Submitted:
22 April 2023
Posted:
23 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Viruses
2.3. Immunofluorescence assay
2.4. PCV2b/SwIV co-infection
2.5. Cell viability assay
2.6. Cytokine mRNAs expression in PCV2b and/or SwIV infected cells
2.6. 3’ mRNA-seq Library Preparation and Sequencing
2.7. Bioinformatic analysis
2.8. Statistical analyses
3. Results
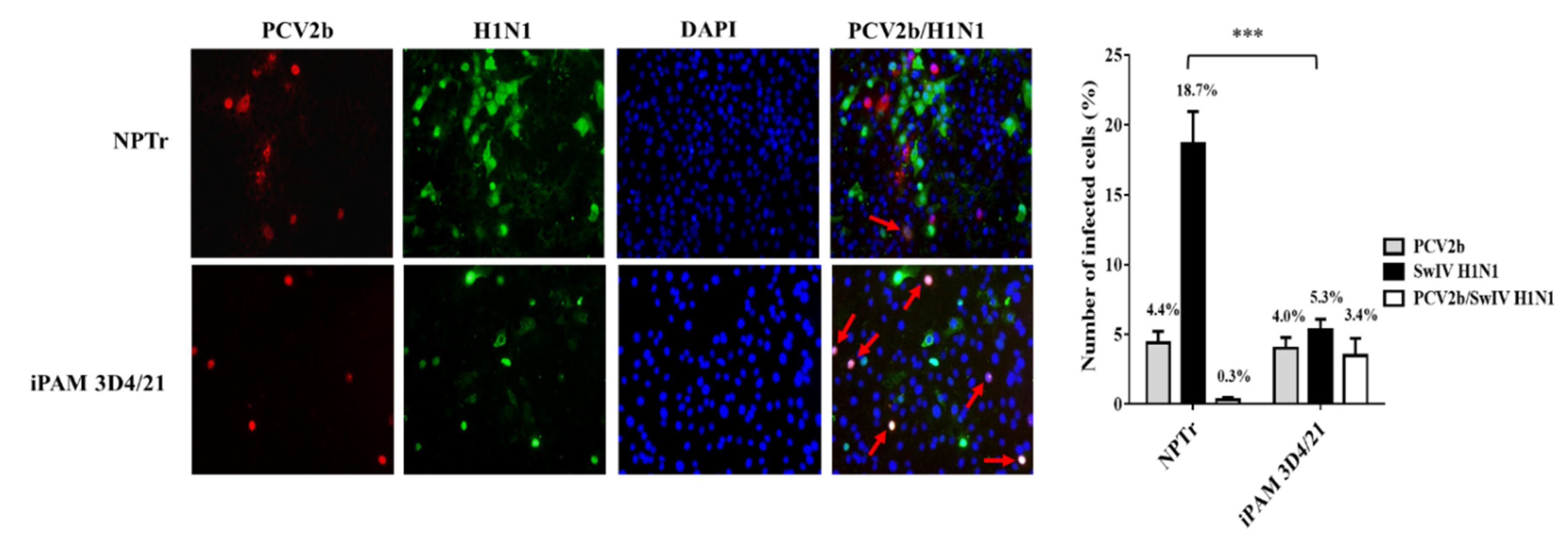
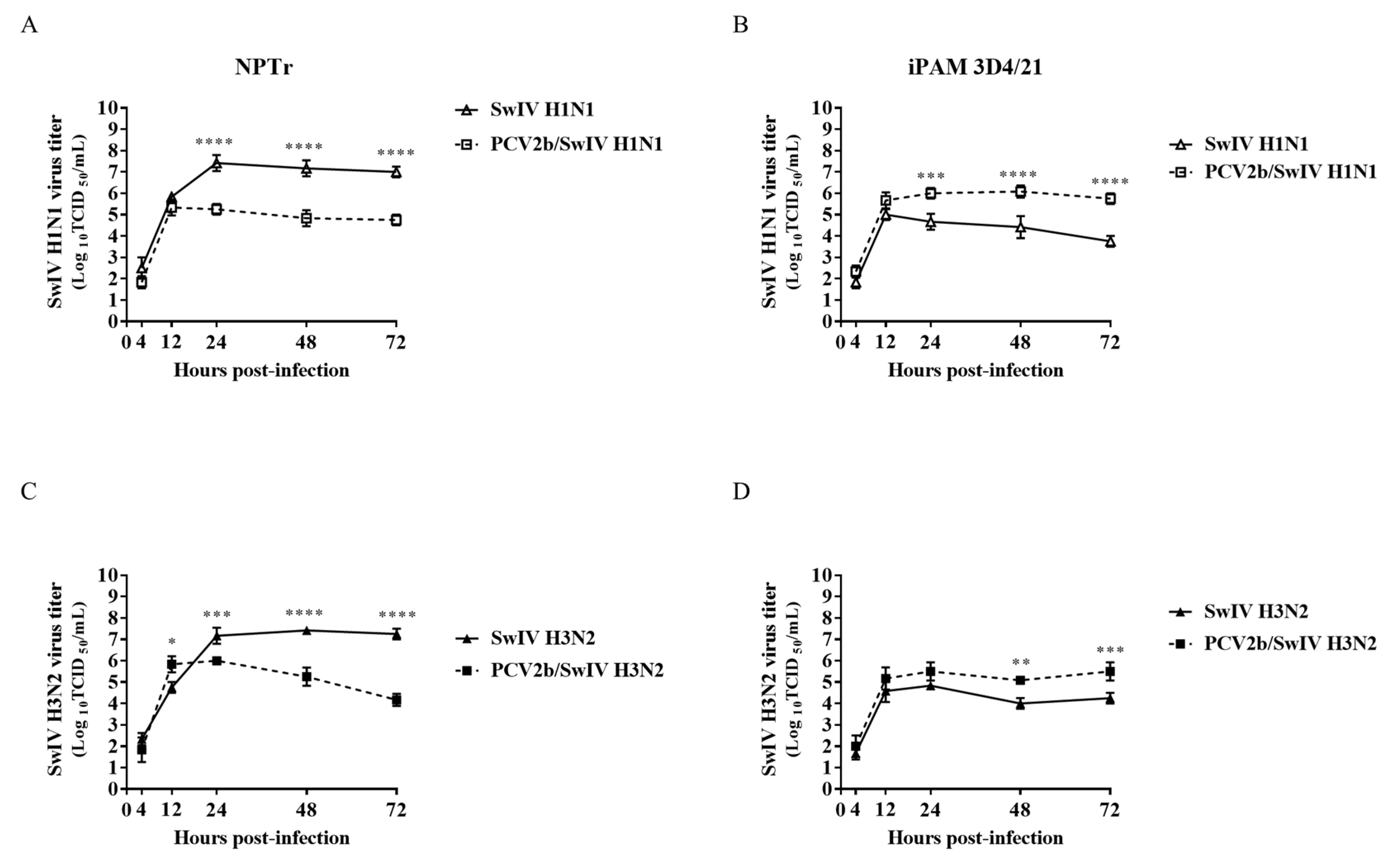
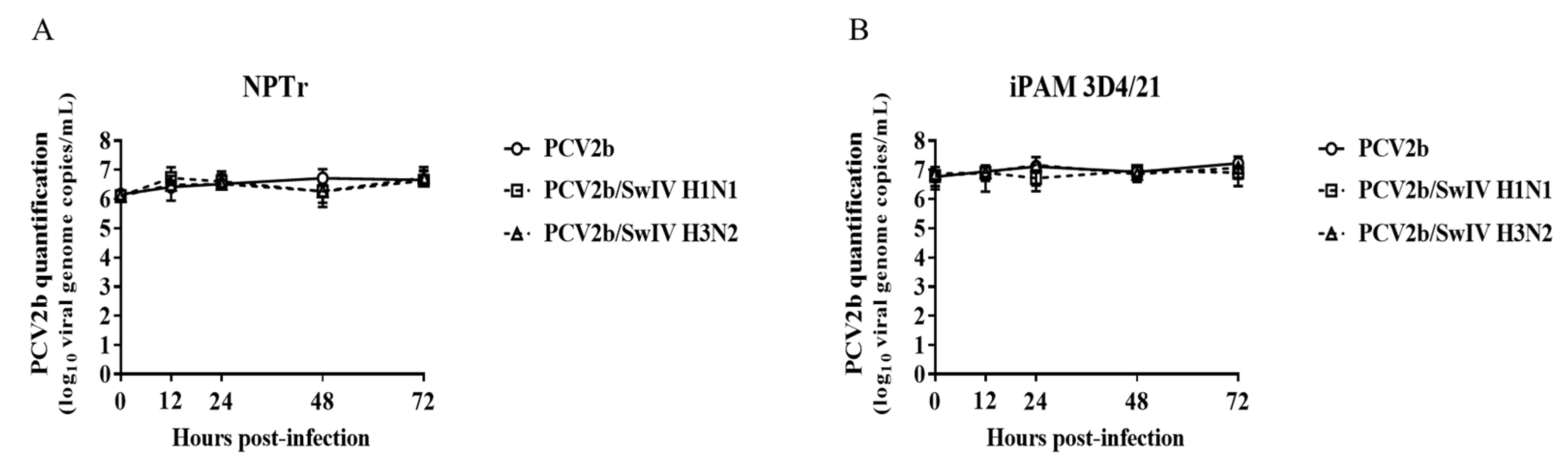
3.1. Modulation of swine influenza A virus replication in co-infected PCV2b/SwIV H1N1 cells
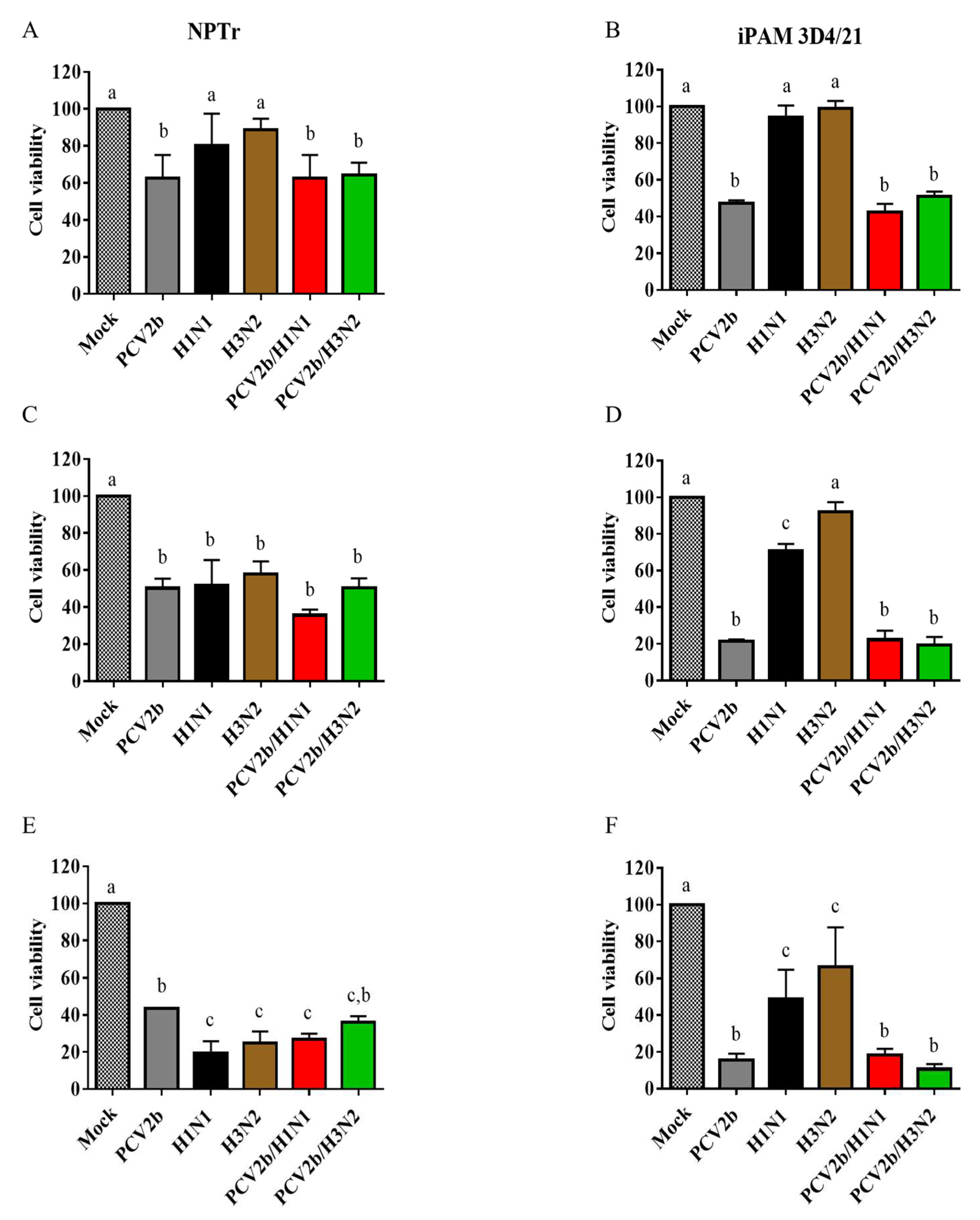
3.2. Cell viability assay
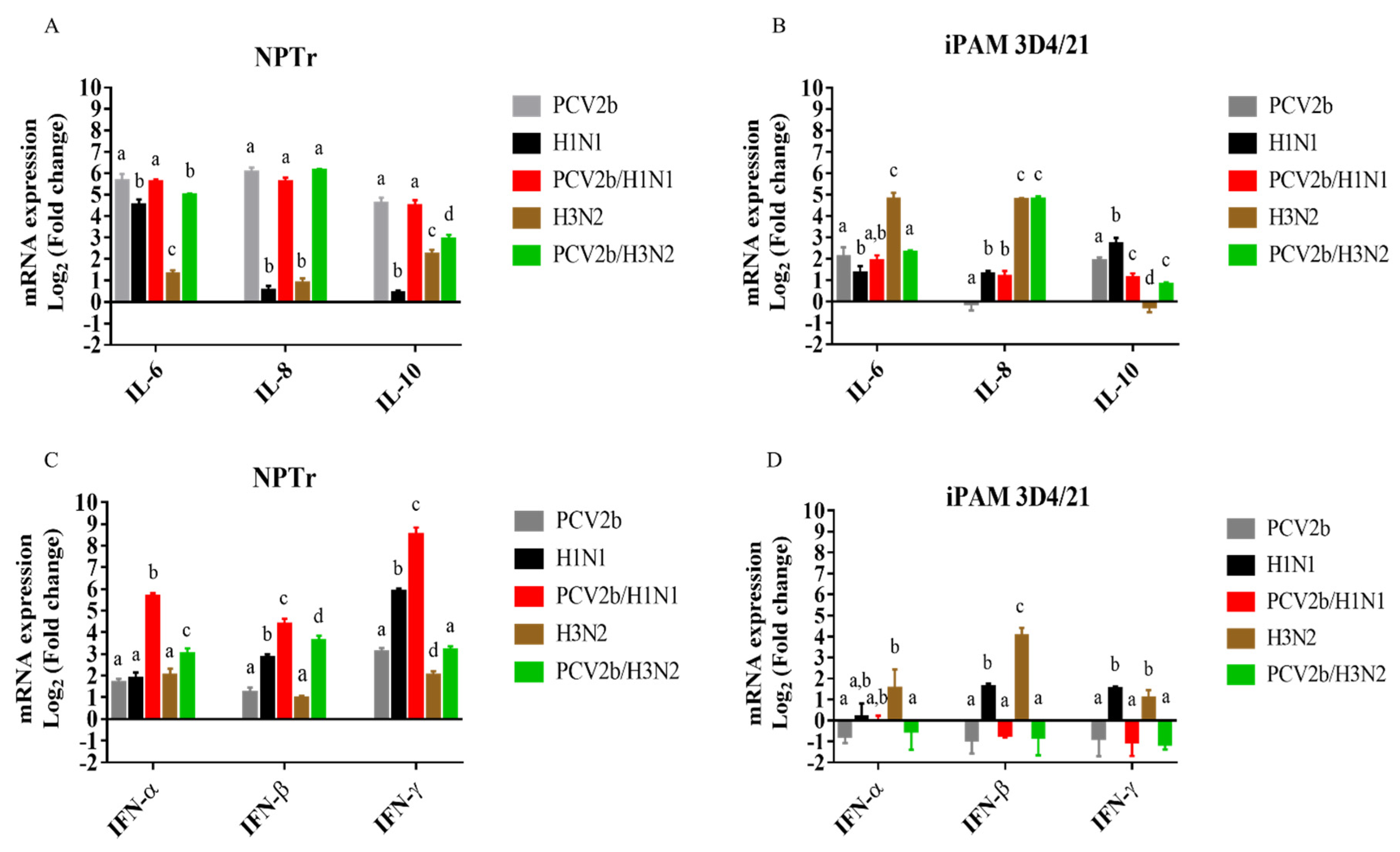
