Submitted:
10 April 2023
Posted:
13 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material & Methods
2.1. Essential oils quantification
2.2. Data source: Protein and ligands molecule files preparation
2.3. Receptor-ligand Docking process
2.4. Drug-likeness prediction for the major EO bioactive compound
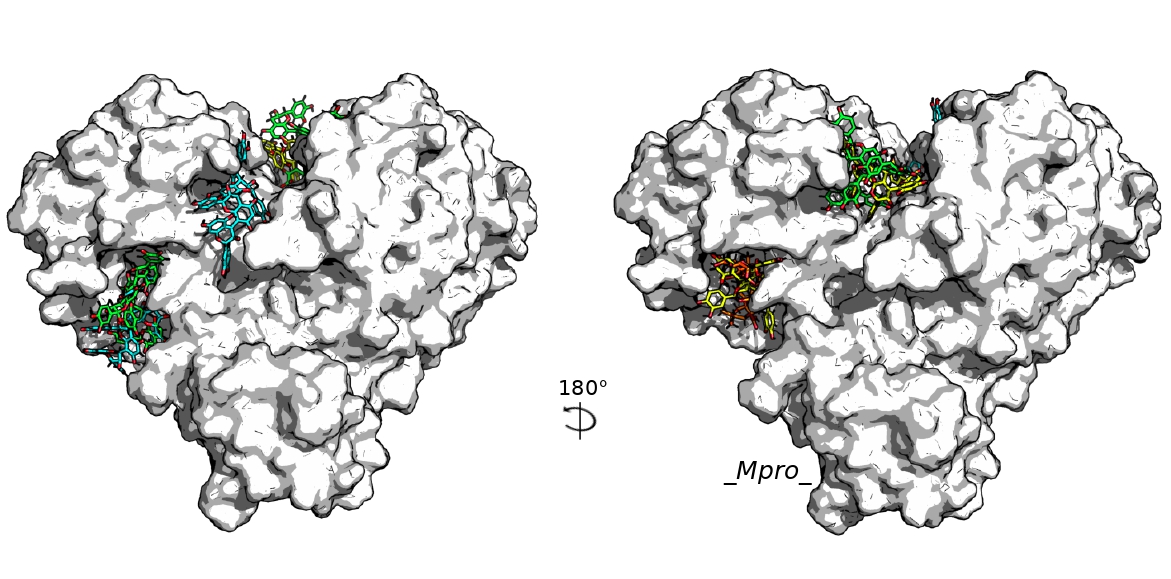
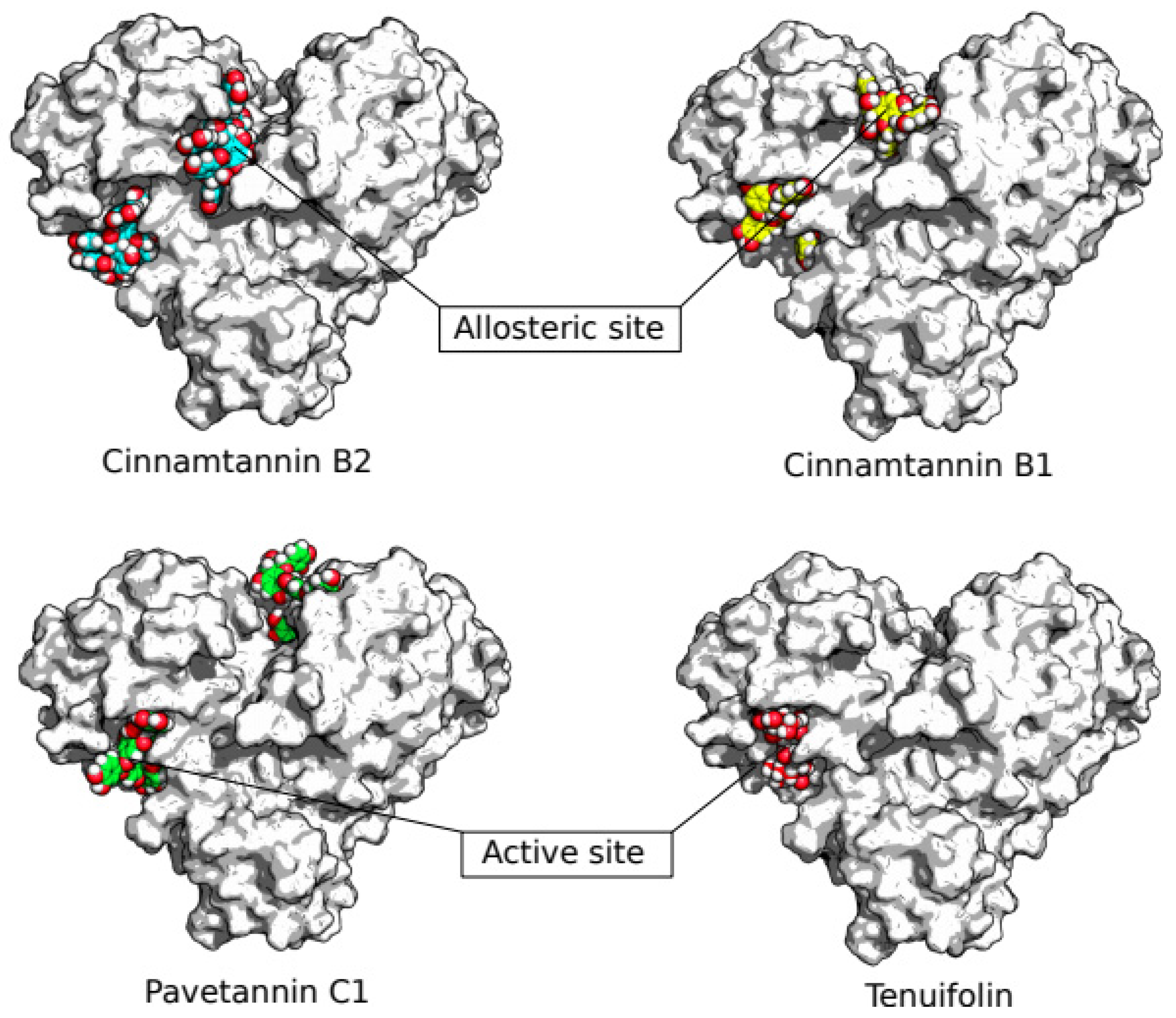
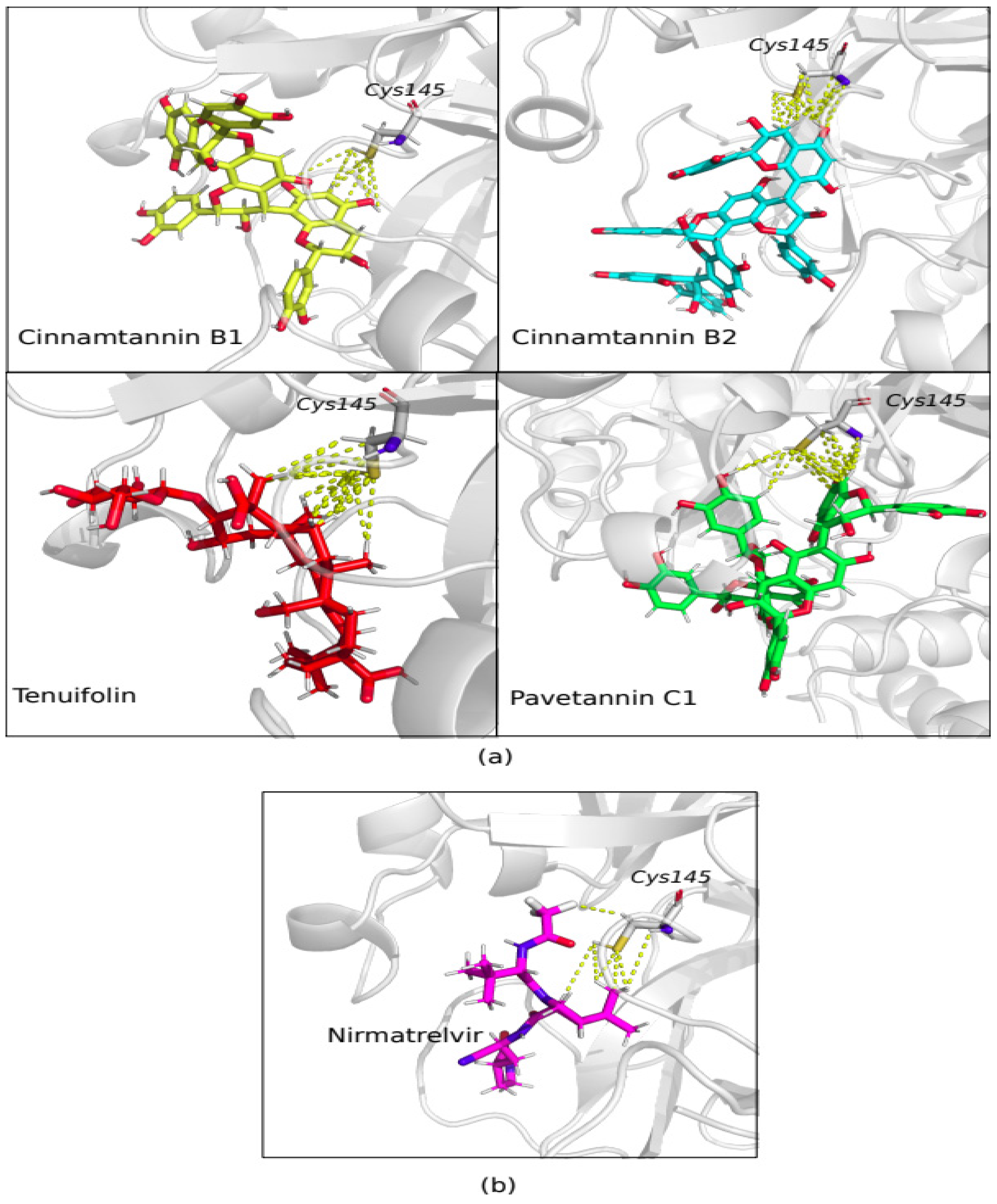
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
References
- World Health, O. (2020). Clinical management of COVID - 19: interim guidance, 27 May 2020 1445 (Geneva: World Health Organization).
- Public Health England. Investigation of novel SARS-CoV-2 variant, Variant of Concern 202012/01 Technical briefing 2- 28 December 2020. PHE: London;2020.
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281-292. [CrossRef]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260-1263. [CrossRef]
