Submitted:
03 April 2023
Posted:
18 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
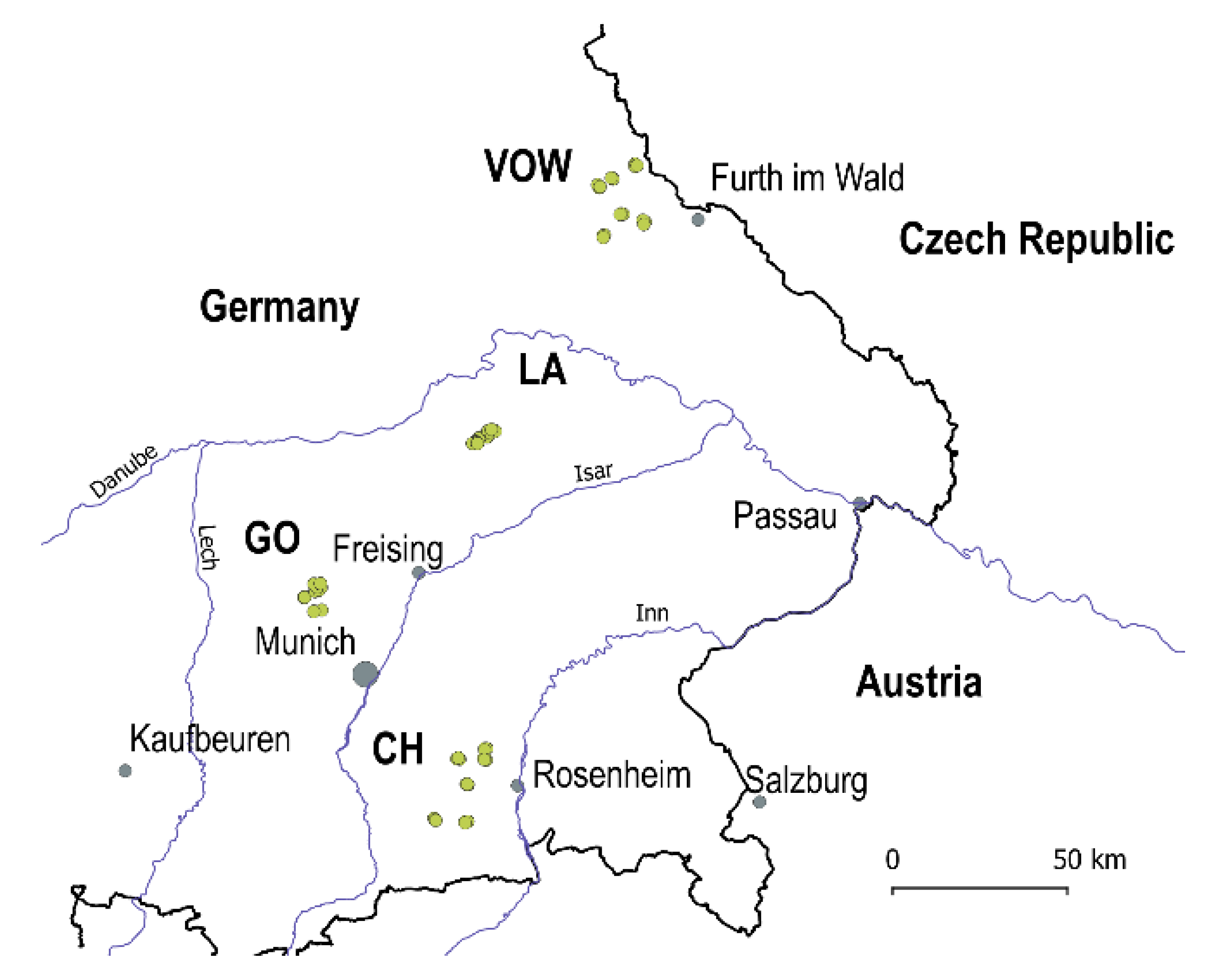
2.1. Study regions and insect collections
2.2. DNA extraction, metabarcoding and insect identification
2.3. Phytoplasma detection and classification
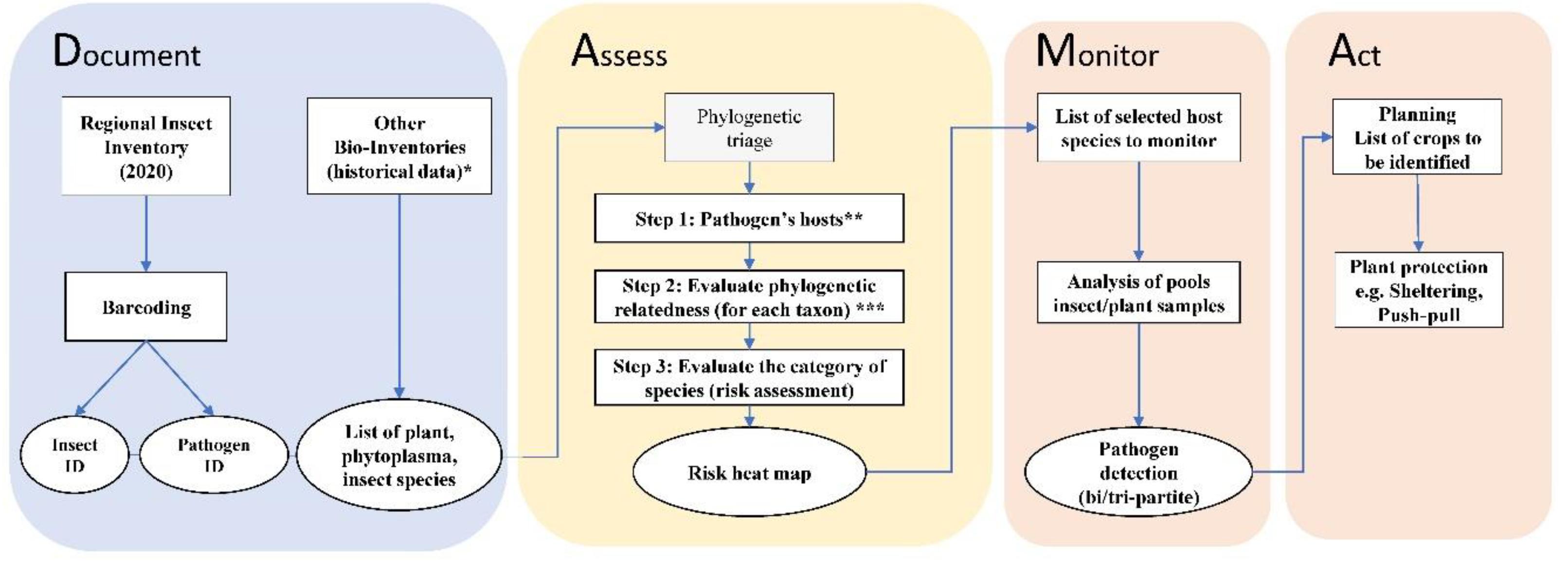
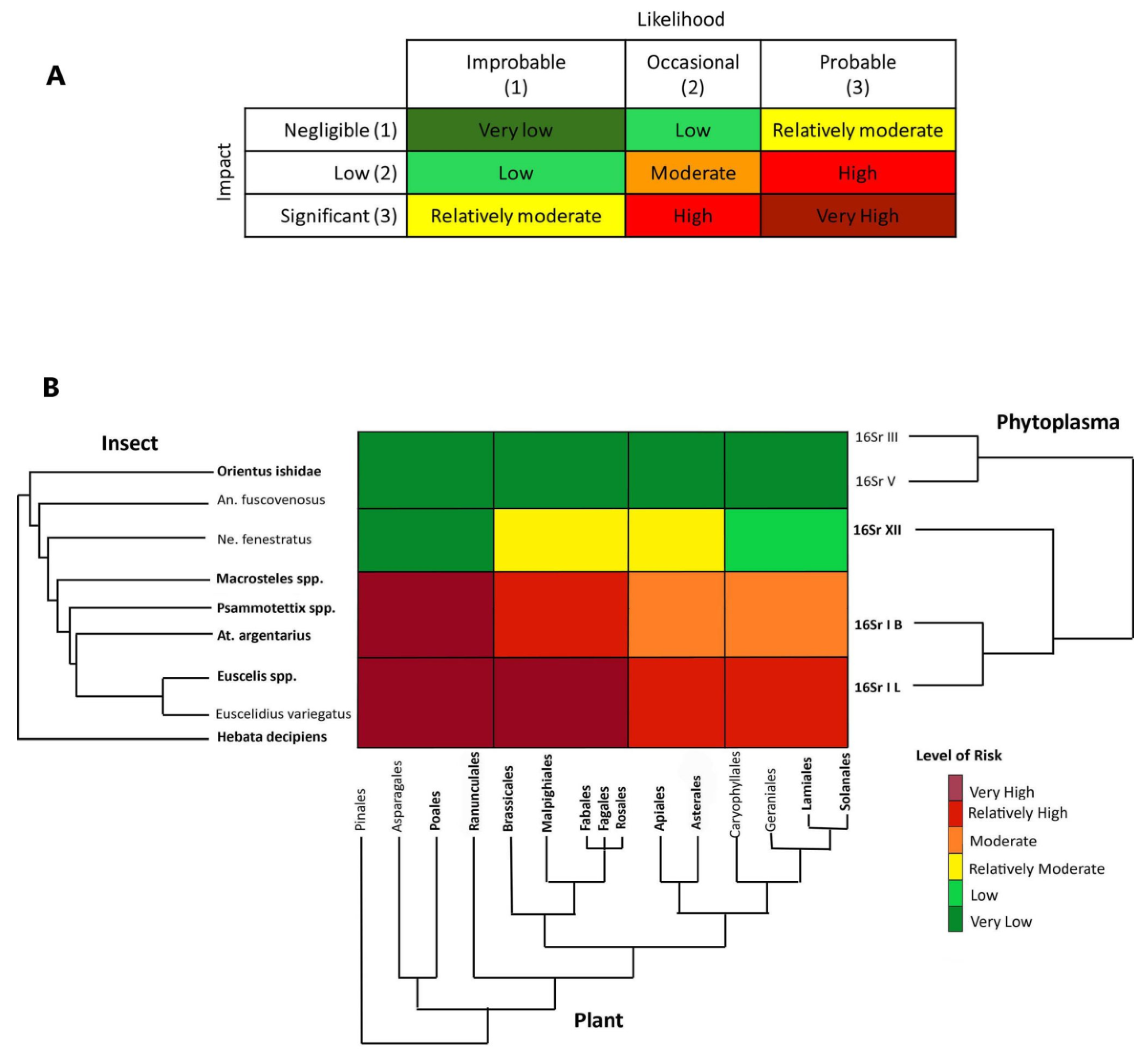
2.4. DAMA protocol, phylogenetic triage and risk assessment
3. Results
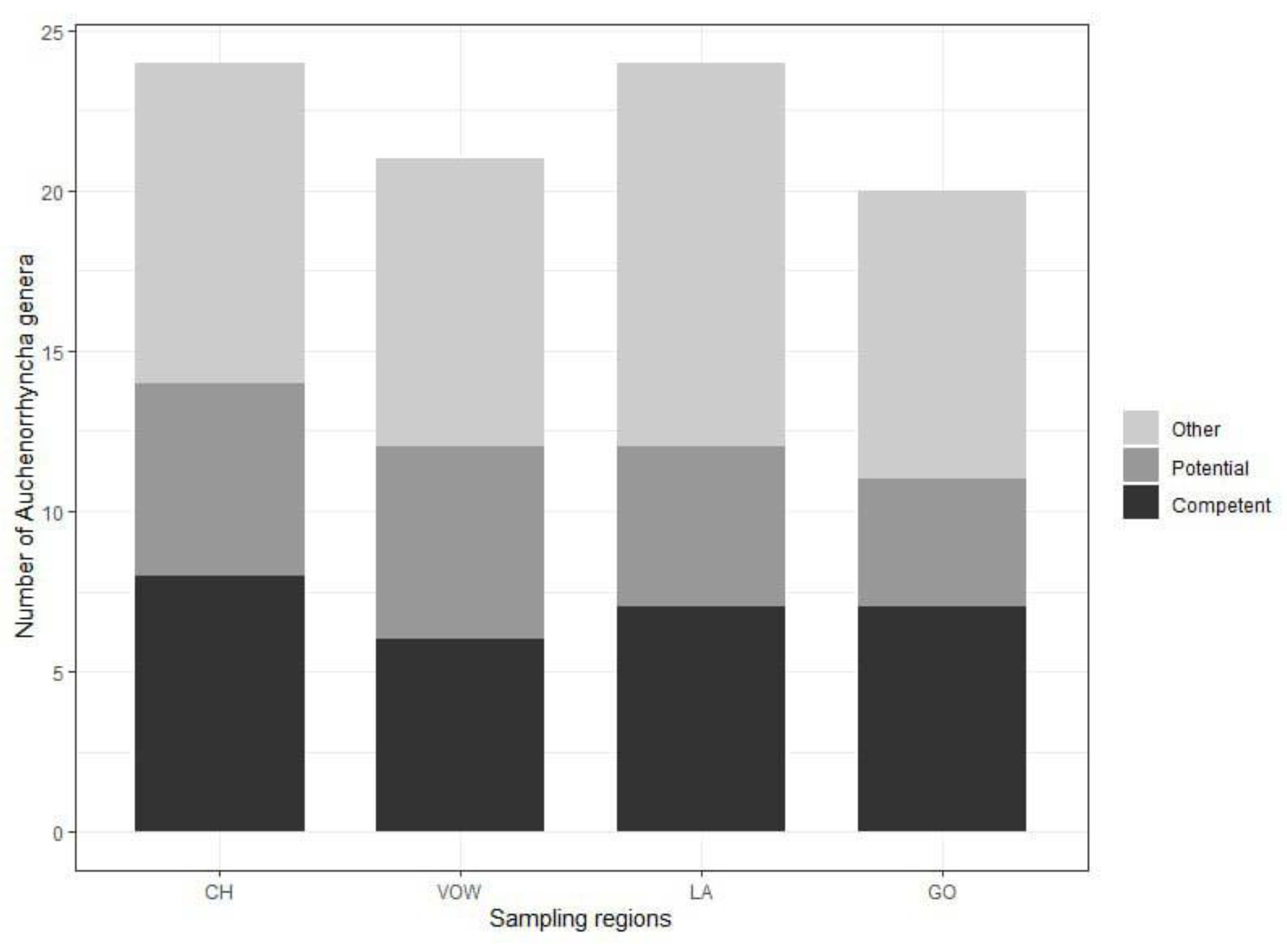
3.1. Metabarcoding insect identification
3.2. Detection of phytoplasmas in the study region
3.3. Phytoplasma-host associations and phylogenetic triage
| Subfamily | Tribe | Species | Habitat | Host Plant | Bavarian region / Germany | RII 2020 |
|---|---|---|---|---|---|---|
| Typhlocybinae | Empoascini | Hebata decipiens* | ruderal areas, fields | weeds, shrubs, various crops | W/W | CH, VOW, GO, LA |
