Submitted:
05 April 2023
Posted:
07 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Proposition
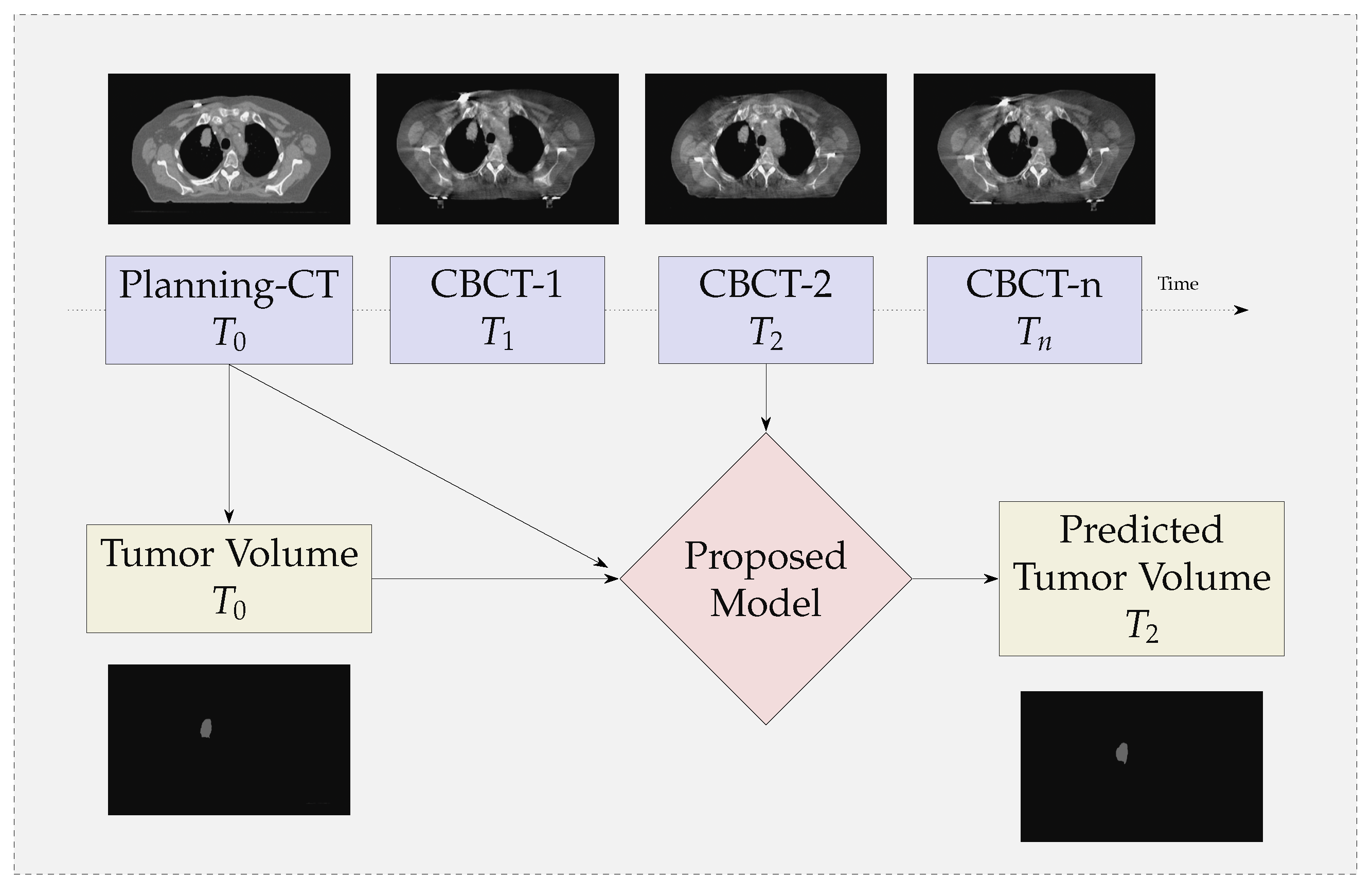
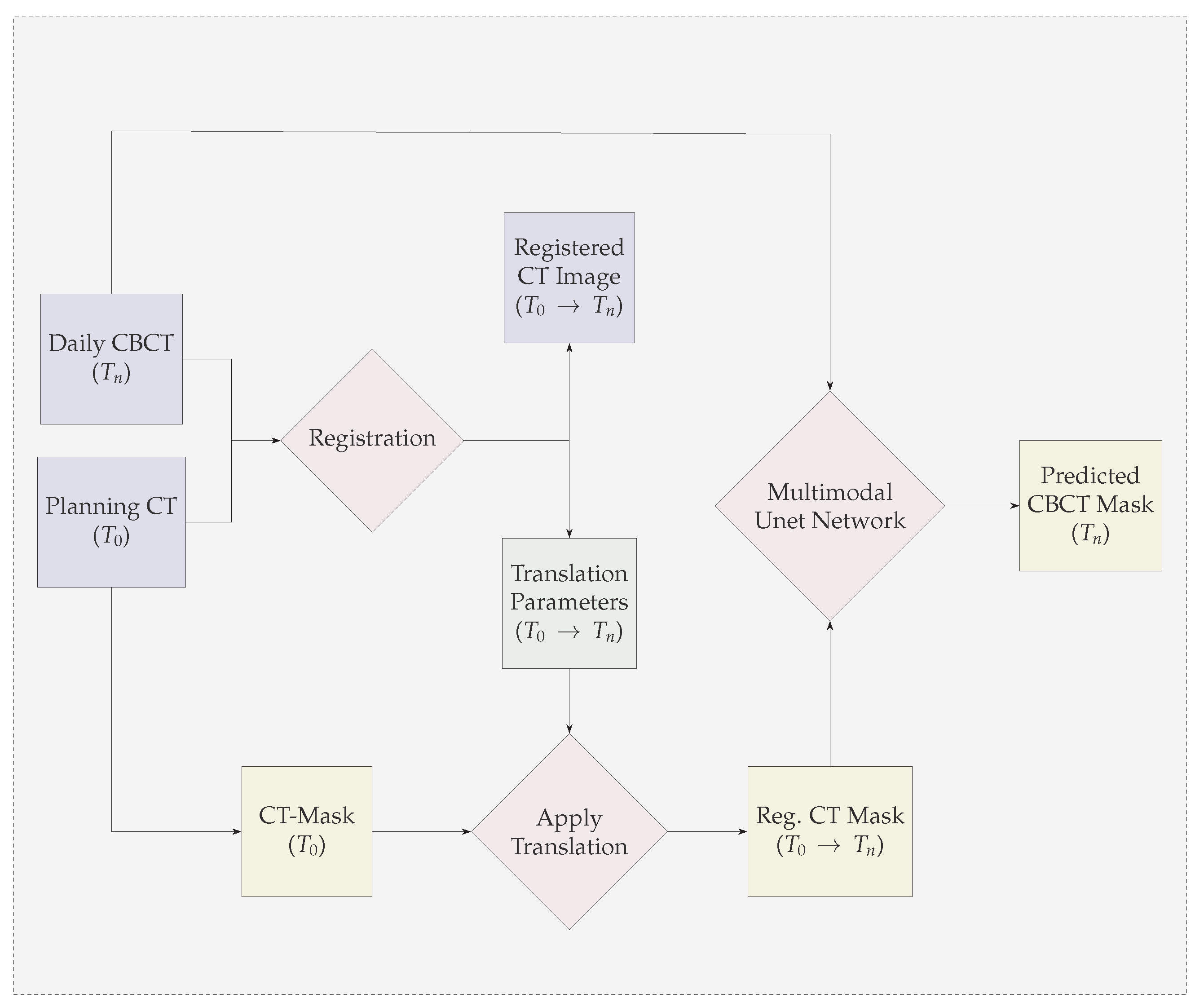
- We propose a multi-modal neural-network which uses CBCT and a registered CT-Mask produced during Planning phase(as shown in Figure 2) to train an end-to-end 3D U-net to automatically delineate the Gross Tumor Volume in the CBCT. It produces reasonably accurate contours of GTV during Radiotherapy.
- We provide a comparison between two types of fusion - Early Fusion and Late Fusion by using different types of imprecise CT-masks. This helps to take a better decision in choosing the architecture.