3.3. Modulation of cytokines mRNAs expression in co-infected cells
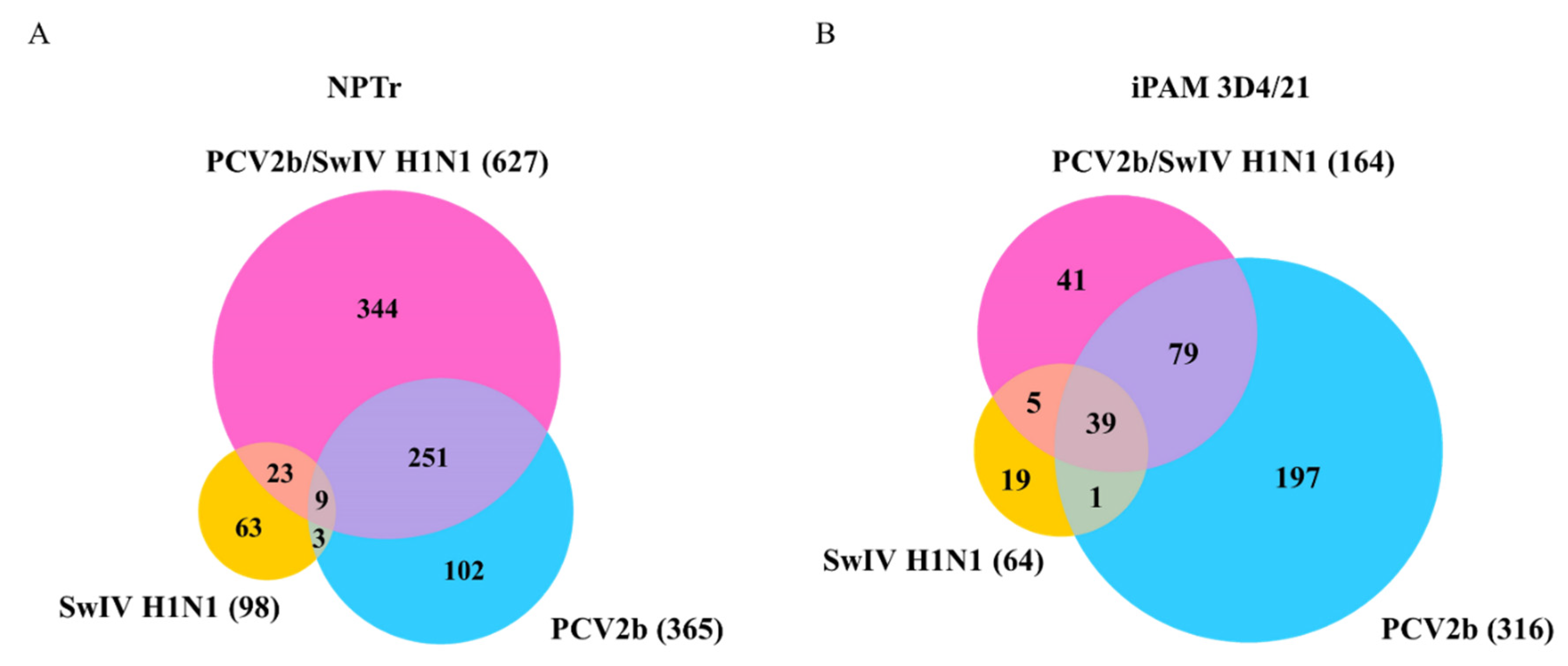
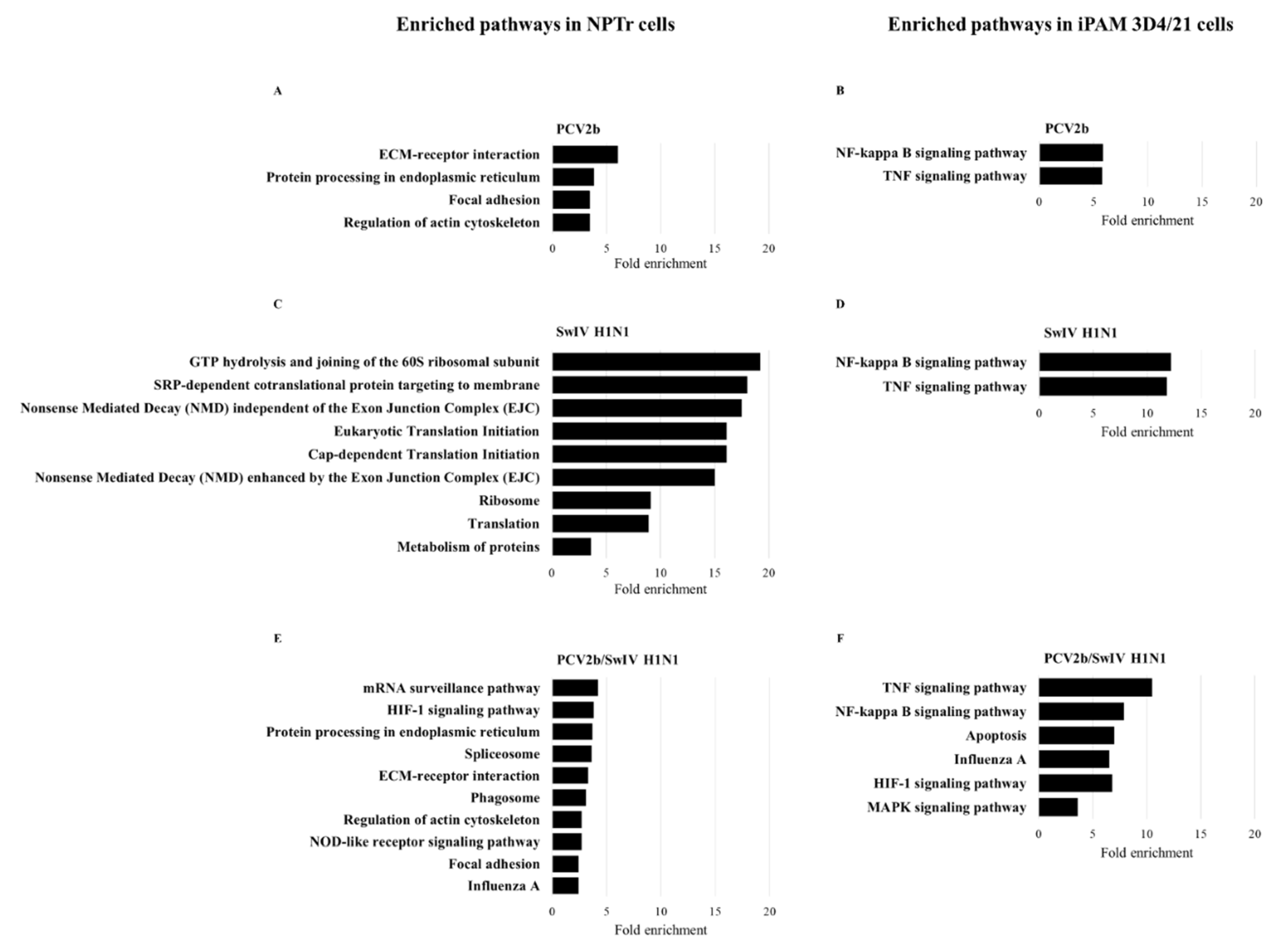
3.4. Differentially Expressed Genes (DEGs) and pathway enrichment analysis in PCV2b, SwIV H1N1 and PCV2b/SwIV H1N1 infected cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Opriessnig, T.; Gimenez-Lirola, L.G.; Halbur, P.G. Polymicrobial respiratory disease in pigs. Anim Health Res Rev 2011, 12, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, S.L.; Halbur, P.G.; Thacker, E.L. Porcine respiratory disease complex. Wiley Online Library, 2002: p. 231-258.
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B.J.N.a.r. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). 2018, 46, D708–D717.