| Deltocephalinae | Athysanini | Euscelidius variegatus* | ruderal areas | forbs | W/W | - |
| Deltocephalinae | Athysanini | Euscelis incisa* | meadows, pastures | grasses and legumes | W/W | GO |
| Deltocephalinae | Athysanini | Euscelis lineolata* | meadows, pastures | grasses and legumes | -/Un | GO |
| Deltocephalinae | Macrostelini | Macrosteles quadripunctulatus* | sandy areas and viticultural region | grasses | Un/Un | CH, VOW, GO, LA |
| Deltocephalinae | Macrostelini | Macrosteles laevis* | ruderal areas | grasses and forbs | W/W | CH, VOW, GO, LA |
| Deltocephalinae | Opsiini | Neoaliturus fenestratus* | dry meadows, disturbed areas, fields | forbs | Un/W | - |
| Deltocephalinae | Paralimnini | Psammotettix alienus* | meadows, disturbed areas, fields | grasses | W/W | CH, GO, LA |
| Deltocephalinae | Athysanini | Athysanus argentarius* | meadows, fields, open forests | grasses | W/W | GO |
| Deltocephalinae | Athysanini | Hardya tenuis* | forest interfaces, grassland | trees and grasses | -/Un | - |
| Deltocephalinae | Paralimnini | Adarrus taurus* | - | - | NR/NR | - |
| Deltocephalinae | Scaphoideini | Osbornellus horvathi* | disturbed areas, fields | forbs and various shrubs | NR/NR | - |
| Deltocephalinae | Scaphoideini | Scaphoideus titanus* | disturbed areas, fields | Vitis | NR/NR | - |
| Deltocephalinae | Scaphoideini | Anoplotettix fuscovenosus** | forest margins, fields | forbs and various shrubs | W/W | - |
| Deltocephalinae | Athysanini | Orientus ishidae** | forest margins, fields | woody and deciduous trees | W/W | CH, GO |
| Typhlocybinae | Erythroneurini | Zyginidia scutellaris** | ruderal, disturbed grassland, maize, winter cereals | grasses | Un/Un | - |
| Deltocephalinae | Athysanini | Laylatina inexpectata** | - | - | NR/NR | - |
| Deltocephalinae | Athysanini | Thamnotettix dilutior** | forests | trees and grasses | NR/W | - |