2. Related Works
3. Materials and Methods
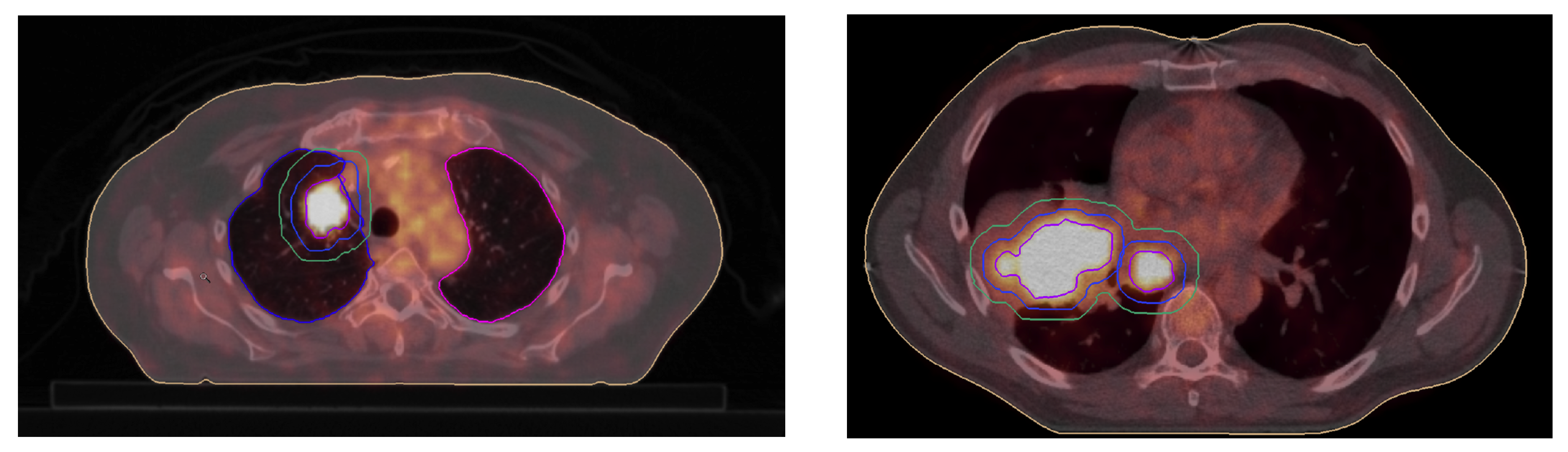
3.1. GTV Seed for Localisation of Tumor
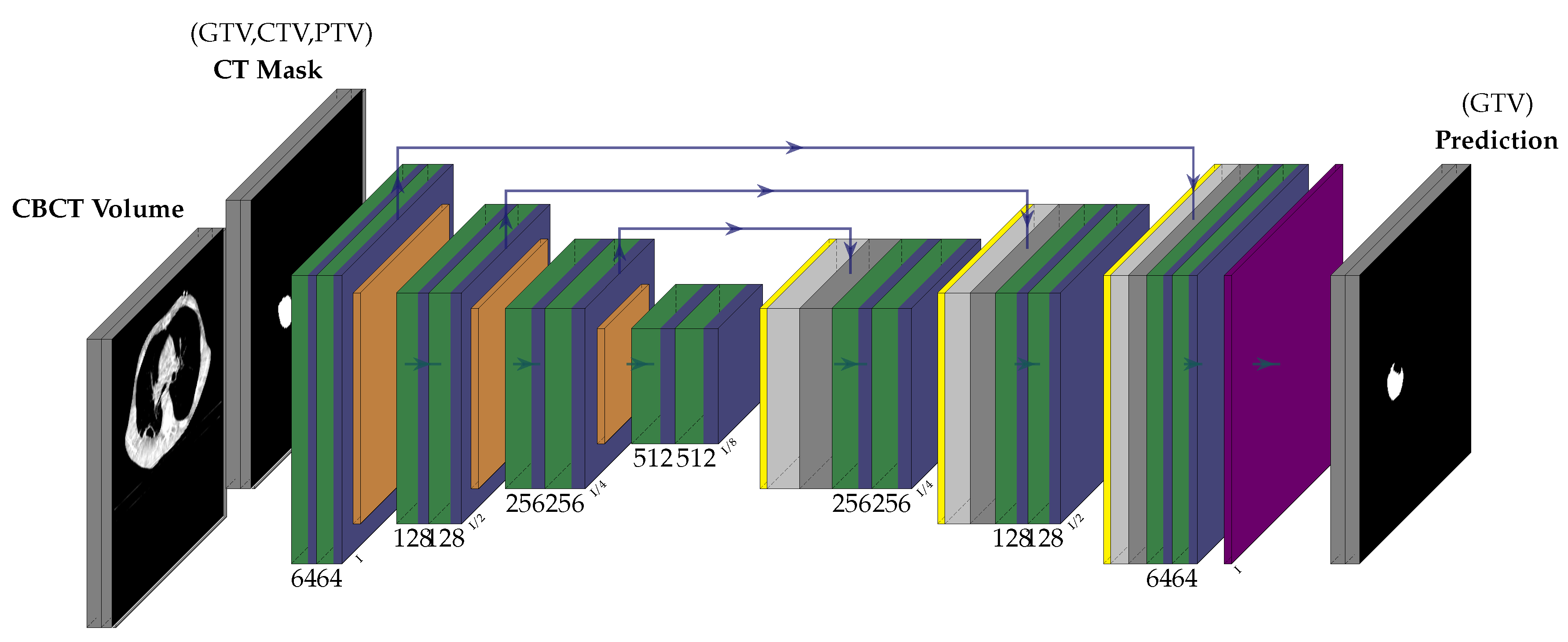
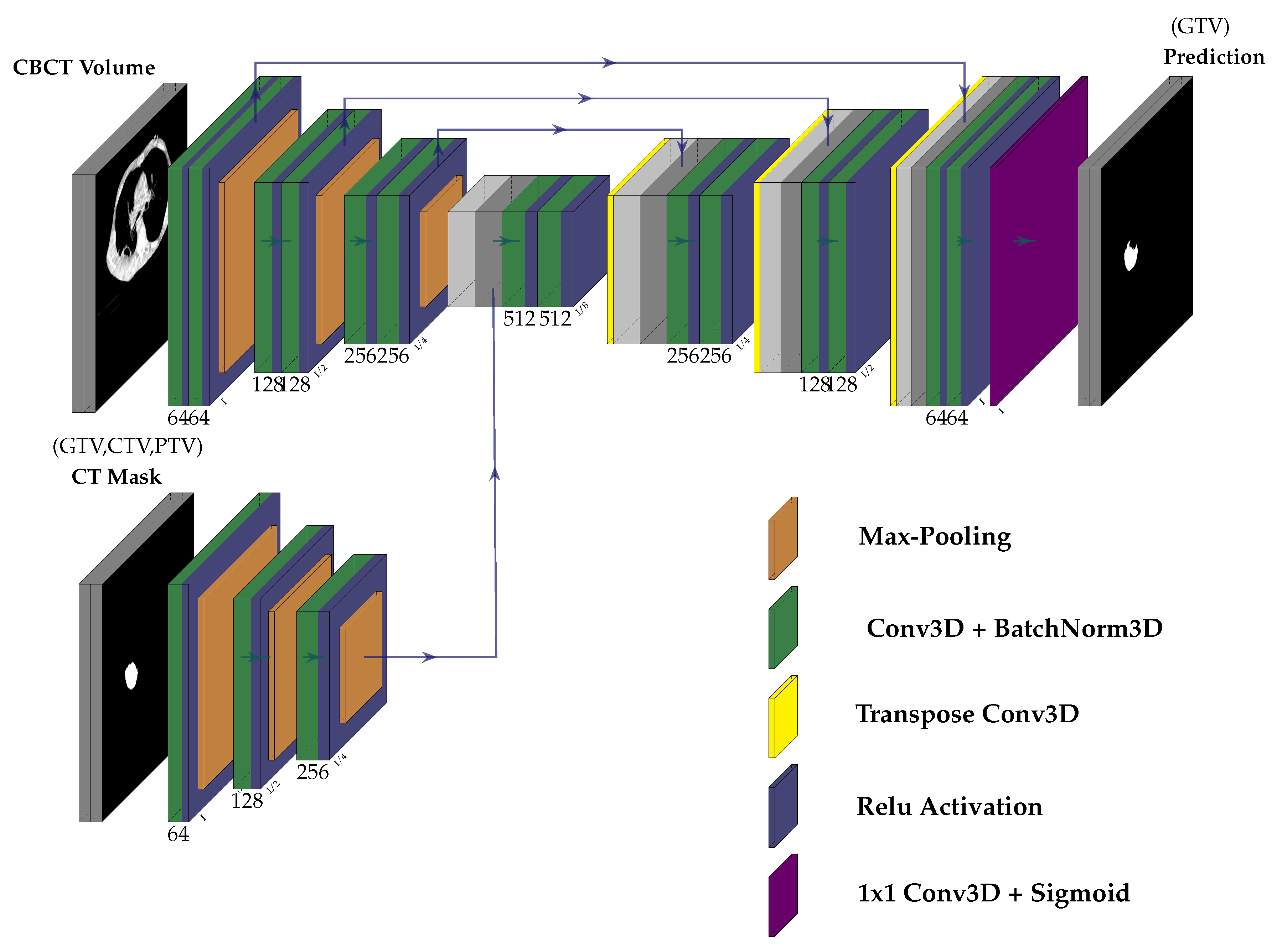
3.2. Network Architecture
3.3. Evaluation Metric and Loss
4. Experiments and Results
4.1. Dataset
4.2. Data Preprocessing
4.3. Training
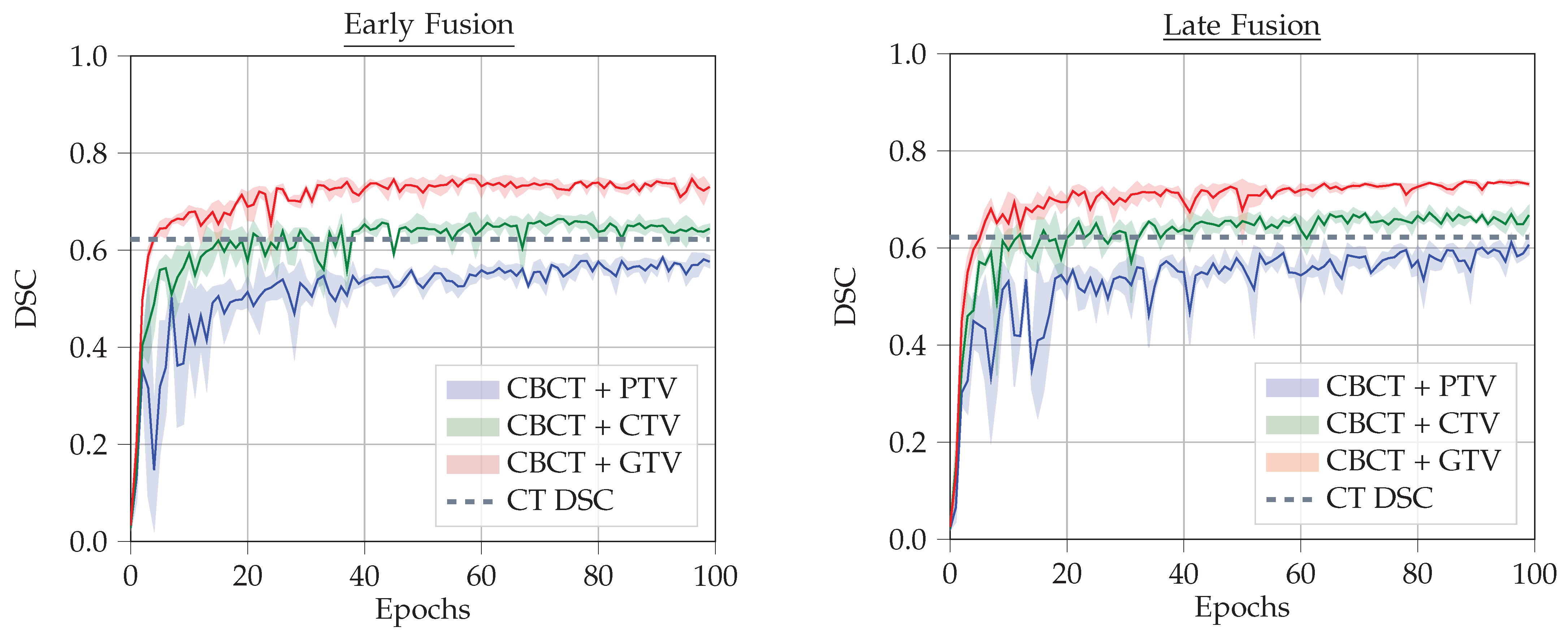
4.4. Results
- CBCT + CT-GTV Mask
- CBCT + CT-CTV Mask
- CBCT + CT-PTV Mask
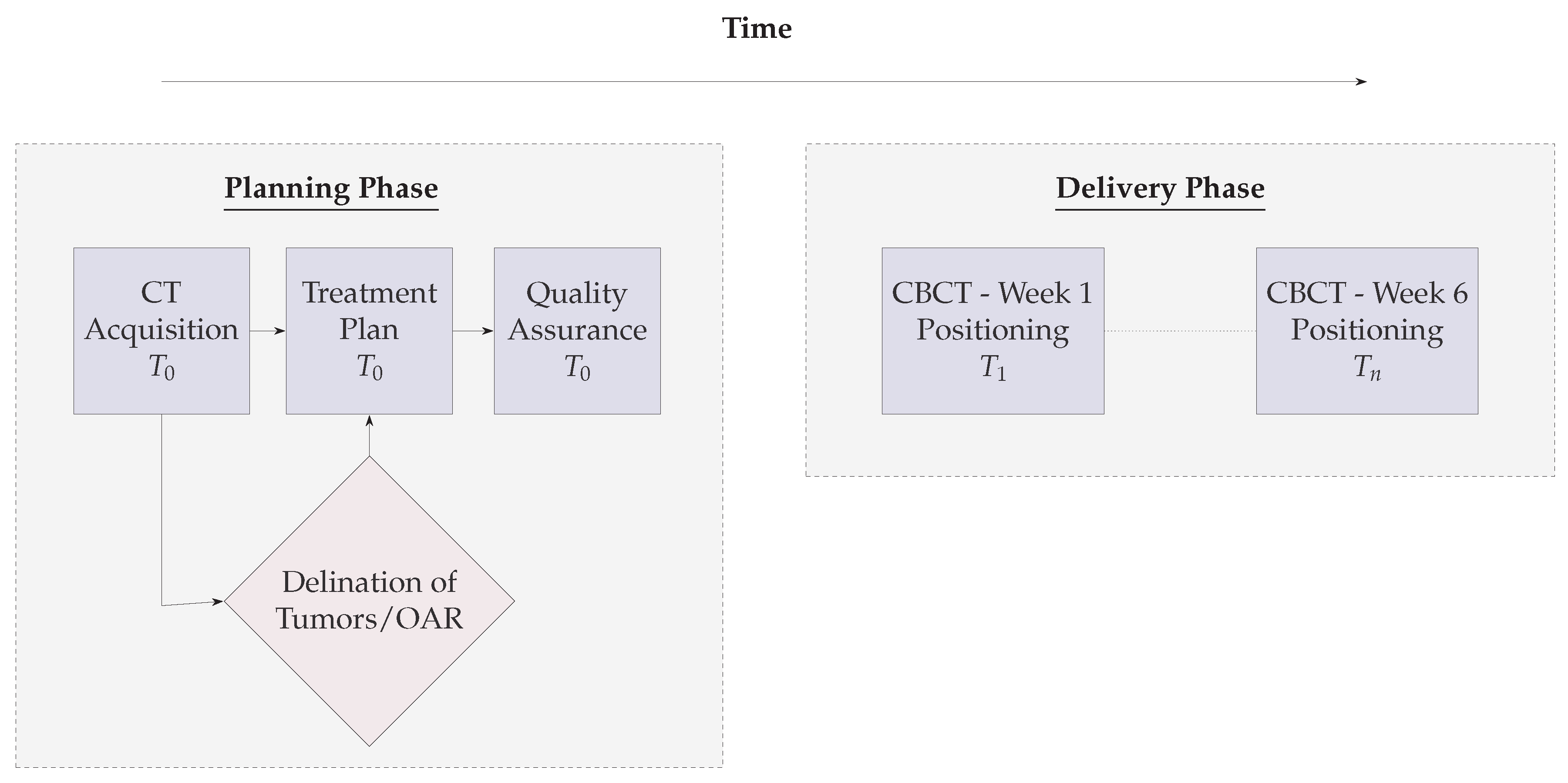
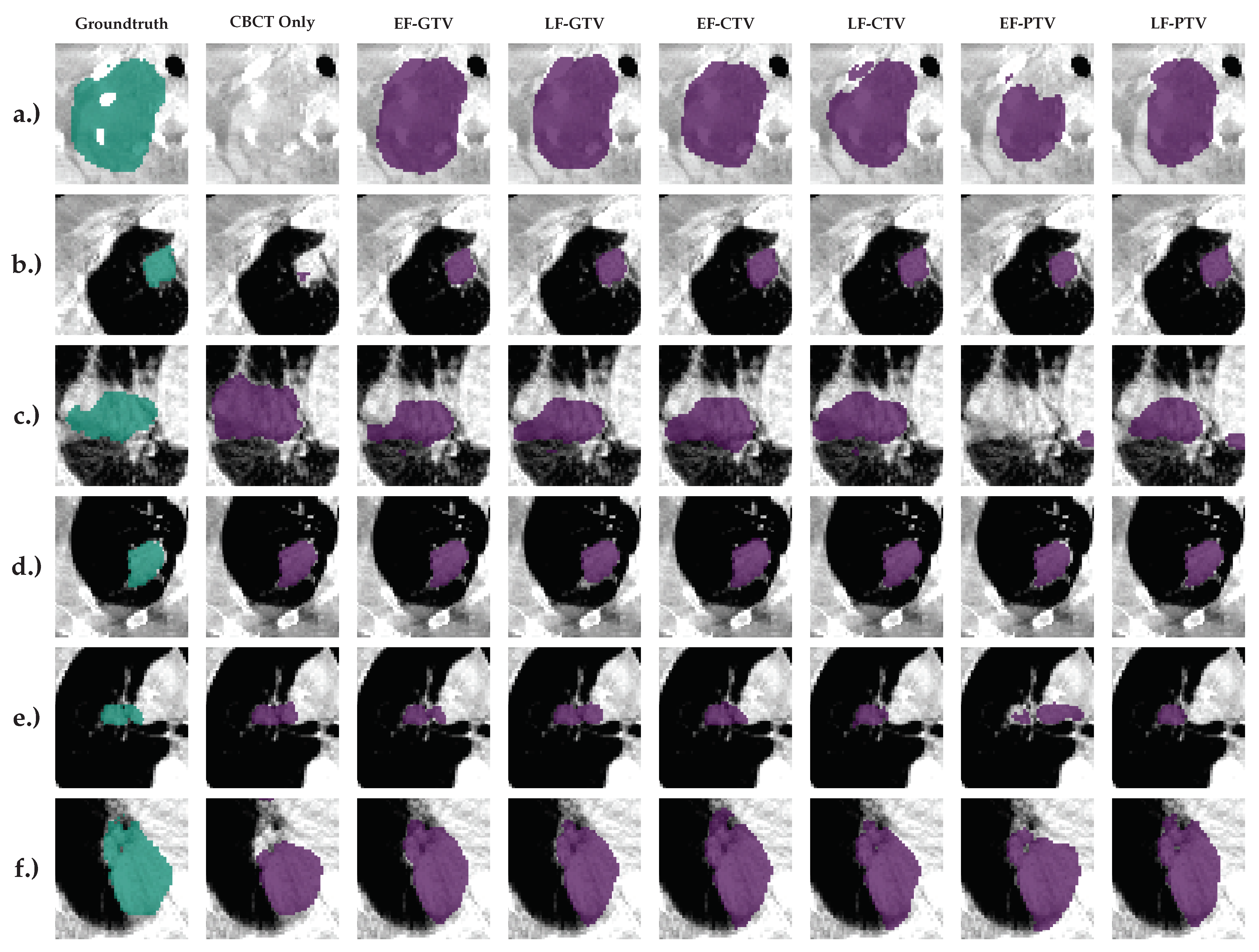
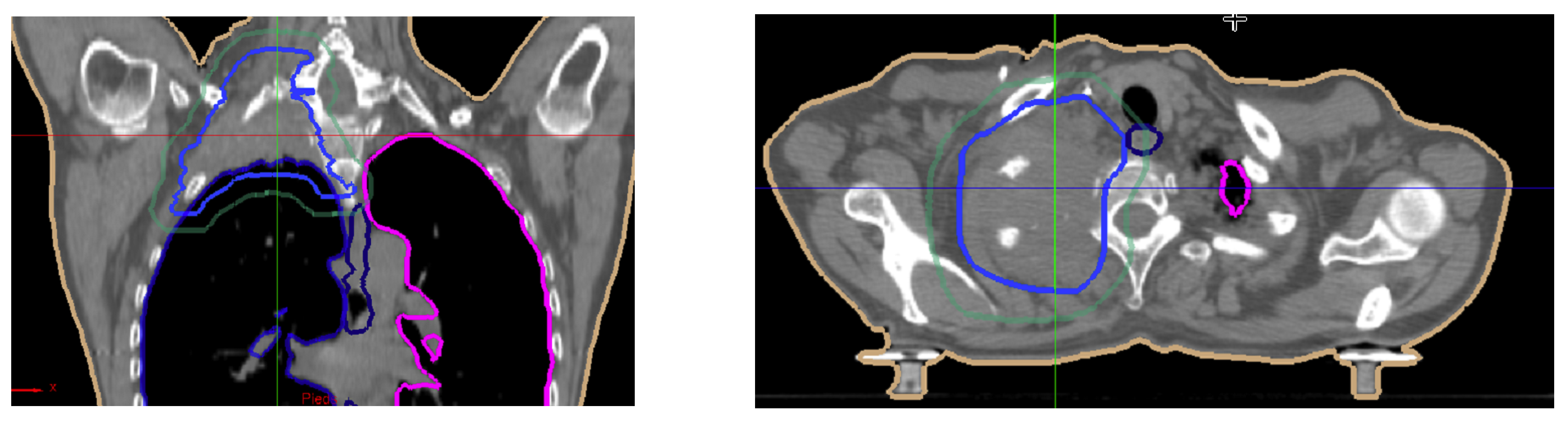
5. Illustrations
6. Conclusion
Acknowledgement
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Dong, X.; Lei, Y.; Wang, T.; Thomas, M.; Tang, L.; Curran, W.J.; Liu, T.; Yang, X. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Medical Physics 2019, 46, 2157–2168. [Google Scholar] [CrossRef]
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.W. Cancer and radiation therapy: Current advances and future directions. International Journal of Medical Sciences 2012, 9, 193–199. [Google Scholar] [CrossRef]