- Chae, C. A review of porcine circovirus 2-associated syndromes and diseases. Vet J 2005, 169, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Segales, J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res 2012, 164, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Hou, L.; Liu, J. Host immune response to infection with porcine circoviruses. Animal Diseases 2021, 1, 1–10. [Google Scholar] [CrossRef]
- Franzo, G.; Segales, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS One 2018, 13, e0208585. [Google Scholar]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016-2018. Transbound Emerg Dis 2020, 67, 1284–1294. [Google Scholar] [CrossRef]
- Link, E.K.; Eddicks, M.; Nan, L.; Ritzmann, M.; Sutter, G.; Fux, R. Discriminating the eight genotypes of the porcine circovirus type 2 with TaqMan-based real-time PCR. Virol J 2021, 18, 70. [Google Scholar] [CrossRef]
- Gagnon, C.A.; Tremblay, D.; Tijssen, P.; Venne, M.H.; Houde, A.; Elahi, S.M. The emergence of porcine circovirus 2b genotype (PCV-2b) in swine in Canada. Can Vet J 2007, 48, 811–819. [Google Scholar]
- Gagnon, C.A.; del Castillo, J.R.; Music, N.; Fontaine, G.; Harel, J.; Tremblay, D. Development and use of a multiplex real-time quantitative polymerase chain reaction assay for detection and differentiation of Porcine circovirus-2 genotypes 2a and 2b in an epidemiological survey. J Vet Diagn Invest 2008, 20, 545–558. [Google Scholar] [CrossRef]
- Louten, J. Essential human virology. 2022: Academic Press.
- Zhuang, Q.; Wang, S.; Liu, S.; Hou, G.; Li, J.; Jiang, W.; Wang, K.; Peng, C.; Liu, D.; Guo, A. Diversity and distribution of type A influenza viruses: an updated panorama analysis based on protein sequences. Virology Journal 2019, 16, 1–38. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Gordon, M.L. A systematic review analyzing the prevalence and circulation of influenza viruses in swine population worldwide. Pathogens 2020, 9, 355. [Google Scholar]
- Sanford, S.; Josephson, G.; Key, D. An epizootic of swine influenza in Ontario. The Canadian Veterinary Journal 1983, 24, 167. [Google Scholar]
- Bikour, M.H.; Frost, E.H.; Deslandes, S.; Talbot, B.; Elazhary, Y. Persistence of a 1930 swine influenza A (H1N1) virus in Quebec. Journal of general virology 1995, 76, 2539–2547. [Google Scholar] [CrossRef] [PubMed]
- Bikour, M.H.; Cornaglia, E.; Weber, J.M.; Elazhary, Y. Antigenic characterization of an H3N2 swine influenza virus isolated from pigs with proliferative and necrotizing pneumonia in Quebec. Canadian Journal of Veterinary Research 1994, 58, 287. [Google Scholar]
- Carman, S.; Stansfield, C.; Weber, J.; Bildfell, R.; Van Dreumel, T. H3N2 influenza A virus recovered from a neonatal pig in Ontario--1997. The Canadian Veterinary Journal 1999, 40, 889. [Google Scholar] [PubMed]
- Grgić, H.; Costa, M.; Friendship, R.M.; Carman, S.; Nagy, É.; Wideman, G.; Weese, S.; Poljak, Z. Molecular characterization of H3N2 influenza A viruses isolated from Ontario swine in 2011 and 2012. Virology journal, 2014. 11: p. 1-14.
- Grgić, H.; Costa, M.; Friendship, R.M.; Carman, S.; Nagy, É.; Poljak, Z. Genetic characterization of H1N1 and H1N2 influenza A viruses circulating in Ontario pigs in 2012. PLoS One 2015, 10, e0127840. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Culhane, M.R.; Trovão, N.S.; Patnayak, D.P.; Halpin, R.A.; Lin, X.; Shilts, M.H.; Das, S.R.; Detmer, S.E. The emergence and evolution of influenza A (H1α) viruses in swine in Canada and the United States. The Journal of general virology 2017, 98, 2663. [Google Scholar] [CrossRef]
- Nfon, C.K.; Berhane, Y.; Hisanaga, T.; Zhang, S.; Handel, K.; Kehler, H.; Labrecque, O.; Lewis, N.S.; Vincent, A.L.; Copps, J. Characterization of H1N1 swine influenza viruses circulating in Canadian pigs in 2009. Journal of virology 2011, 85, 8667–8679. [Google Scholar] [CrossRef]
- Ma, W. Swine influenza virus: Current status and challenge. Virus research, 2020. 288: p. 198118.
- La Gruta, N.L.; Kedzierska, K.; Stambas, J.; Doherty, P.C. A question of self-preservation: immunopathology in influenza virus infection. Immunology and cell biology 2007, 85, 85–92. [Google Scholar] [CrossRef]
- Sangpratum, N.; Kedkovid, R.; Woonwong, Y.; Sirisereewan, C.; Kesdangsakonwut, S.; Thanawongnuwech, R.; Arunorat, J. Dual infection of a Thai isolate HP-PRRSV and the pdmH1N1 2009 SIV in weanling pigs. The Thai Journal of Veterinary Medicine 2019, 49, 71–79. [Google Scholar] [CrossRef]
- Kitikoon, P.; Vincent, A.L.; Jones, K.R.; Nilubol, D.; Yu, S.; Janke, B.H.; Thacker, B.J.; Thacker, E.L. Vaccine efficacy and immune response to swine influenza virus challenge in pigs infected with porcine reproductive and respiratory syndrome virus at the time of SIV vaccination. Vet Microbiol, 2009. 139(3-4): p. 235-44.
- Van Reeth, K.; Nauwynck, H.; Pensaert, M. Clinical effects of experimental dual infections with porcine reproductive and respiratory syndrome virus followed by swine influenza virus in conventional and colostrum-deprived pigs. J Vet Med B Infect Dis Vet Public Health 2001, 48, 283–292. [Google Scholar] [CrossRef]
- Van Reeth, K.; Nauwynck, H.; Pensaert, M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol, 1996. 48(3-4): p. 325-35.
- Czyzewska-Dors, E.; Pomorska-Mol, M.; Dors, A.; Pluta, A.; Podgorska, K.; Kwit, K.; Stasiak, E.; Lukomska, A. Proinflammatory Cytokine Changes in Bronchoalveolar Lavage Fluid Cells Isolated from Pigs Infected Solely with Porcine Reproductive and Respiratory Syndrome Virus or Co-infected with Swine Influenza Virus. J Vet Res 2019, 63, 489–495. [Google Scholar] [CrossRef]
- Niederwerder, M.C.; Jaing, C.J.; Thissen, J.B.; Cino-Ozuna, A.G.; McLoughlin, K.S.; Rowland, R.R. Microbiome associations in pigs with the best and worst clinical outcomes following co-infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2). Vet Microbiol, 2016. 188: p. 1-11.