4. Discussion
4.1. Why do we need a proactive approach for phytoplasma diseases?
4.2. Applying the DAMA protocol to reduce the costs of potential EIPD
4.3. Economic importance of aster yellows phytoplasma in the study region
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brooks, D.R.; Hoberg, E.P.; Boeger, W.A.; Trivellone, V. Emerging Infectious Disease: An Underappreciated Area of Strategic Concern for Food Security. Transbound Emerg Dis 2021, tbed.14009. [CrossRef]
- Trivellone, V.; Hoberg, E.P.; Boeger, W.A.; Brooks, D.R. Food Security and Emerging Infectious Disease: Risk Assessment and Risk Management. R. Soc. open sci. 2022, 9, 211687. [Google Scholar] [CrossRef] [PubMed]
- Trivellone, V. Let Emerging Plant Diseases Be Predictable. MANTER: Journal of Parasite Biodiversity 2022, 30, 1–8. [Google Scholar] [CrossRef]
- Agosta, S.J. The Stockholm Paradigm Explains the Dynamics of Darwin’s Entangled Bank, Including Emerging Infectious Disease. MANTER: Journal of Parasite Biodiversity 2022, 27, 1–17. [Google Scholar] [CrossRef]
- Molnár, O.; Hoberg, E.; Trivellone, V.; Földvári, G.; Brooks, D.R. The 3P Framework: A Comprehensive Approach to Coping with the Emerging Infectious Disease Crisis. MANTER: Journal of Parasite Biodiversity 2022, 23, 1–14. [Google Scholar] [CrossRef]
- Brooks, D.R.; Hoberg, E.P.; Boeger, W.A. The Stockholm Paradigm: Climate Change and Emerging Disease; The University of Chicago Press: Chicago, 2019; ISBN 978-0-226-63230-8. [Google Scholar]
- Hoberg, E.P.; Boeger, W.A.; Molnár, O.; Gábor, F.; Gardner, S.; Juarrero, A.; Kharchenko, V.A.; Ortiz, E.; Trivellone, V.; Brooks, D.R. The DAMA Protocol, an Introduction: Finding Pathogens before They Find Us. MANTER: Journal of Parasite Biodiversity 2022, 21, 1–20. [Google Scholar] [CrossRef]
- Molnár, O.; Knickel, M.; Marizzi, C. Taking Action: Turning Evolutionary Theory into Preventive Taking Action: Turning Evolutionary Theory into Preventive Policies Policies. MANTER: Journal of Parasite Biodiversity 2022, 28, 1–20. [Google Scholar] [CrossRef]
- Brooks, D.R.; Hoberg, E.P.; Boeger, W.A.; Gardner, S.L.; Galbreath, K.E.; Herczeg, D.; Mejía-Madrid, H.H.; Rácz, S.E.; Dursahinhan, A.T. Finding Them Before They Find Us: Informatics, Parasites, and Environments in Accelerating Climate Change. Comparative Parasitology 2014, 81, 155–164. [Google Scholar] [CrossRef]
- Földvári, G.; Jahfari, S.; Rigó, K.; Jablonszky, M.; Szekeres, S.; Majoros, G.; Tóth, M.; Molnár, V.; Coipan, E.C.; Sprong, H. Candidatus Neoehrlichia Mikurensis and Anaplasma Phagocytophilum in Urban Hedgehogs. Emerging Infectious Diseases 2014, 20, 496–498. [Google Scholar] [CrossRef]
- Földvári, G.; Rigó, K.; Jablonszky, M.; Biró, N.; Majoros, G.; Molnár, V.; Tóth, M. Ticks and the City: Ectoparasites of the Northern White-Breasted Hedgehog (Erinaceus Roumanicus) in an Urban Park. Ticks and Tick-borne Diseases 2011, 2, 231–234. [Google Scholar] [CrossRef]
- Szekeres, S.; Docters van Leeuwen, A.; Tóth, E.; Majoros, G.; Sprong, H.; Földvári, G. Road-killed Mammals Provide Insight into Tick-borne Bacterial Pathogen Communities within Urban Habitats. Transbound Emerg Dis 2019, 66, 277–286. [Google Scholar] [CrossRef]
- Brooks, D.R.; Boeger, W.A.; Hoberg, E. The Stockholm Paradigm: Lessons for the Emerging Infectious The Stockholm Paradigm: Lessons for the Emerging Infectious Disease Crisis Disease Crisis. MANTER: Journal of Parasite Biodiversity 2022, 22, 1–10. [Google Scholar] [CrossRef]
- Hoberg, E.P.; Trivellone, V.; Cook, J.; Dunnum, J.L.; Boeger, W.A.; Brooks, D.R.; Agosta, S.J.; Colella, J.C. Knowing the Biosphere: Documentation, Specimens, Archives, and Names Reveal Environmental Change and Emerging Pathogens. MANTER: Journal of Parasite Biodiversity 2022, 26. [Google Scholar] [CrossRef]
- Bertaccini, A.; Duduk, B.; Paltrinieri, S.; Contaldo, N. Phytoplasmas and Phytoplasma Diseases: A Severe Threat to Agriculture. AJPS 2014, 05, 1763–1788. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Trivellone, V.; Krüger, K. The Biology and Ecology of Leafhopper Transmission of Phytoplasmas. In Phytoplasmas: Plant Pathogenic Bacteria - II: Transmission and Management of Phytoplasma - Associated Diseases; Bertaccini, A., Weintraub, P.G., Rao, G.P., Mori, N., Eds.; Springer: Singapore, 2019; pp. 27–51. ISBN 9789811328329. [Google Scholar]
- Trivellone, V.; Wei, W.; Filippin, L.; Dietrich, C.H. Screening Potential Insect Vectors in a Museum Biorepository Reveals Undiscovered Diversity of Plant Pathogens in Natural Areas. Ecol. Evol. 2021, 11, 6493–6503. [Google Scholar] [CrossRef] [PubMed]
- Trivellone, V.; Cao, Y.; Dietrich, C.H. Comparison of Traditional and Next-Generation Approaches for Uncovering Phytoplasma Diversity, with Discovery of New Groups, Subgroups and Potential Vectors. Biology 2022, 11, 977. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Trivellone, V.; Dietrich, C.H.; Zhao, Y.; Bottner-Parker, K.D.; Ivanauskas, A. Identification of Phytoplasmas Representing Multiple New Genetic Lineages from Phloem-Feeding Leafhoppers Highlights the Diversity of Phytoplasmas and Their Potential Vectors. Pathogens 2021, 10, 352. [Google Scholar] [CrossRef] [PubMed]
- Agosta, S.J. On Ecological Fitting, Plant–Insect Associations, Herbivore Host Shifts, and Host Plant Selection. Oikos 2006, 114, 556–565. [Google Scholar] [CrossRef]
- Agosta, S.J.; Janz, N.; Brooks, D.R. How Specialists Can Be Generalists: Resolving the “Parasite Paradox” and Implications for Emerging Infectious Disease. Zoologia (Curitiba) 2010, 27, 151–162. [Google Scholar] [CrossRef]
- Hallmann, C.A.; Sorg, M.; Jongejans, E.; Siepel, H.; Hofland, N.; Schwan, H.; Stenmans, W.; Müller, A.; Sumser, H.; Hörren, T.; et al. More than 75 Percent Decline over 27 Years in Total Flying Insect Biomass in Protected Areas. PLoS ONE 2017, 12, e0185809. [Google Scholar] [CrossRef]
- Ronquist, F. 250 Years of Swedish Taxonomy. In Systema Naturae 250 - The Linnaean Ark; Polaszek, A., Ed.; 2010; pp. 241–252.
- Uhler, J.; Redlich, S.; Zhang, J.; Hothorn, T.; Tobisch, C.; Ewald, J.; Thorn, S.; Seibold, S.; Mitesser, O.; Morinière, J.; et al. Relationship of Insect Biomass and Richness with Land Use along a Climate Gradient. Nat Commun 2021, 12, 5946. [Google Scholar] [CrossRef]
- Ssymank, A.; Sorg, M.; Doczkal, D.; Rulik, B.; Merkel-Wallner, G.; Vischer-Leopold, M. Praktische Hinweise Und Empfehlungen Zur Anwendung von Malaisefallen Für Insekten in Der Biodiversitätserfassung Und Im Monitoring; Entomologischer Verein Krefeld, 2018;
- Bioform Malaise Trap Type Bartak Supplied by the Company Bioform. Item No: A84b. Available online: https://www.bioform.de/shop.php?action=tree&wg=1&pid=706&treeid=225 (accessed on 18 November 2022).