- Tian, X., Li, C., Huang, A., Xia, S., Lu, S., Shi, Z., & Ying, T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging microbes & infections, 9(1), 382-385. [CrossRef]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses, 1-23. [CrossRef]
- Oudshoorn, D., Rijs, K., Limpens, R. W., Groen, K., Koster, A. J., Snijder, E. J., & Bárcena, M. (2017). Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. MBio, 8(6), e01658-17. [CrossRef]
- Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020). Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences, 117(21), 11727-11734. [CrossRef]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–10. [CrossRef]
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., & Bolton, E. E. (2021). PubChem in 2021: new data content and improved web interfaces. Nucleic acids research, 49(D1), D1388-D1395. [CrossRef]
- O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of cheminformatics, 3(1), 1-14. [CrossRef]
- Rose, P. W., Prlić, A., Altunkaya, A., Bi, C., Bradley, A. R., Christie, C. H., & Burley, S. K. (2016). The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic acids research, gkw1000. [CrossRef]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., & Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289-293. [CrossRef]
- Schrodinger, L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1.
- Bowman, G. R., & Pande, V. S. (2009). Simulated tempering yields insight into the low-resolution Rosetta scoring functions. Proteins: Structure, Function, and Bioinformatics, 74(3), 777-788. [CrossRef]
- Unni, S., Huang, Y., Hanson, R. M., Tobias, M., Krishnan, S., Li, W. W., & Baker, N. A. (2011). Web servers and services for electrostatics calculations with APBS and PDB2PQR. Journal of computational chemistry, 32(7), 1488-1491. [CrossRef]
- M. Sanner. 1999. Python: A Programming Language for Software Integration and Development. J. Mol. Graph. Model. 17(1): 57-61.
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 31(2), 455-461. [CrossRef]
- Bitencourt-Ferreira, G., & Azevedo, W. F. D. (2019). Docking with SwissDock. In Docking Screens for Drug Discovery (pp. 189-202). Humana, New York, NY.