- Fan, P.; Wei, Y.; Guo, L.; Wu, H.; Huang, L.; Liu, J.; Liu, C. Synergistic effects of sequential infection with highly pathogenic porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Virol J, 2013. 10: p. 265.
- Opriessnig, T.; Gauger, P.C.; Faaberg, K.S.; Shen, H.; Beach, N.M.; Meng, X.J.; Wang, C.; Halbur, P.G. Effect of porcine circovirus type 2a or 2b on infection kinetics and pathogenicity of two genetically divergent strains of porcine reproductive and respiratory syndrome virus in the conventional pig model. Vet Microbiol, 2012. 158(1-2): p. 69-81.
- Sinha, A.; Shen, H.G.; Schalk, S.; Beach, N.M.; Huang, Y.W.; Meng, X.J.; Halbur, P.G.; Opriessnig, T. Porcine reproductive and respiratory syndrome virus (PRRSV) influences infection dynamics of porcine circovirus type 2 (PCV2) subtypes PCV2a and PCV2b by prolonging PCV2 viremia and shedding. Vet Microbiol, 2011. 152(3-4): p. 235-46.
- Provost, C.; Hamonic, G.; Gagnon, C.A.; Meurens, F. Dual infections of CD163 expressing NPTr epithelial cells with influenza A virus and PRRSV. Veterinary microbiology, 2017. 207: p. 143-148.
- Dobrescu, I.; Levast, B.; Lai, K.; Delgado-Ortega, M.; Walker, S.; Banman, S.; Townsend, H.; Simon, G.; Zhou, Y.; Gerdts, V. In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses. Veterinary microbiology, 2014. 169(1-2): p. 18-32.
- Richmond, O.; Cecere, T.E.; Erdogan, E.; Meng, X.J.; Pineyro, P.; Subramaniam, S.; Todd, S.M.; LeRoith, T. The PD-L1/CD86 ratio is increased in dendritic cells co-infected with porcine circovirus type 2 and porcine reproductive and respiratory syndrome virus, and the PD-L1/PD-1 axis is associated with anergy, apoptosis, and the induction of regulatory T-cells in porcine lymphocytes. Vet Microbiol, 2015. 180(3-4): p. 223-9.
- Cecere, T.E.; Meng, X.J.; Pelzer, K.; Todd, S.M.; Beach, N.M.; Ni, Y.Y.; Leroith, T. Co-infection of porcine dendritic cells with porcine circovirus type 2a (PCV2a) and genotype II porcine reproductive and respiratory syndrome virus (PRRSV) induces CD4(+)CD25(+)FoxP3(+) T cells in vitro. Vet Microbiol, 2012. 160(1-2): p. 233-9.
- Dorr, P.M.; Baker, R.B.; Almond, G.W.; Wayne, S.R.; Gebreyes, W.A. Epidemiologic assessment of porcine circovirus type 2 coinfection with other pathogens in swine. J Am Vet Med Assoc 2007, 230, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Lenz, S.D.; Van Alstine, W.G.; Stevenson, G.W.; Langohr, I.M.; Pogranichniy, R.M. Infection of cesarean-derived colostrum-deprived pigs with porcine circovirus type 2 and Swine influenza virus. Comp Med 2010, 60, 45–50. [Google Scholar] [PubMed]
- Misinzo, G.; Meerts, P.; Bublot, M.; Mast, J.; Weingartl, H.M.; Nauwynck, H.J. Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31. J Gen Virol, 2005. 86(Pt 7): p. 2057-2068.
- Sanchez, R.E., Jr. Meerts, P.; Nauwynck, H.J.; Pensaert, M.B.; Change of porcine circovirus 2 target cells in pigs during development from fetal to early postnatal life. Vet Microbiol, 2003. 95(1-2): p. 15-25.
- Wu, N.-H.; Yang, W.; Beineke, A.; Dijkman, R.; Matrosovich, M.; Baumgärtner, W.; Thiel, V.; Valentin-Weigand, P.; Meng, F.; Herrler, G. The differentiated airway epithelium infected by influenza viruses maintains the barrier function despite a dramatic loss of ciliated cells. Scientific reports 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nicol, M.Q.; Dutia, B.M. The role of macrophages in influenza A virus infection. Future Virology 2014, 9, 847–862. [Google Scholar]
- Yu, W.C.; Chan, R.W.; Wang, J.; Travanty, E.A.; Nicholls, J.M.; Peiris, J.M.; Mason, R.J.; Chan, M.C. Viral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 viruses. Journal of virology 2011, 85, 6844–6855. [Google Scholar] [CrossRef] [PubMed]
- Günther, J. and Seyfert, H.-M. The first line of defence: insights into mechanisms and relevance of phagocytosis in epithelial cells. in Seminars in immunopathology. 2018. Springer .
- Vareille, M.; Kieninger, E.; Edwards, M.R.; Regamey, N. The airway epithelium: soldier in the fight against respiratory viruses. Clinical microbiology reviews 2011, 24, 210–229. [Google Scholar] [CrossRef] [PubMed]
- Cline, T.D.; Beck, D.; Bianchini, E. Influenza virus replication in macrophages: balancing protection and pathogenesis. The Journal of general virology 2017, 98, 2401. [Google Scholar] [CrossRef]
- Ferrari, M.; Scalvini, A.; Losio, M.N.; Corradi, A.; Soncini, M.; Bignotti, E.; Milanesi, E.; Ajmone-Marsan, P.; Barlati, S.; Bellotti, D.; et al. Establishment and characterization of two new pig cell lines for use in virological diagnostic laboratories. J Virol Methods 2003, 107, 205–212. [Google Scholar] [CrossRef]
- Kärber, G. Tabellen zur näherungsweisen Bestimmung von Titern. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 1931. 162: p. 480.