- Lecchi, M.; Fossati, P.; Elisei, F.; Orecchia, R.; Lucignani, G. Current concepts on imaging in radiotherapy. European Journal of Nuclear Medicine and Molecular Imaging 2008, 35, 821–837. [Google Scholar] [CrossRef]
- Liu, X.; Li, K.W.; Yang, R.; Geng, L.S. Review of Deep Learning Based Automatic Segmentation for Lung Cancer Radiotherapy. Frontiers in Oncology 2021, 11. [Google Scholar] [CrossRef]
- Zhen, X.; Gu, X.; Yan, H.; Zhou, L.; Jia, X.; Jiang, S.B. CT to cone-beam CT deformable registration with simultaneous intensity correction. Physics in Medicine and Biology 2012, 57, 6807–6826. [Google Scholar] [CrossRef]
- Suzuki, K. Overview of deep learning in medical imaging. Radiological Physics and Technology 2017, 10, 257–273. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Neural Information Processing Systems 2012. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Springer International Publishing 2015. [Google Scholar] [CrossRef]
- Özgün, Ç.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Medical Image Computing and Computer-Assisted Intervention 2016. [Google Scholar] [CrossRef]
- Ge, R.; Cai, H.; Yuan, X.; Qin, F.; Huang, Y.; Wang, P.; Lyu, L. MD-UNET: Multi-input dilated U-shape neural network for segmentation of bladder cancer. Computational Biology and Chemistry 2021, 93. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tyagi, N.; Rimner, A.; Hu, Y.C.; Veeraraghavan, H.; Li, G.; Hunt, M.; Mageras, G.; Zhang, P. Segmenting lung tumors on longitudinal imaging studies via a patient-specific adaptive convolutional neural network. Radiotherapy and Oncology 2019, 131, 101–107. [Google Scholar] [CrossRef]
- Li, L.; Zhao, X.; Lu, W.; Tan, S. Deep learning for variational multimodality tumor segmentation in PET/CT. Neurocomputing 2020, 392, 277–295. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L.; Lu, W.; Tan, S. Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Physics in Medicine and Biology 2019, 64. [Google Scholar] [CrossRef]
- Jin, D.; Guo, D.; Ho, T.Y.; Harrison, A.P.; Xiao, J.; kan Tseng, C.; Lu, L. DeepTarget: Gross tumor and clinical target volume segmentation in esophageal cancer radiotherapy. Medical Image Analysis 2021, 68. [Google Scholar] [CrossRef]