- Grosdidier, A., Zoete, V., & Michielin, O. (2011). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic acids research, 39(suppl_2), W270-W277. [CrossRef]
- Goddard, T. D., Huang, C. C., Meng, E. C., Pettersen, E. F., Couch, G. S., Morris, J. H., & Ferrin, T. E. (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science, 27(1), 14-25. [CrossRef]
- Sivropoulou, A., Nikolaou, C., Papanikolaou, E., Kokkini, S., Lanaras, T., & Arsenakis, M. (1997). Antimicrobial, cytotoxic, and antiviral activities of Salvia fructicosa essential oil. Journal of Agricultural and Food Chemistry, 45(8), 3197-3201. [CrossRef]
- Benencia, F., & Courreges, M. C. (2000). In vitro and in vivo activity of eugenol on human herpesvirus. Phytotherapy Research: An International Journal Devoted to Pharmacological and Toxicological Evaluation of Natural Product Derivatives, 14(7), 495-500. [CrossRef]
- Hayashi, K., Imanishi, N., Kashiwayama, Y., Kawano, A., Terasawa, K., Shimada, Y., & Ochiai, H. (2007). Inhibitory effect of cinnamaldehyde, derived from Cinnamomi cortex, on the growth of influenza A/PR/8 virus in vitro and in vivo. Antiviral Research, 74(1), 1-8. [CrossRef]
- Khan, A., Heng, W., Wang, Y., Qiu, J., Wei, X., Peng, S., & Wei, D. Q. (2021). In silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro). Phytotherapy Research. [CrossRef]
- Rajendran, M., Roy, S., Ravichandran, K., Mishra, B., Gupta, D. K., Nagarajan, S., & Provaznik, I. (2022). In silico screening and molecular dynamics of phytochemicals from Indian cuisine against SARS-CoV-2 MPro. Journal of Biomolecular Structure and Dynamics, 40(7), 3155-. [CrossRef]
- Galeotti N, Di Cesare Mannelli L, Mazzanti G, Bartolini A, Ghelardini C. Menthol: a natural analgesic compound. Neurosci Lett. 2002 Apr 12;322(3):145-8. [CrossRef]
- World Health Organization. (2019)"Evaluations of the Joint FAO/WHO Expert Committee on Food Additives (JECFA).".
- MCCASKILL, David et CROTEAU, Rodney. Monoterpene and sesquiterpene biosynthesis in glandular trichomes of peppermint (Mentha x piperita) rely exclusively on plastid-derived isopentenyl diphosphate. Planta, 1995, vol. 197, no 1, p. 49-56. [CrossRef]
- Vijayasteltar, L., Nair, G. G., Maliakel, B., Kuttan, R., & Krishnakumar, I. M. (2016). Safety assessment of a standardized polyphenolic extract of clove buds: Subchronic toxicity and mutagenicity studies. Toxicology reports, 3, 439-449. [CrossRef]
- NAIR, B. Final report on the safety assessment of Mentha Piperita (Peppermint) Oil, Mentha Piperita (Peppermint) Leaf Extract, Mentha Piperita (Peppermint) Leaf, and Mentha Piperita (Peppermint) Leaf Water. International Journal of Toxicology, 2001, vol. 20, p. 61.
- BHOWAL, Mridul et GOPAL, Murugananthan. Eucalyptol: Safety and pharmacological profile. J. Pharm. Sci, 2015, vol. 5, p. 125-131. [CrossRef]
- LAEKEMAN, G. Assessment report on Cinnamomum verum JS Presl, cortex and corticisaetheroleum. London: European Medicines Agency, 2011.
- CRAVOTTO, Giancarlo et CINTAS, P. Extraction of flavourings from natural sources. In : Modifying flavour in food. Woodhead Publishing, 2007. p. 41-63.