- Spearman, C. Review of The Method of ‘Right and Wrong Cases’(‘Constant Stimuli’) without Gauss’s Formula. Psychol Bull 1909, 6, 27–28. [Google Scholar]
- Racine, S.; Kheyar, A.; Gagnon, C.A.; Charbonneau, B.; Dea, S. Eucaryotic expression of the nucleocapsid protein gene of porcine circovirus type 2 and use of the protein in an indirect immunofluorescence assay for serological diagnosis of postweaning multisystemic wasting syndrome in pigs. Clin Diagn Lab Immunol 2004, 11, 736–741. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nature methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jakab, G.J. Immune impairment of alveolar macrophage phagocytosis during influenza virus pneumonia. American Review of Respiratory Disease 1982, 126, 778–782. [Google Scholar] [PubMed]
- Du, Q.; Huang, Y.; Wang, T.; Zhang, X.; Chen, Y.; Cui, B.; Li, D.; Zhao, X.; Zhang, W.; Chang, L.; et al. Porcine circovirus type 2 activates PI3K/Akt and p38 MAPK pathways to promote interleukin-10 production in macrophages via Cap interaction of gC1qR. Oncotarget 2016, 7, 17492–17507. [Google Scholar] [CrossRef]
- Liu, S.; Li, Q.; Qiao, J.; Wang, J.; Cui, D.; Gu, K.; Zhou, S.; Li, H. Endothelial IL-8 induced by porcine circovirus type 2 affects dendritic cell maturation and antigen-presenting function. Virol J 2019, 16, 154. [Google Scholar] [CrossRef]
- Gu, C.; Gao, X.; Guo, D.; Wang, J.; Wu, Q.; Nepovimova, E.; Wu, W.; Kuca, K. Combined Effect of Deoxynivalenol (DON) and Porcine Circovirus Type 2 (Pcv2) on Inflammatory Cytokine mRNA Expression. Toxins 2021, 13. [Google Scholar] [CrossRef]
- Yang, S.; Liu, B.; Yin, S.; Shang, Y.; Zhang, X.; Khan, M.U.Z.; Liu, X.; Cai, J. Porcine Circovirus Type 2 Induces Single Immunoglobulin Interleukin-1 Related Receptor (SIGIRR) Downregulation to Promote Interleukin-1beta Upregulation in Porcine Alveolar Macrophage. Viruses 2019, 11. [Google Scholar] [CrossRef]
- Wu, X.; Wang, X.; Shi, T.; Luo, L.; Qiao, D.; Wang, Z.; Han, C.; Du, Q.; Tong, D.; Huang, Y. Porcine Circovirus Type 2 Rep Enhances IL-10 Production in Macrophages via Activation of p38-MAPK Pathway. Viruses 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Jeng, C.R.; Lin, T.L.; Liu, J.J.; Chiou, M.T.; Tsai, Y.C.; Chia, M.Y.; Jan, T.R.; Pang, V.F. Immunopathological effects of porcine circovirus type 2 (PCV2) on swine alveolar macrophages by in vitro inoculation. Vet Immunol Immunopathol, 2006. 110(3-4): p. 207-19.
- Li, W.; Liu, S.; Wang, Y.; Deng, F.; Yan, W.; Yang, K.; Chen, H.; He, Q.; Charreyre, C.; Audoneet, J.C. Transcription analysis of the porcine alveolar macrophage response to porcine circovirus type 2. BMC Genomics, 2013. 14: p. 353.
- Van Reeth, K.; Van Gucht, S.; Pensaert, M. Correlations between lung proinflammatory cytokine levels, virus replication, and disease after swine influenza virus challenge of vaccination-immune pigs. Viral immunology 2002, 15, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, J.; Zhang, Q.-g.; Huang, K.; Ma, D.; Du, Q.; Tong, D.; Huang, Y. Porcine circovirus type 2 infection inhibits the activation of type I interferon signaling via capsid protein and host gC1qR. Veterinary Microbiology, 2022. 266: p. 109354.
- Hasslung, F.C.; Berg, M.; Allan, G.M.; Meehan, B.M.; McNeilly, F.; Fossum, C. Identification of a sequence from the genome of porcine circovirus type 2 with an inhibitory effect on IFN-alpha production by porcine PBMCs. J Gen Virol 2003, 84, 2937–2945. [Google Scholar] [CrossRef] [PubMed]
- Vincent, I.E.; Balmelli, C.; Meehan, B.; Allan, G.; Summerfield, A.; McCullough, K.C. Silencing of natural interferon producing cell activation by porcine circovirus type 2 DNA. Immunology 2007, 120, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.Y.; Choi, Y.C.; Park, I.B.; Lee, C.H.; Kang, S.J.; Chun, T. The ORF5 protein of porcine circovirus type 2 enhances viral replication by dampening type I interferon expression in porcine epithelial cells. Vet Microbiol, 2018, 226, 50-58.
- Li, J.; Lu, M.; Huang, B.; Lv, Y. Porcine circovirus type 2 inhibits interferon-β expression by targeting Karyopherin alpha-3 in PK-15 cells. Virology 2018, 520, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Xie, J.L.; Jia, C.W.; Ren, H.Y.; Zhou, S.H. Effects of porcine circovirus type 2 and pseudorabies vaccine co-inoculation on regulatory cytokine mRNA expression in pig peripheral blood mononuclear cells. Genet Mol Res 2014, 13, 1540–1547. [Google Scholar] [CrossRef]
- Chen, M.; Han, J.; Zhang, Y.; Duan, D.; Zhang, S. Porcine circovirus type 2 induces type I interferon production via MyD88–IKKα–IRFs signaling rather than NF-κB in porcine alveolar macrophages in vitro. Res. Vet. Sci. 2016, 104, 188–194. [Google Scholar] [CrossRef]
- Huang, B.; Zhang, L.; Lu, M.; Li, J.; Lv, Y. PCV2 infection activates the cGAS/STING signaling pathway to promote IFN-beta production and viral replication in PK-15 cells. Vet Microbiol 2018, 227, 34–40. [Google Scholar] [CrossRef]
- Wikstrom, F.H.; Meehan, B.M.; Berg, M.; Timmusk, S.; Elving, J.; Fuxler, L.; Magnusson, M.; Allan, G.M.; McNeilly, F.; Fossum, C. Structure-dependent modulation of alpha interferon production by porcine circovirus 2 oligodeoxyribonucleotide and CpG DNAs in porcine peripheral blood mononuclear cells. J. Virol. 2007, 81, 4919–4927. [Google Scholar] [CrossRef]
- Kekarainen, T.; Montoya, M.; Dominguez, J.; Mateu, E.; Segales, J. Porcine circovirus type 2 (PCV2) viral components immunomodulate recall antigen responses. Vet Immunol Immunopathol, 2008. 124(1-2), 41-9.
