Submitted:
08 March 2023
Posted:
09 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Method
2.1. Bacteria Isolation and Biochemical Characterization
2.2. DNA Extraction, PCR and Sequence Analysis
2.3. Genome Sequencing, Assembly and Identification
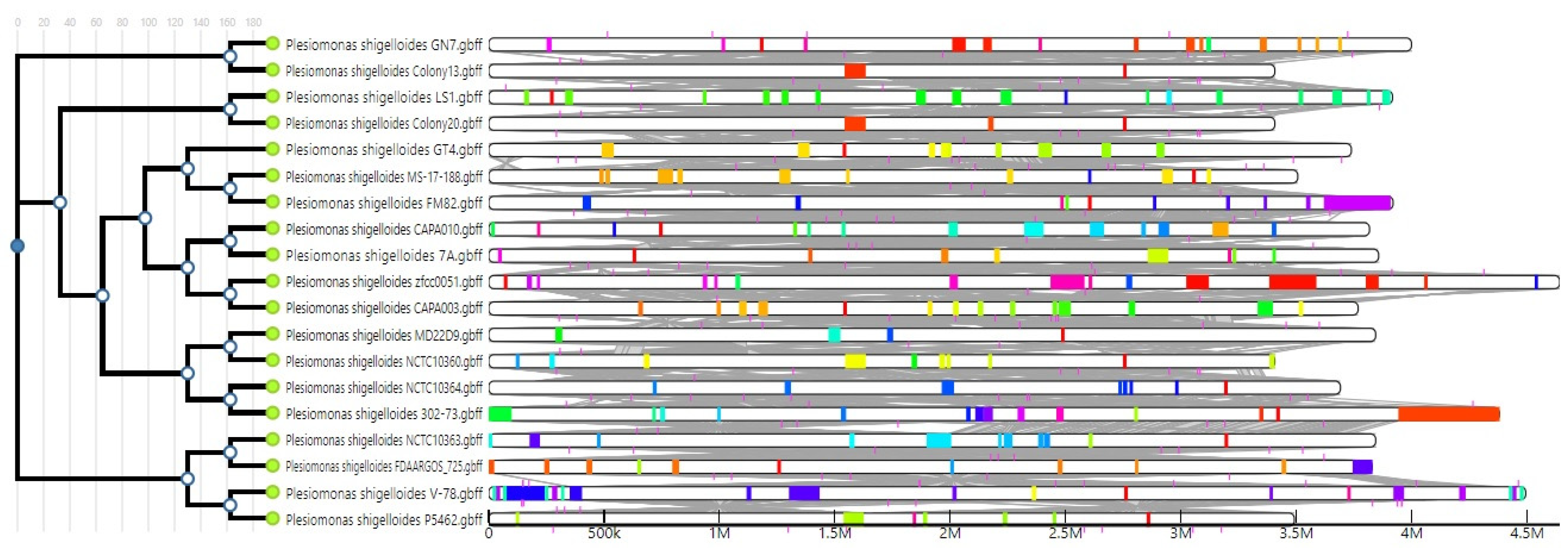