- EUROPEAN FOOD SAFETY AUTHORITY (EFSA). Camphor in flavourings and other food ingredients with flavouring properties-Opinion of the Scientific Panel on Food Additives, Flavourings, Processing Aids and Materials in Contact with Food on a request from the Commission. EFSA Journal, 2008, vol. 6, no 7, p. 729. [CrossRef]
- Zebib, H., Bultosa, G., & Abera, S. (2015). Physico-chemical properties of sesame (Sesamum indicum L.) varieties grown in Northern Area, Ethiopia. Agricultural Sciences, 6(02), 238. [CrossRef]
- FAO/WHO (1967) Toxicological Evaluation of Some Flavouring Substances and Non-Nutritive Sweetening Agents, FAO Nutrition Meetings Report, Series No. 44a; WHO/Food Add./68.33.
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., & Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289-293. [CrossRef]
- Ercan, S., & Çınar, E. (2021). A molecular docking study of potential inhibitors and repurposed drugs against SARS-CoV-2 main protease enzyme. Journal of the Indian Chemical Society, 98(3), 100041. [CrossRef]
- Prasanth, D. S. N. B. K., Murahari, M., Chandramohan, V., Panda, S. P., Atmakuri, L. R., & Guntupalli, C. (2021). In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. Journal of Biomolecular Structure and Dynamics, 39(13), 4618-4632. [CrossRef]
- Chandra Manivannan, A., Malaisamy, A. K., Eswaran, M., Meyyazhagan, A., Arumugam, V. A., Rengasamy, K. R., & Liu, W. C. (2022). Evaluation of clove phytochemicals as potential antiviral drug candidate targeting SARS-CoV-2 Main Protease: An computational docking, molecular dynamics simulation and pharmacokinetic profiling. Frontiers in Molecular Biosciences, 604. [CrossRef]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409-412. [CrossRef]