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A New Versatile Primer Set Targeting a Short Fragment of the Mitochondrial COI Region for Metabarcoding Metazoan Diversity: Application for Characterizing Coral Reef Fish Gut Contents. Front Zool 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Morinière, J.; Cancian de Araujo, B.; Lam, A.W.; Hausmann, A.; Balke, M.; Schmidt, S.; Hendrich, L.; Doczkal, D.; Fartmann, B.; Arvidsson, S.; et al. Species Identification in Malaise Trap Samples by DNA Barcoding Based on NGS Technologies and a Scoring Matrix. PLoS ONE 2016, 11, e0155497. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet j. 2011, 17, 10. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BARCODING: Bold: The Barcode of Life Data System (Http://Www.Barcodinglife.Org): BARCODING. Molecular Ecology Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Morinière, J.; Balke, M.; Doczkal, D.; Geiger, M.F.; Hardulak, L.A.; Haszprunar, G.; Hausmann, A.; Hendrich, L.; Regalado, L.; Rulik, B.; et al. A DNA Barcode Library for 5,200 German Flies and Midges (Insecta: Diptera) and Its Implications for Metabarcoding-based Biomonitoring. Mol Ecol Resour 2019, 19, 900–928. [Google Scholar] [CrossRef] [PubMed]
- NCBI National Center for Biotechnology Information (NCBI). Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 29 November 2022).
- Porter, T.M.; Hajibabaei, M. Automated High Throughput Animal CO1 Metabarcode Classification. Sci Rep 2018, 8, 4226. [Google Scholar] [CrossRef]
- Burmeister, J.; Panassiti, B. Sample Species Richness Accounting for Different Determination Depth; figshare, 2022; p. 13656 Bytes.
- Gundersen, D.E.; Lee, I.-M. Ultrasensitive Detection of Phytoplasmas by Nested-PCR Assays Using Two Universal Primer Pairs. Phytopathologia Mediterranea 1996, 35, 144–151. [Google Scholar]
- Zhao, Y.; Wei, W.; Lee, I.-M.; Shao, J.; Suo, X.; Davis, R.E. Construction of an Interactive Online Phytoplasma Classification Tool, IPhyClassifier, and Its Application in Analysis of the Peach X-Disease Phytoplasma Group (16SrIII). Int J Syst Evol Microbiol 2009, 59, 2582–2593. [Google Scholar] [CrossRef]
- BIB Botanischer Informationsknoten Bayern - Online Database of Bavarian Flora Available online:. Available online: https://daten.bayernflora.de/ (accessed on 18 November 2022).
- Bayerisches Landesamt für Statistik Getreide Auf Über Der Hälfte Des Ackerlandes in Bayern Available online:. Available online: https://www.statistik.bayern.de/presse/mitteilungen/2022/pm116/ (accessed on 26 November 2022).
- Biedermann, R.; Niedringhaus, R. The Plant-and Leafhoppers of Germany: Identification Key to All Species.; WABV Fründ, 2009.
- Nickel, H. The Leafhoppers and Planthoppers of Germany (Hemiptera Auchenorrhyncha): Patterns and Strategies in a Highly Diverse Group of Phytophageous Insects.; Pensoft Publishers, Sofia-Moscow and Goecke & Evers, Keltern, 2003; Vol. 50.
- Nickel, H.; Remane, R. Artenliste Der Zikaden Deutschlands, Mit Angabe von Nährpflanzen, Nahrungsbreite, Lebenszyklus, Areal Und Gefährdung (Hemiptera, Fulgoromorpha et Cicadomorpha). Beiträge zur Zikadenkunde 2002, 5, 27–64. [Google Scholar]
- Trivellone, V. An Online Global Database of Hemiptera-Phytoplasma-Plant Biological Interactions. Biodivers Data J 2019. [Google Scholar] [CrossRef] [PubMed]
- QGIS QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundatio Project. 2019. Available online: http://qgis.osgeo.org (accessed on 29 November 2022).
- Trivellone, V. Hemiptera-Phytoplasma-Plant Database Links Available online:. Available online: http://trivellone.speciesfile.org/ (accessed on 15 March 2020).
- Cao, Y.; Dietrich, C.H.; Zahniser, J.N.; Dmitriev, D.A. Dense Sampling of Taxa and Characters Improves Phylogenetic Resolution among Deltocephaline Leafhoppers (Hemiptera: Cicadellidae: Deltocephalinae). Systematic Entomology 2022, 47, 430–444. [Google Scholar] [CrossRef]
- Kumar, S.; Suleski, M.; Craig, J.M.; Kasprowicz, A.E.; Sanderford, M.; Li, M.; Stecher, G.; Hedges, S.B. TimeTree 5: An Expanded Resource for Species Divergence Times. Molecular Biology and Evolution 2022, 39, msac174. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Trivellone, V.; Dietrich, C.H. A Timetree for Phytoplasmas (Mollicutes) with New Insights on Patterns of Evolution and Diversification. Molecular Phylogenetics and Evolution 2020, 149, 106826. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.; Kätzel, R.; Kube, M. Widespread Occurrence of ‘Candidatus Phytoplasma Ulmi’ in Elm Species in Germany. BMC Microbiol 2020, 20, 74. [Google Scholar] [CrossRef]
- Schneider, B.; Seemüller, E. Strain Differentiation of Candidatus Phytoplasma Mali by SSCP-and Sequence Analyses of the HflB Gene. Journal of Plant Pathology 2009, 91, 103–112. [Google Scholar] [CrossRef]
- Bavarian State Research Center for Agriculture Jahresbericht 2020 Institut Für Pflanzenschutz; Bavarian State Research Center for Agriculture [Bayerische Landesanstalt fur Landwirtschaft], 2020.