- Harrison, A.P.; Xu, Z.; George, K.; Lu, L.; Summers, R.M.; Mollura, D.J. Progressive and Multi-Path Holistically Nested Neural Networks for Pathological Lung Segmentation from CT Images. International Conference on Medical Image Computing and Computer-Assisted Intervention 2017. [Google Scholar] [CrossRef]
- Wang, H.; Hu, J.; Song, Y.; Zhang, L.; Bai, S.; Yi, Z. Multi-view fusion segmentation for brain glioma on CT images. Applied Intelligence 2022, 52, 7890–7904. [Google Scholar] [CrossRef]
- Ma, L.; Chi, W.; Morgan, H.E.; Lin, M.H.; Chen, M.; Sher, D.; Moon, D.; Vo, D.T.; Avkshtol, V.; Lu, W.; et al. Registration-Guided Deep Learning Image Segmentation for Cone Beam CT-based Online Adaptive Radiotherapy. Medical Physics 2022. [Google Scholar] [CrossRef]
- Fu, Y.; Lei, Y.; Wang, T.; Tian, S.; Patel, P.; Jani, A.B.; Curran, W.J.; Liu, T.; Yang, X. Pelvic multi-organ segmentation on cone-beam CT for prostate adaptive radiotherapy. Medical Physics 2020, 47, 3415–3422. [Google Scholar] [CrossRef]