- Bakre, A.A.; Jones, L.P.; Murray, J.; Reneer, Z.B.; Meliopoulos, V.A.; Cherry, S.; Schultz-Cherry, S.; Tripp, R.A. Innate Antiviral Cytokine Response to Swine Influenza Virus by Swine Respiratory Epithelial Cells. J. Virol. 2021, 95, e00692-21. [Google Scholar] [CrossRef]
- Crosse, K.M.; Monson, E.A.; Beard, M.R.; Helbig, K.J. Interferon-stimulated genes as enhancers of antiviral innate immune signaling. J. Innate Immun. 2018, 10, 85–93. [Google Scholar] [CrossRef] [PubMed]
- De Veer, M.J.; Holko, M.; Frevel, M.; Walker, E.; Der, S.; Paranjape, J.M.; Silverman, R.H.; Williams, B.R. Functional classification of interferon-stimulated genes identified using microarrays. Journal of leukocyte biology 2001, 69, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Current opinion in virology 2011, 1, 519–525. [Google Scholar]
- Wu, W.; Wang, C.; Xia, C.; Liu, S.; Mei, Q. MicroRNA let-7 Suppresses Influenza A Virus Infection by Targeting RPS16 and Enhancing Type I Interferon Response. Front. Cell. Infect. Microbiol. 2022, 937. [Google Scholar] [CrossRef]
- Fong, C.H.-Y.; Lu, L.; Chen, L.-L.; Yeung, M.-L.; Zhang, A.J.; Zhao, H.; Yuen, K.-Y.; To, K.K.-W. Interferon-gamma inhibits influenza A virus cellular attachment by reducing sialic acid cluster size. Iscience 2022, 25, 104037. [Google Scholar] [CrossRef]
- Garcia-Sastre, A. Induction and evasion of type I interferon responses by influenza viruses. Virus research 2011, 162, 12–18. [Google Scholar] [CrossRef]
- Pauli, E.-K.; Schmolke, M.; Wolff, T.; Viemann, D.; Roth, J.; Bode, J.G.; Ludwig, S. Influenza A virus inhibits type I IFN signaling via NF-κB-dependent induction of SOCS-3 expression. PLoS pathogens 2008, 4, e1000196. [Google Scholar] [CrossRef]
- Czerkies, M.; Kochańczyk, M.; Korwek, Z.; Prus, W.; Lipniacki, T. RSV protects bystander cells against IAV infection by triggering secretion of type I and type III interferons. bioRxiv 2022, 11, 463877. [Google Scholar]
- Gaur, P.; Munjal, A.; Lal, S.K. Influenza virus and cell signaling pathways. Med. Sci. Monit. : Int. Med. J. Exp. Clin. Res. 2011, 17, RA148. [Google Scholar] [CrossRef]
- Kanehisa, M. and Goto, S.; KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Coutermarsh-Ott, S.; Eden, K.; Allen, I.C. Beyond the inflammasome: regulatory NOD-like receptor modulation of the host immune response following virus exposure. J. Gen. Virol. 2016, 97 Pt 4, 825. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.P.; Zhou, Y. Influenza A Virus Infection Activates NLRP3 Inflammasome through Trans-Golgi Network Dispersion. Viruses 2022, 14, 88. [Google Scholar] [PubMed]
- Tate, M.D.; Mansell, A. An update on the NLRP3 inflammasome and influenza: the road to redemption or perdition? Current opinion in immunology 2018, 54, 80–85. [Google Scholar]
- Malik, G.; Zhou, Y. Innate immune sensing of influenza A virus. Viruses 2020, 12, 755. [Google Scholar] [PubMed]
- Tate, M.D.; Ong, J.D.; Dowling, J.K.; McAuley, J.L.; Robertson, A.B.; Latz, E.; Drummond, G.R.; Cooper, M.A.; Hertzog, P.J.; Mansell, A. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Scientific reports 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kesavardhana, S.; Samir, P.; Zheng, M.; Malireddi, R.S.; Karki, R.; Sharma, B.R.; Place, D.E.; Briard, B.; Vogel, P.; Kanneganti, T.-D. DDX3X coordinates host defense against influenza virus by activating the NLRP3 inflammasome and type I interferon response. Journal of Biological Chemistry, 2021. 296.
- Thulasi Raman, S.N.; Liu, G.; Pyo, H.M.; Cui, Y.C.; Xu, F.; Ayalew, L.E.; Tikoo, S.K.; Zhou, Y. DDX3 interacts with influenza A virus NS1 and NP proteins and exerts antiviral function through regulation of stress granule formation. Journal of Virology 2016, 90, 3661–3675. [Google Scholar] [CrossRef] [PubMed]
- Wurzer, W.J.; Planz, O.; Ehrhardt, C.; Giner, M.; Silberzahn, T.; Pleschka, S.; Ludwig, S. Caspase 3 activation is essential for efficient influenza virus propagation. The EMBO journal 2003, 22, 2717–2728. [Google Scholar] [CrossRef]
- Kumar, R.; Khandelwal, N.; Thachamvally, R.; Tripathi, B.N.; Barua, S.; Kashyap, S.K.; Maherchandani, S.; Kumar, N. Role of MAPK/MNK1 signaling in virus replication. Virus research, 2018. 253, 48-61.
- Reyes, A.; Corrales, N.; Gálvez, N.M.; Bueno, S.M.; Kalergis, A.M.; González, P.A. Contribution of hypoxia inducible factor-1 during viral infections. Virulence 2020, 11, 1482–1500. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, W.; Zhang, J.; Zhang, J.; Zhang, H.; Zhu, Y.; Meng, X.; Yi, Z.; Wang, R. Influenza A virus (H1N1) infection induces glycolysis to facilitate viral replication. Virologica Sinica 2021, 36, 1532–1542. [Google Scholar] [CrossRef]
- Zhao, C.; Chen, J.; Cheng, L.; Xu, K.; Yang, Y.; Su, X. Deficiency of HIF-1α enhances influenza A virus replication by promoting autophagy in alveolar type II epithelial cells. Emerging Microbes Infections 2020, 9, 691–706. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