- Monien, S.; Willmer, C.; Kaland, B. Monitoring of Cacopsylla Species in Schleswig-Holstein 2009-2011. Julius-Kühn-Archiv 2012, 438, 354. [Google Scholar]
- Heinze, K.; Kunze, L. Die Europaische Asterngelbsucht Und Ihre Ubertragung Durch Zwergzikaden. Nachrichtenblatt des deutschen Pflanzenschutzdienstes 1955, 7, 161–164. [Google Scholar]
- Seemüller, E.; Marcone, C.; Lauer, U.; Ragozzino, A.; Göschl, M. Current Status of Molecular Classification of the Phytoplasmas. Journal of Plant Pathology 1998, 24. [Google Scholar]
- Berges, R.; Cousin, M.-T.; Roux, J.; Mäurer, R.; Seemüller, E. Detection of Phytoplasma Infections in Declining Populus Nigra ‘Italica’ Trees and Molecular Differentiation of the Aster Yellows Phytoplasmas Identified in Various Populus Species. European Journal of Forest Pathology 1997, 27, 33–43. [Google Scholar] [CrossRef]
- Ipach, U.; Klingl, L.; Maixner, M. First Occurrence of Aster Yellows Disease on Grapevine in the Palatinate Area. In Proceedings of the 57 Deutsche Pflanzenschutztagung; Julius Kühn-Archiv: Berlin, Germany, 2010; Vol. 428, pp. 315–316. [Google Scholar]
- Lee, I.-M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bottner, K.D.; Marcone, C.; Seemüller, E. “Candidatus Phytoplasma Asteris”, a Novel Phytoplasma Taxon Associated with Aster Yellows and Related Diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Maixner, M. Comparative Experimental Transmission of Grapevine Yellows Phytoplasmas to Plants and Artificial Feeding Medium. In Proceedings of the Proceedings of the 14th Conference of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine; Locorotondo, Italy, 2003; pp. 109–110.
- Lederer, W.; Seemüller, E. Occurrence of Mycoplasma-like Organisms in Diseased and Non-Symptomatic Alder Trees (Alnus Spp.). European Journal of Forest Pathology 1991, 21, 90–96. [Google Scholar] [CrossRef]
- Maixner, M.; Reinert, W. Oncopsis Alni (Schrank) (Auchenorrhyncha: Cicadellidae) as a Vector of the Alder Yellows Phytoplasma of Alnus Glutinosa (L.) Gaertn. European Journal of Plant Pathology 1999, 105, 87–94. [Google Scholar] [CrossRef]
- Maixner, M.; Reinert, W.; Darimont, H. Transmission of Grapevine Yellows by Oncopsis Alni (Schrank) (Auchenorrhyncha: Macropsinae). VITIS - Journal of Grapevine Research 2000, 39, 83–83. [Google Scholar] [CrossRef]
- Maurer, R.; Seemuller, E. Nature and Genetic Relatedness of the Mycoplasma-like Organism Causing Rubus Stunt in Europe. Plant Pathology 1994, 44, 244–249. [Google Scholar] [CrossRef]
- Krczal, G.; Krczal, H.; Kunze, L. Fieberiella Florii (Stal), a Vector of Apple Proliferation Agent.; Acta Horticulturae (Wageningen): Salonika, Greece, June 12-18, 1988, 1989; Vol. 235, pp. 99–106.
- Jarausch, B.; Schwind, N.; Jarausch, W.; Krczal, G.; Dickler, E.; Seemüller, E. First Report of Cacopsylla Picta as a Vector of Apple Proliferation Phytoplasma in Germany. Plant Dis 2003, 87, 101. [Google Scholar] [CrossRef] [PubMed]
- Jarausch, B.; Fuchs, A.; Muhlenz, I.; Lampe, I.; Harzer, U.; Jarausch, W. Research on European Stone Fruit Yellows (ESFY) in Germany. Bulletin of Insectology 2007, 60, 389–390. [Google Scholar]
- Jarausch, B.; Maixner, M.; Kehrli, P.; Schaerer, S. Presence and Distribution of Phytoplasma Diseases and Vectors in Germany and Switzerland – Current State of the Art. In Proceedings of the Integrated Management of Phytoplasma Epidemics in different crop systems; Portugal, Lisbon; 2013; pp. 6–7. [Google Scholar]
- Seemüller, E.; Schneider, B.; Maurer, R.; Ahrens, U.; Daire, X.; Kison, H.; Lorenz, K.-H.; Firrao, G.; Avinent, L.; Sears, B.B.; et al. Phylogenetic Classification of Phytopathogenic Mollicutes by Sequence Analysis of 16S Ribosomal DNA. Int. J. Syst. Bacteriol. 1994, 44, 440–446. [Google Scholar] [CrossRef]
- Langer, M.; Maixner, M. Molecular Characterisation of Grapevine Yellows Associated Phytoplasmas of the Stolbur-Group Based on RFLP-Analysis of Non-Ribosomal DNA. VITIS - Journal of Grapevine Research 2004, 43, 191–191. [Google Scholar] [CrossRef]
- Schneider, B.; Ahrens, U.; Kirkpatrick, B.C.; SeemüLler, E. 1993 Classification of Plant-Pathogenic Mycoplasma-like Organisms Using Restriction-Site Analysis of PCR-Amplified 16S RDNA. Microbiology 1993, 139, 519–527. [Google Scholar] [CrossRef]
- Darimont, H.; Maixner, M. Actual Distribution of Hyalesthes Obsoletus Signoret (Auchenorrhyncha: Cixiidae) in German Viticulture and Its Significance as a Vector of Bois Noir.; 2001; Vol. 24, pp. 199–202.