- Lei, Y.; Wang, T.; Tian, S.; Dong, X.; Jani, A.B.; Schuster, D.; Curran, W.J.; Patel, P.; Liu, T.; Yang, X. Male pelvic multi-organ segmentation aided by CBCT-based synthetic MRI. Physics in Medicine and Biology 2020, 65. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wang, S.; Liang, X.; Balagopal, A.; Nguyen, D.; Yang, M.; Wang, Z.; Ji, J.X.; Qian, X.; Jiang, S. Cone-Beam Computed Tomography (CBCT) segmentation by adversarial learning domain adaptation. Medical Image Computing and Computer Assisted Intervention 2017. [Google Scholar]
- Brion, E.; Léger, J.; Barragán-Montero, A.M.; Meert, N.; Lee, J.A.; Macq, B. Domain adversarial networks and intensity-based data augmentation for male pelvic organ segmentation in cone beam CT. Computers in Biology and Medicine 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Lau, T.; Luo, J.; Chang, E.I.C.; Xu, Y. Unsupervised 3D End-to-End Medical Image Registration with Volume Tweening Network. IEEE Journal of Biomedical and Health Informatics 2019. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Medical Imaging 2015, 15. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; Facebook, Z.D.; Research, A.I.; Lin, Z.; Desmaison, A.; Antiga, L.; et al. Automatic differentiation in PyTorch. Neural Information Processing Systems Workshop 2017. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. International Conference on Learning Representations 2014. [Google Scholar] [CrossRef]
| 1 | Centre des Ressources Informatiques et Applications Numérique de Normandie, France |
| CBCT | Fusion | Tumor Mask | DSC | VS | Recall | Precision |
|---|---|---|---|---|---|---|
| Yes | EF | GTV | 0.702±0.015 | 0.837±0.037 | 0.845±0.007 | 0.853±0.010 |
| Yes | LF | GTV | 0.706±0.002 | 0.859±0.018 | 0.824±0.003 | 0.818±0.006 |
| Yes | EF | CTV | 0.680±0.017 | 0.839±0.022 | 0.804±0.013 | 0.735±0.057 |
| Yes | LF | CTV | 0.708±0.028 | 0.850±0.052 | 0.822±0.011 | 0.740±0.022 |
| Yes | EF | PTV | 0.460±0.016 | 0.667±0.113 | 0.788±0.019 | 0.465±0.089 |
| Yes | LF | PTV | 0.665±0.012 | 0.860±0.028 | 0.787±0.009 | 0.686±0.033 |
| Yes | NA | NA | 0.425±0.025 | 0.574±0.020 | 0.608±0.037 | 0.266±0.041 |
| No | NA | GTV | 0.577 | |||
| No | NA | CTV | 0.378 | |||
| No | NA | PTV | 0.189 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