- Günther, S., Reinke, P. Y., Fernández-García, Y., Lieske, J., Lane, T. J., Ginn, H. M., & Meents, A. (2021). X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science, 372(6542), 642-646. [CrossRef]
- Alzyoud, L., Ghattas, M. A., & Atatreh, N. (2022). Allosteric binding sites of the SARS-CoV-2 main protease: Potential targets for broad-spectrum anti-coronavirus agents. Drug Design, Development and Therapy, 2463-2478. [CrossRef]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2021). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 39(9), 3347-3357. [CrossRef]
- Pfizer. (2021). Pfizer’s Novel COVID-19 Oral Antiviral Treatment Candidate Reduced Risk of Hospitalization or Death by 89% in Interim Analysis of Phase 2/3 EPIC-HR Study. Nota de prensa.
- Edwards, J. (2000). The practice of aromatherapy. Churchill Livingstone.
- Tisserand, R., & Young, R. (2013). Essential oil safety: a guide for health care professionals. Elsevier Health Sciences.
- Buchbauer, G., Jirovetz, L., & Jager, W. (1993). Aromatherapy: evidence for sedative effects of the essential oil of lavender after inhalation. Zeitschrift fur Naturforschung. C, Journal of biosciences, 48(11-12), 796-801. [CrossRef]
| Plants | Essential Oil (EO) | Acceptable daily intake (mg/kg body weight / day) | References |
|---|---|---|---|
| Mentha spicata | Spearmint EO | 40 | [27] |
| Mentha | Menthol (crystals) | 4 | [28] |
| Mentha aquatica | Watermint EO | 5 | [29] |
| Syzygium aromaticum | Cloves EO | 2.5 | [30] |
| Mentha piperita | peppermint EO | 200 | [31] |
| Mentha poulegium | Pennyroyal EO | 2.3 | [32] |