- Marcone, C.; Gibb, K.S.; Streten, C.; Schneider, B. “Candidatus Phytoplasma Spartii”, “Candidatus Phytoplasma Rhamni” and “Candidatus Phytoplasma Allocasuarinae”, Respectively Associated with Spartium Witches’-Broom, Buckthorn Witches’-Broom and Allocasuarina Yellows Diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.; Torres, E.; Martin, M.P.; Schröder, M.; Behnke, H.-D.; Seemüller, E. “Candidatus Phytoplasma Pini”, a Novel Taxon from Pinus Silvestris and Pinus Halepensis. Int. J. Syst. Evol. Microbiol. 2005, 55, 303–307. [Google Scholar] [CrossRef]
- Panassiti, B.; Hartig, F.; Breuer, M.; Biedermann, R. Bayesian Inference of Environmental and Biotic Factors Determining the Occurrence of the Grapevine Disease ‘bois Noir’. Ecosphere 2015, 6, art143. [Google Scholar] [CrossRef]
- Weber, A.; Maixner, M. Survey of Populations of the Planthopper Hyalesthes Obsoletus Sign. (Auchenorrhyncha, Cixiidae) for Infection with the Phytoplasma Causing Grapevine Yellows in Germany. Journal of Applied Entomology 1998, 122, 375–381. [Google Scholar] [CrossRef]
- Araujo, S.B.L.; Braga, M.P.; Brooks, D.R.; Agosta, S.J.; Hoberg, E.P.; von Hartenthal, F.W.; Boeger, W.A. Understanding Host-Switching by Ecological Fitting. PLOS ONE 2015, 10, e0139225. [Google Scholar] [CrossRef] [PubMed]
- Hoberg, E.P.; Brooks, D.R. Evolution in Action: Climate Change, Biodiversity Dynamics and Emerging Infectious Disease. Philos Trans R Soc Lond B Biol Sci 2015, 370, 20130553. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H. On Ecological Fitting. Oikos 1985, 45, 308–310. [Google Scholar] [CrossRef]
- Yan, B.; Dietrich, C.H.; Yu, X.; Jiao, M.; Dai, R.; Yang, M. Mitogenomic Phylogeny of Typhlocybinae (Hemiptera: Cicadellidae) Reveals Homoplasy in Tribal Diagnostic Morphological Traits. Ecology and Evolution 2022, 12, e8982. [Google Scholar] [CrossRef]
- Brooks, D.R. Historical Ecology: A New Approach to Studying the Evolution of Ecological Associations. Annals of the Missouri Botanical Garden 1985, 72, 660–680. [Google Scholar] [CrossRef]
- Mitrovic, M.; Trivellone, V.; Cvrkovic, T.; Jakovljevic, M.; Krstic, O.; Jovic, J.; Toševski, I. Experimental and Molecular Evidence of Neoaliturus Fenestratus Role in the Transmission of “Stolbur” Phytoplasma to Lettuce and Carrot Plants. Phyt. Moll. 2019, 9, 109. [Google Scholar] [CrossRef]
- Al-Habshy, A.; Amer, S.; Hashem, M. Population Fluctuations of Aphid and Leafhopper Insects Infesting Barely Plants and Some Predators, Egypt. Journal of Plant Protection and Pathology 2019, 10, 451–457. [Google Scholar] [CrossRef]
- Galetto, L.; Marzachì, C.; Demichelis, S.; Bosco, D. Host Plant Determines the Phytoplasma Transmission Competence of Empoasca Decipiens (Hemiptera: Cicadellidae). jnl. econ. entom. 2011, 104, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Jakovljević, M.; Jović, J.; Krstić, O.; Mitrović, M.; Marinković, S.; Toševski, I.; Cvrković, T. Diversity of Phytoplasmas Identified in the Polyphagous Leafhopper Euscelis Incisus (Cicadellidae, Deltocephalinae) in Serbia: Pathogen Inventory, Epidemiological Significance and Vectoring Potential. Eur J Plant Pathol 2020, 156, 201–221. [Google Scholar] [CrossRef]
- Olivier, C.Y.; Séguin-Swartz, G.; Galka, B.; Olfert, O.O. Aster Yellows in Leafhoppers and Field Crops in Saskatchewan, Canada, 2001–2008. The Americas Journal of Plant Science and Biotechnology 2011, 5, 88–94. [Google Scholar]
- Vilbaste, J. Preliminary List of Flomoptera-Cicadinea of Latvia and Lithuania. Toim, Biol Eesti NSV Tead Akad 1974, 23, 131–163. [Google Scholar]
- Kunkel, L.O. Studies on Aster Yellows. American Journal of Botany 1926, 13, 646–705. [Google Scholar] [CrossRef]
- Olivier, C.Y.; Lowery, D.T.; Stobbs, L.W. Phytoplasma Diseases and Their Relationships with Insect and Plant Hosts in Canadian Horticultural and Field Crops. Can Entomol 2009, 141, 425–462. [Google Scholar] [CrossRef]
- Lehmann, W. Blu Tenvergrunung Des Rapes — Eine Virose. Naturwissenschaften 1969, 56, 94–95. [Google Scholar] [CrossRef]
- Landi, L.; Isidoro, N.; Riolo, P. Natural Phytoplasma Infection of Four Phloem-Feeding Auchenorrhyncha Across Vineyard Agroecosystems in Central–Eastern Italy. Journal of Economic Entomology 2013, 106, 604–613. [Google Scholar] [CrossRef]
- Raccah, B.; Klein, M. Transmission of the Safflower Phyllody Mollicute by Neolaiturusfenestratus. Phytopathology 1982, 72, 230–232. [Google Scholar] [CrossRef]
- Salehi, M.; Izadpanah, K.; Nejat, N. A New Phytoplasma Infecting Lettuce in Iran. Plant Disease 2006, 90, 247–247. [Google Scholar] [CrossRef] [PubMed]
- Parnell, S.; van den Bosch, F.; Gottwald, T.; Gilligan, C.A. Surveillance to Inform Control of Emerging Plant Diseases: An Epidemiological Perspective. Annu Rev Phytopathol 2017, 55, 591–610. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.R.; Hoberg, E.P.; Boeger, W.A.; Trivellone, V. Assess: Using Evolution to Save Time and Resources. In An Evolutionary Pathway for Coping with Emerging Infectious Disease.; Gardner, S.L., Brooks, D.R., Boeger, W.A., Hoberg, E.P., Eds.; Zea Press: Lincoln, Nebraska, 2023. [Google Scholar]
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The Persistent Threat of Emerging Plant Disease Pandemics to Global Food Security. Proc Natl Acad Sci USA 2021, 118, e2022239118. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.R. Triage for the Biosphere; The Brundtland Commission’s Report – 10 Years; Scandinavian Univ. Press: Oslo, 1998; pp. 71–80. [Google Scholar]
- Brooks, D.R.; Hoberg, E.P. Triage for the Biosphere: The Need and Rationale for Taxonomic Inventories and Phylogenetic Studies of Parasites. Comparative Parasitology 2000, 68, 1–25. [Google Scholar]
- Gurr, G.M.; Johnson, A.C.; Ash, G.J.; Wilson, B.A.L.; Ero, M.M.; Pilotti, C.A.; Dewhurst, C.F.; You, M.S. Coconut Lethal Yellowing Diseases: A Phytoplasma Threat to Palms of Global Economic and Social Significance. Frontiers in Plant Science 2016, 7. [Google Scholar] [CrossRef]
- Kumari, S.; Nagendran, K.; Rai, A.B.; Singh, B.; Rao, G.P.; Bertaccini, A. Global Status of Phytoplasma Diseases in Vegetable Crops. Front Microbiol 2019, 10, 1349. [Google Scholar] [CrossRef]
- Audy, J.R. The Localization of Disease with Special Reference to the Zoonoses. Transactions of the Royal Society of Tropical Medicine and Hygiene 1958, 52, 309–328. [Google Scholar]
- Zwolińska, A.; Krawczyk, K.; Borodynko-Filas, N.; Pospieszny, H. Non-Crop Sources of Rapeseed Phyllody Phytoplasma (‘Candidatus Phytoplasma Asteris’: 16SrI-B and 16SrI-(B/L)L), and Closely Related Strains. Crop Protection 2019, 119, 59–68. [Google Scholar] [CrossRef]
- Foldvari, G.; Szabó, É.; Tóth, G.E.; Lanszki, Z.; Zana, B.; Varga, Z.; Kemenesi, G. Emergence of Hyalomma Marginatum and Hyalomma Rufipes Adults Revealed by Citizen Science Tick Monitoring in Hungary. Authorea 2022. [Google Scholar] [CrossRef]
- Hoberg, E.P.; Boeger, W.A.; Molnár, O.; Földvári, G.; Gardner, S.L.; Juarrero, A.; Kharchenko, V.; Ortiz, E.; Preiser, W.; Trivellone, V. The DAMA Protocol: Anticipating to Prevent and Mitigate Emerging Infectious Diseases. In: ; S.L. Gardner, D.R. Brooks, W.A. Boeger, E.P. Hoberg, Eds. . Current Volume. In An Evolutionary Pathway for Coping with Emerging Infectious Disease; Gardner, S.L., Brooks, D.R., Boeger, W.A., Hoberg, E.P., Eds.; Zea (Press): Lincoln, Nebraska, 2023.
