirus isolates. J. Gen. Virol. 2016, 77, 1953–1965. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Poplawsky, A.R.; Nikolaeva, O.V.; Myers, J.R.; Karasev, A.V. Recombinants of Bean common mosaic virus (BCMV) and genetic determinants of BCMV involved in overcoming resistance in common bean. Phytopathology, 2014, 104, 786–793. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, P.N.; Kapil, R.; Sharma, S.K.; Sharma, O.P. Analysis of 3′-terminal region of Bean common mosaic virus strains infecting common bean in India. Indian J. Virol. 2011, 22, 37–43. [Google Scholar] [CrossRef]
- Johary, T.; Dizadji, A.; Naderpour, M. Biological and molecular characteristics of Bean common mosaic virus isolates circulating in common bean in Iran. J. Plant Pathol. 2016, 98, 301–310. [Google Scholar] [CrossRef]
- Abadkhah, M.; Hajizadeh, M.; Koolivand, D. Global population genetic structure of Bean common mosaic virus. Arch. Phytopathol. Plant Prot. 2020, 53, 266–281. [Google Scholar] [CrossRef]
- Spence, N.J.; Walkey, D.G.A. Variation for pathogenicity among isolates of bean common mosaic virus in Africa and a reinterpretation of the genetic relationship between cultivars of Phaseolus vulgaris and pathotypes of BCMV. Plant Pathol. 1995, 44, 527–546. [Google Scholar] [CrossRef]
- Sengooba, T.N.; Spence, N.J.; Walkey, D.G.A.; Allen, D.J.; Femi Lana, A. The occurrence of bean common mosaic necrosis virus in wildand forage legumes in Uganda. Plant Pathol. 1997, 46, 95–103. [Google Scholar] [CrossRef]
- Coutts, B.A.; Kehoe, M.A.; Webster, C.G.; Wylie, S.J.; Jones, R.A.C. Indigenous and introduced potyviruses of legumes and Passiflora spp. from Australia: Biological properties and comparison of coat protein nucleotide sequences. Arch. Virol. 2011, 156, 1757–1774. [Google Scholar] [CrossRef] [PubMed]
- Worrall, E.A.; Hayward, A.C.; Fletcher, S.J.; Mitter, N. Molecular characterization and analysis of conserved potyviral motifs in bean common mosaic virus (BCMV) for RNAi-mediated protection. Arch. Virol. 2019, 164, 181–194. [Google Scholar] [CrossRef]
- Moradi, Z.; Mehrvar, M. Genetic variability and molecular evolution of Bean common mosaic virus populations in Iran: Comparison with the populations in the world. Eur. J. Plant Pathol. 2019, 154, 673–690. [Google Scholar] [CrossRef]
- Sangeeta, S.B.; Ramappa, H.K.; Aishwarya, P. Biological and serological confirmation of host range of bean common mosaic virus (BCMV) infecting field bean. Pharma Innovation 2022, 11, 2046–2049. [Google Scholar]
- DPV. Available online: https://www.dpvweb.net/dpv/showdpv/?dpvno=337 (accessed on 14 March 2023).
- PVD. Available online: http://47.90.94.155/PlantVirusBase/#/search?val=Bean%20common%20mosaic%20virus (accessed on 14 March 2023).
- Ţîţei, V. The prospect of cultivation and utilization of Coronilla varia L. in Moldova. Oltenia-Stud. Nat. Sci. 2021, 37, 46–52. [Google Scholar]
- Shanjani, P.S.; Rasoulzadeh, L.; Javadi, H. Evaluation of morphological traits in the populations of Coronilla varia L. J. Rangel. Sci., 2023, 13, 52–71. [Google Scholar] [CrossRef]
- Worrall, E.A.; Wamonje, F.O.; Mukeshimana, G.; Harvey, J.J.W.; Carr, J.P.; Mitter, N. Bean common mosaic virus and Bean common mosaic necrosis virus: Relationships, biology, and prospects for control. Adv. Virus Res. 2015, 93, 1–46. [Google Scholar] [CrossRef]
- García-Arenal, F. , Fraile A., Malpica, J.M. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 2001, 39, 157–186. [Google Scholar] [CrossRef]
- Roossinck, M.J. Plant RNA virus evolution. Curr. Opin. Microbiol. 2003, 6, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Nigam, D.; LaTourrette, K.; Souza, P.F.N.; Garcia-Ruiz, H. Genome-wide variation in Potyviruses. Front. Plant Sci. 2019, 10, 1439. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.C.; Miklas, P.N.; Druffel, K.L.; Wyatt, S.D. NL-3 K strain is a stable and naturally occurring interspecific recombinant derived from Bean common mosaic necrosis virus and Bean common mosaic virus. Phytopathology 2005, 95, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.I.; Salvador, B.; Carbonell, A.; Saenz, P.; Leon, D.S.; Oliveros, J.C.; Delgadillo, M.O.; Garcia, J.A.; Simonmateo, C. Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host-dependent pathogenicity features. Mol. Plant Pathol. 2012, 13, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Shan, S.; Huang, Y.; Jiang, C.; Zhang, H.; Li, Y.; Chen, J.; Wei, Z.; Sun, Z. The hypervariable N-terminal of soybean mosaic virus P1 protein influences its pathogenicity and host defense responses. Phytopathol. Res. 2022, 4, 10. [Google Scholar] [CrossRef]
- Cejnar, P.; Kučková, Š.; Šantrůček, J.; Glasa, M.; Komínek, P.; Mihálik, D.; Slavíková, L.; Leišová-Svobodová, L.; Smirnova, T.; Hynek, R.; Kundu, J.K.; Ryšánek, P. Efficient confirmation of plant viral proteins and identification of specific viral strains by nanoLC-ESI-Q-TOF using single-leaf-tissue samples. Pathogens 2020, 9, 966. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucl. Acid Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- NCBI Protein Database. Available online: https://www.ncbi.nlm.nih.gov/protein/ (accessed on 14 April 2023).
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucl. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Feng, X.; Orellana, G.E.; Myers, J.R.; Karasev, A.V. Recessive resistance to Bean common mosaic virus conferred by the bc-1 and bc-2 genes in common bean (Phaseolus vulgaris) affects long-distance movement of the virus. Phytopathology 2018, 108, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- BLAST (The Basic Local Alignment Search Tool). Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome (accessed on 10 March 2023).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).