| Eucalyptus globulus | Eucalyptus EO | 4.28 | [33] |
| Cannula | Cinnamon EO | 0.1 | [34] |
| Melaleuca leucadendron cajuputii | Cajeput EO | 0.17 | [35] |
| Cinnamomum camphora | Camphor | 50 | [36] |
| Sesama | Sesame Oil | 15000 | [37] |
| No | Ligands | Ligands Topological Polar Surface Area (Ų) | Binding affinity (kcal/mol) “Active site” |
Binding affinity (kcal/mol) “Allosteric site” |
|---|---|---|---|---|
| 1 | Cinnamtannin B1 | 320 | -9.56 | -11.12 |
| 2 | Cinnamtannin B2 | 431 | -9.40 | -10.74 |
| 3 | Pavetannin C1 | 431 | -10.30 | -10.79 |
| 4 | Syzyginin B | 349 | -10.10 | - |
| 5 | Procyanidin C1 | 331 | -9.05 | - |
| 6 | Tenuifolin | 214 | -8.75 | - |
| 7 | Nirmatrelvir | 131 | -9.24 | - |
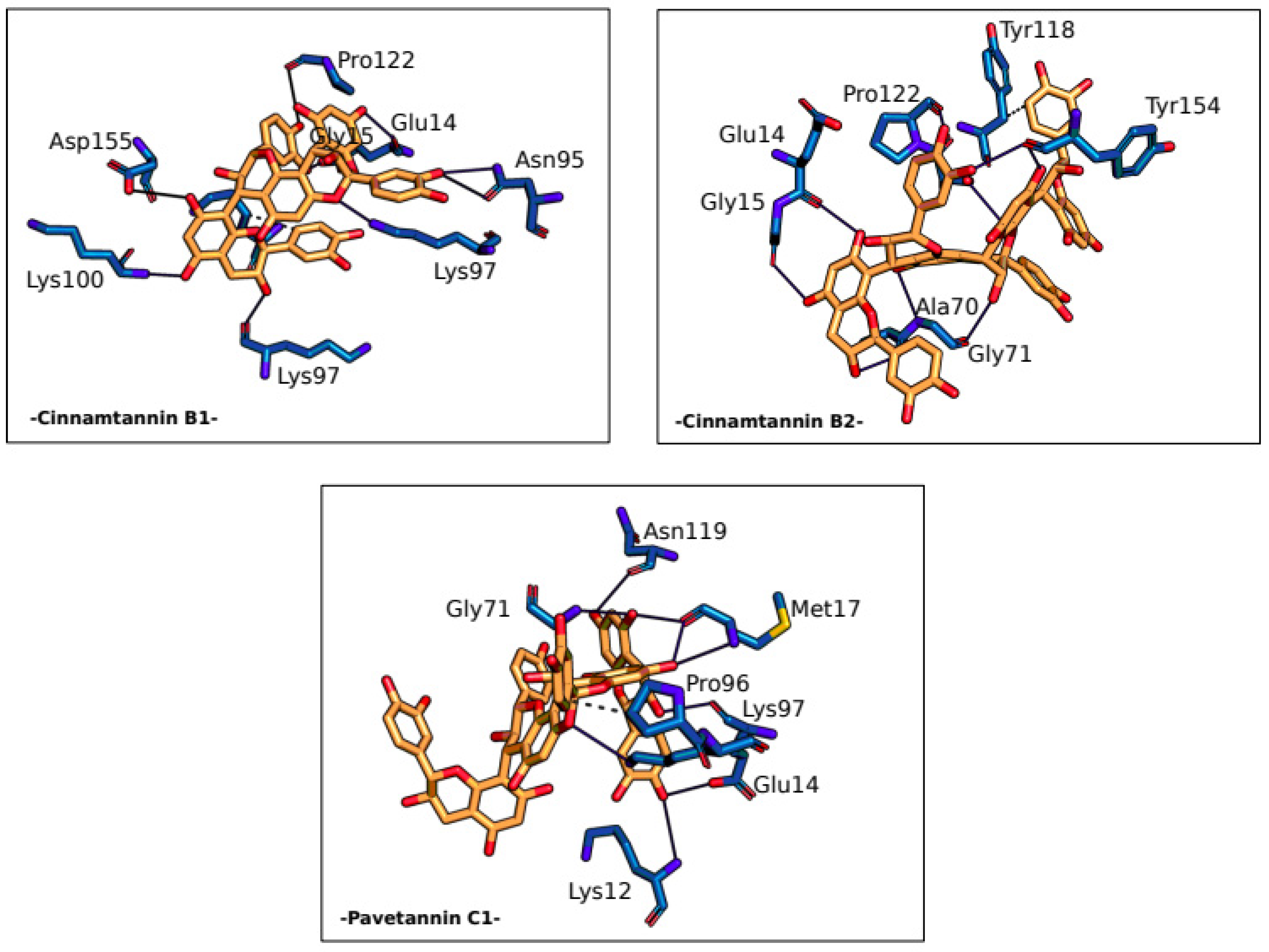
| Residue | AA | Distance H-A |
Distance D-A | Donor angle | Donor Atom | Acceptor Atom |
|---|---|---|---|---|---|---|
| -Cinnamtannin B1- | ||||||
| 14A | GLU | 2.18 | 3.07 | 150.97 | 28 [O2] | 311 [O-] |
| 14A | GLU | 1.74 | 2.69 | 163.65 | 20 [O2] | 306 [O2] |
| 95A | ASN | 3.53 | 3.94 | 106.70 | 1568 [Nam] | 24 [O2] |
| 95A | ASN | 2.51 | 3.10 | 118.88 | 24 [O2] | 1569 [O2] |
| 97A | LYS | 2.40 | 3.13 | 131.54 | 18 [O3] | 6275 [O2] |
| 97A | LYS | 2.37 | 3.31 | 150.71 | 1598 [N3+] | 63 [O2] |
| 100A | LYS | 3.22 | 3.75 | 115.47 | 6322 [Nam] | 22 [O2] |
| 122A | PRO | 2.91 | 3.86 | 162.78 | 30 [O2] | 1996 [O2] |
| 155A | ASP | 3.13 | 3.82 | 129.57 | 16 [O2] | 7160 [O-] |
| -Cinnamtannin B2- | ||||||
| 14A | GLU | 2.04 | 2.80 | 132.84 | 9387 [O2] | 4889 [O2] |
| 15A | GLY | 2.00 | 2.83 | 141.70 | 9395 [O2] | 4904 [O2] |
| 70A | ALA | 2.11 | 3.04 | 159.71 | 9389 [O3] | 5762 [O2] |
| 71A | GLY | 2.65 | 3.20 | 114.54 | 5769 [Nam] | 9381 [O2] |