- Galetto, L.; Nardi, M.; Saracco, P.; Bressan, A.; Marzachì, C.; Bosco, D. Variation in Vector Competency Depends on Chrysanthemum Yellows Phytoplasma Distribution within Euscelidius Variegatus. Entomologia Experimentalis et Applicata 2009, 131, 200–207. [Google Scholar] [CrossRef]
- EU Commission Implementing Regulation (EU) 2019/2072 of 28 November 2019 Establishing Uniform Conditions for the Implementation of Regulation (EU) 2016/2031 of the European Parliament and the Council, as Regards Protective Measures against Pests of Plants, and Repealing Commission Regulation (EC) No 690/2008 and Amending Commission Implementing Regulation (EU) 2018/2019; 2019; Vol. 319. 28 November.
- Chiykowski, L.N. REACTION OF SOME WHEAT VARIETIES TO ASTER YELLOWS VIRUS. Can. J. Plant Sci. 1967, 47, 149–151. [Google Scholar] [CrossRef]
- Isaacs, J. Heading off Aster Yellows in Field Crops. Country Guide 2022.
- Stillson, P.T.; Szendrei, Z. Identifying Leafhopper Targets for Controlling Aster Yellows in Carrots and Celery. Insects 2020, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- Chrustek, A.; Hołyńska-Iwan, I.; Dziembowska, I.; Bogusiewicz, J.; Wróblewski, M.; Cwynar, A.; Olszewska-Słonina, D. Current Research on the Safety of Pyrethroids Used as Insecticides. Medicina (Kaunas) 2018, 54, 61. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Relyea, R. The Effect of a Common Pyrethroid Insecticide on Wetland Communities. Environ. Res. Commun. 2019, 1, 015003. [Google Scholar] [CrossRef]
- Wu, Y.; Hao, X.; Li, Z.; Gu, P.; An, F.; Xiang, J.; Wang, H.; Luo, Z.; Liu, J.; Xiang, Y. Identification of the Phytoplasma Associated with Wheat Blue Dwarf Disease in China. Plant Disease 2010, 94, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.F.; Xiang, J.Y.; Yang, Y.; Zhang, R. The Primary Infection of Wheat My-Coplasmalike Organism Blue Dwarf Disease (WMBD). Acta Phytophylac. Sin. 1996, 11, 107–110. [Google Scholar]
- Marcone, C.; Ragozzino, A.; Seemüller, E. Detection and Identification of Phytoplasmas Infecting Vegetable, Ornamental and Forage Crops in Southern Italy. Journal of Plant Pathology 1997. [Google Scholar]
- Schneider, B.; Ahrens, U.; Kirkpatrick, B.C.; SeemüLler, E. 1993 Classification of Plant-Pathogenic Mycoplasma-like Organisms Using Restriction-Site Analysis of PCR-Amplified 16S RDNA. Microbiology 1993, 139, 519–527. [Google Scholar] [CrossRef]
- Valiunas, D. Identification of Phytoplasmas in Lithuania and Estimation of Their Biodiversity and Molecular Evolutionary Relationships. PhD thesis, Institute of Botany: Vilnius, Lithuania., 2003.
- Zwolińska, A.; Krawczyk, K.; Klejdysz, T.; Pospieszny, H. First Report of “Candidatus Phytoplasma Asteris” Associated with Oilseed Rape Phyllody in Poland. Plant Dis 2011, 95, 1475. [Google Scholar] [CrossRef]
- Jurga, M.; Zwolińska, A. Phytoplasmas in Poaceae Species: A Threat to the Most Important Cereal Crops in Europe. J Plant Pathol 2020, 102, 287–297. [Google Scholar] [CrossRef]
- Mumford, R.A.; Potyondi, L.; Harju, V.A.; Henry, C.M. The Identification of a Phytoplasma from the Aster Yellows Group Infecting Sugar Beet in Hungary. Plant Pathology 2000, 49, 806–806. [Google Scholar] [CrossRef]
- Zwolińska, A.; Borodynko-Filas, N. Intra and Extragenomic Variation between 16S RRNA Genes Found in 16SrI-B-Related Phytopathogenic Phytoplasma Strains. Annals of Applied Biology 2021, 179, 368–381. [Google Scholar] [CrossRef]
- Mitrović, M.; Marinković, S.; Cvrković, T.; Jović, J.; Krstić, O.; Jakovljević, M. Framework for Risk Assessment of ‘Candidatus Phytoplasma Solani’ Associated Diseases Outbreaks in Agroecosystems in Serbia. J Plant Pathol 2022, 104, 537–552. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