| 71A | GLY | 3.19 | 3.64 | 110.53 | 9371 [O3] | 5772 [O2] |
| 121A | SER | 3.38 | 3.89 | 115.24 | 6570 [O3] | 9449 [O2] |
| 122A | PRO | 2.86 | 3.58 | 131.51 | 9403 [O2] | 6579 [O2] |
| 154A | TYR | 2.74 | 3.15 | 105.73 | 9401 [O2] | 2354 [O2] |
| 154A | TYR | 2.86 | 3.81 | 165.39 | 9385 [O2] | 2354 [O2] |
| -Pavetannin C1- | ||||||
| 14A | GLU | 1.90 | 2.82 | 156.43 | 9409 [O2] | 212 [O-] |
| 14A | GLU | 1.87 | 2.80 | 159.59 | 9385 [O3] | 207 [O2] |
| 17A | MET | 2.13 | 2.77 | 122.36 | 9397 [O2] | 240 [O2] |
| 17A | MET | 3.17 | 3.48 | 100.00 | 237 [Nam] | 9387 [O2] |
| 17A | MET | 2.07 | 2.82 | 131.63 | 9387 [O2] | 240 [O2] |
| 71A | GLY | 2.29 | 3.04 | 130.92 | 1087 [Nam] | 9397 [O2] |
| 97A | LYS | 3.14 | 3.87 | 128.25 | 1499 [N3+] | 9367 [O2] |
| 119A | ASN | 2.57 | 3.19 | 121.41 | 9403 [O2] | 1865 [O2] |
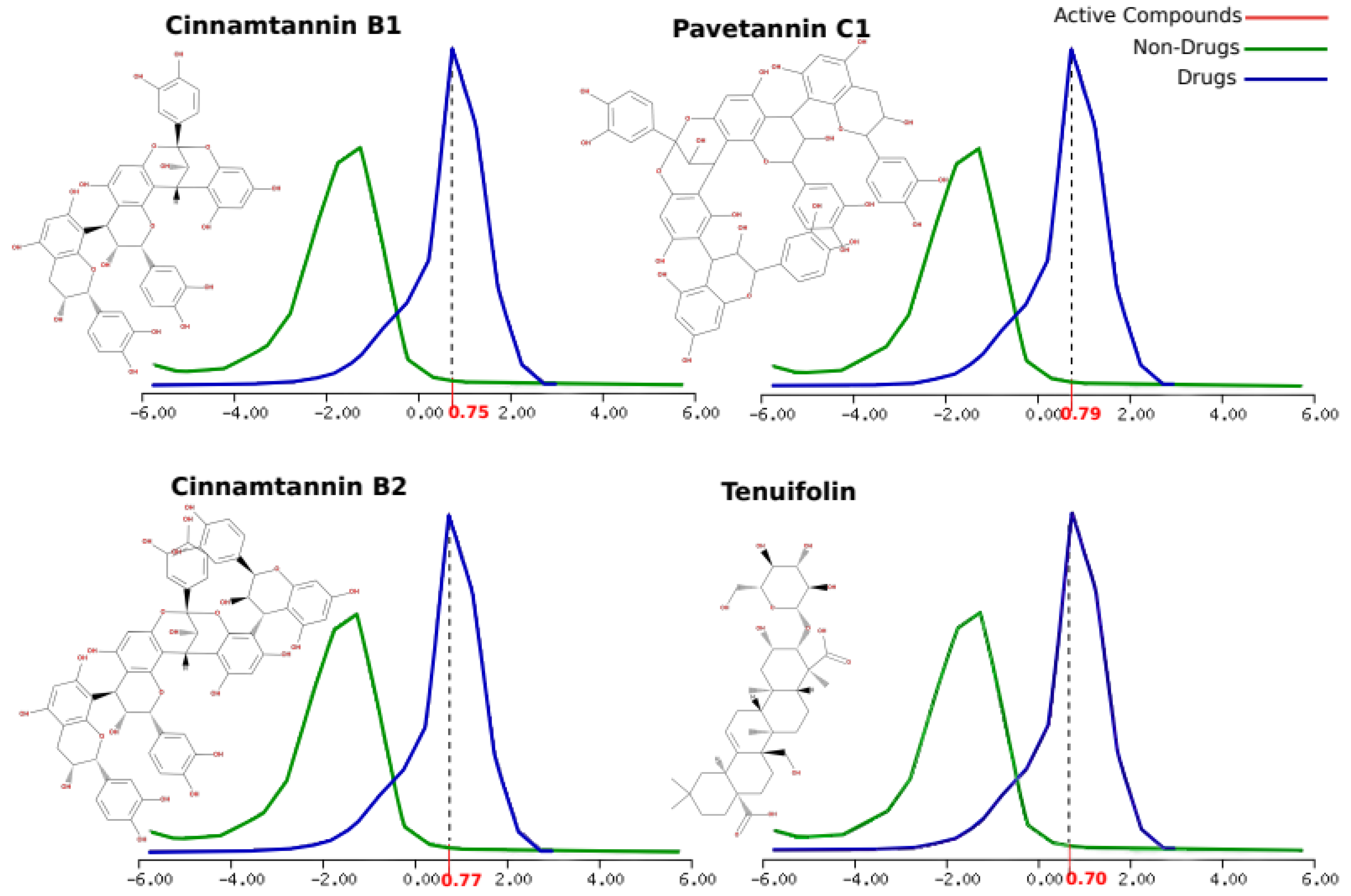
| Active compounds | Molecular formula | Molecular weight (KDa) |
Number of HBA | Number of HBD | MolLogP | MolLogS Log(moles/L) |
MolPSA (Ų) |
MolVol (ų) |
pKa | BBB Score | Number of stereo centers | Drug-likeness model score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pavetannin C1 | C60H48O24 | 1152.25 | 24 | 19 | 3.46 | -3.04 | 344.95 | 1039.90 | <0. / 9.52 | 0 | 11 | 0.79 |
|
Tenuifolin |
C36H56O12 | 680.38 | 12 | 8 | 1.02 | -1.30 | 168.92 | 737.65 | <0. / 5.17 | 0.34 | 15 | 0.70 |
| Cinnamtannin B1 | C45H36 O18 | 864.19 | 18 | 14 | 2.40 | -2.60 | 257.26 | 782.13 | <0. / 9.52 | 0 | 8 | 0.75 |
| Cinnamtannin B2 | C60H48O24 | 1152.25 | 14 | 19 | 3.12 | -3.06 | 345.71 | 1039.96 | <0. / 9.52 | 0 | 11 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