2.4. Comparative Genome Analysis
2.5. Antimicrobial Susceptibility and Resistance Gene Characteristics
3. Results
3.1. Biochemical Characterization
3.2. PCR and Sequence Analysis
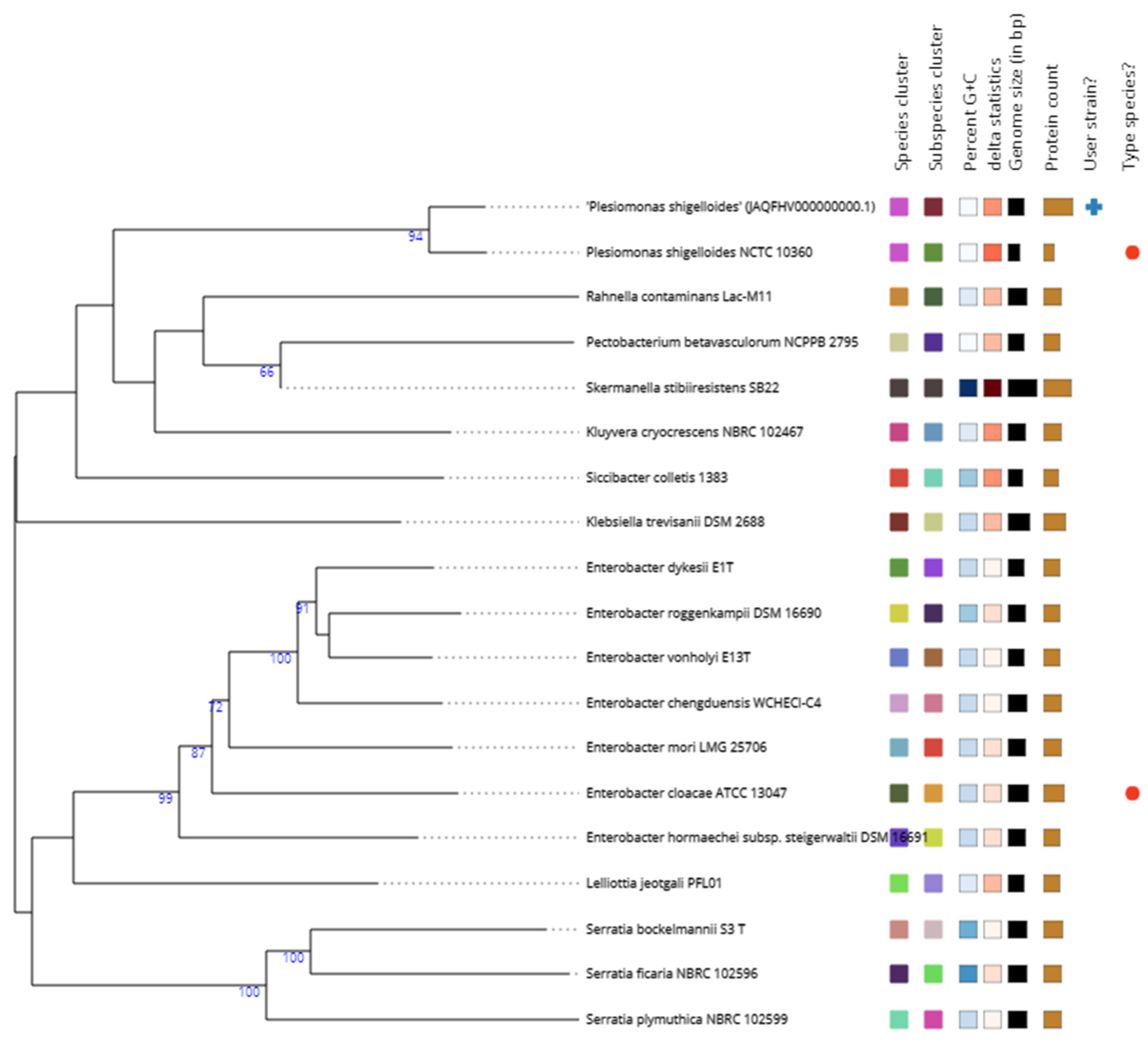
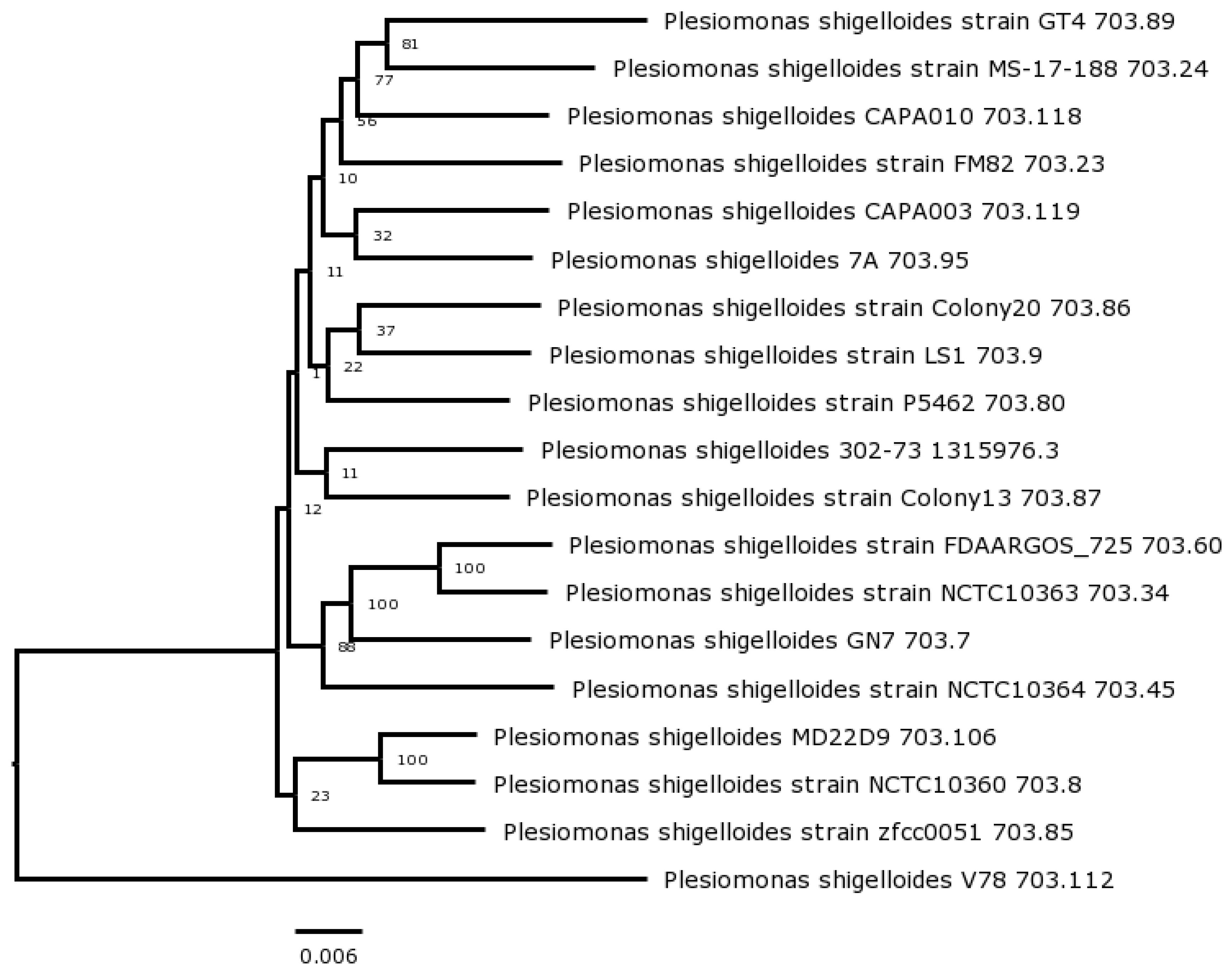
3.3. Phylogenomics
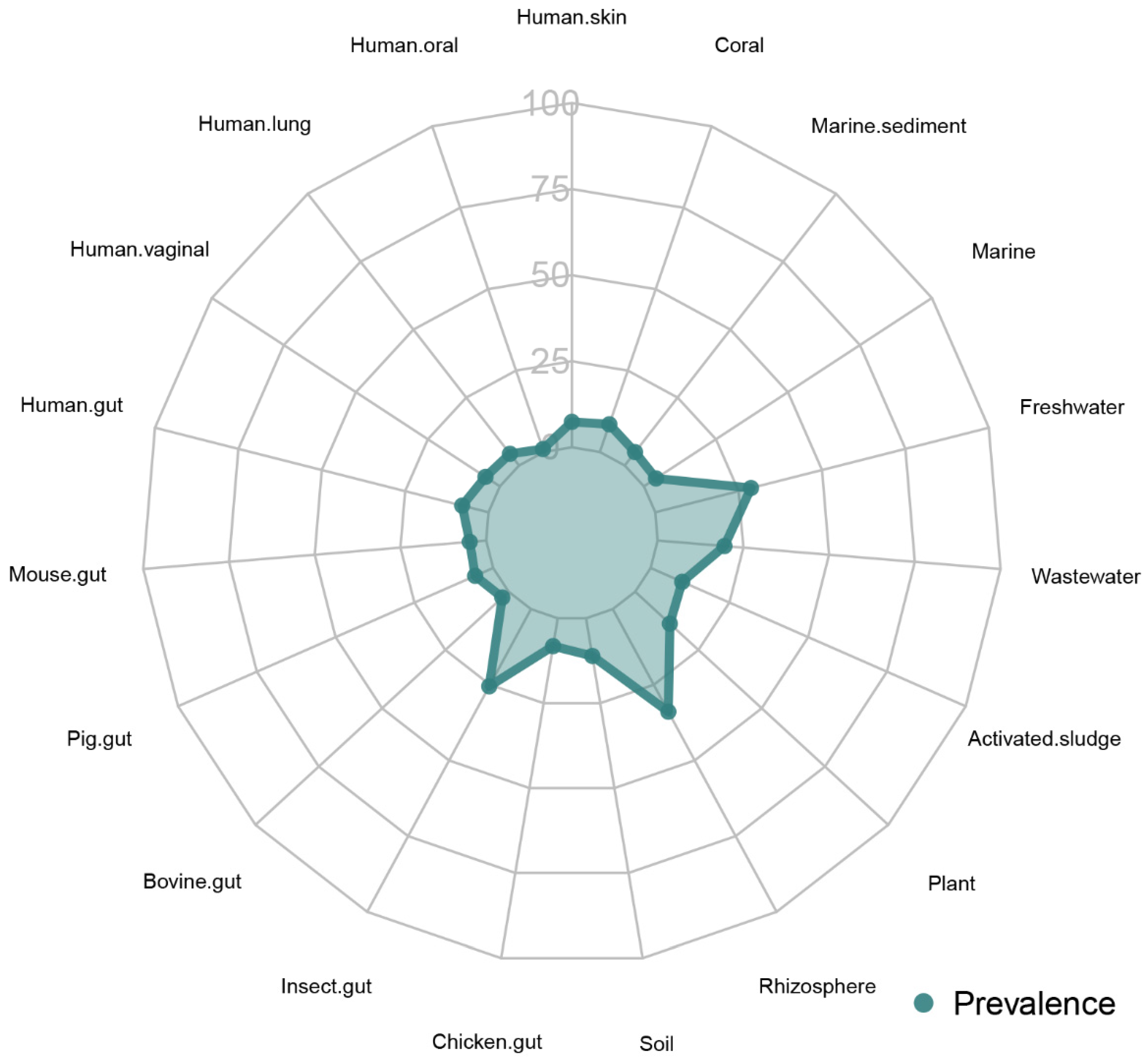
3.4. Functional and Ecological Analyses
3.5. Secondary Metabolites
3.6. Antimicrobial Susceptibility and Resistance Gene Characteristics
3.7. Antimicrobial Resistance Genes and Virulence
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
- Protologue: Emended description of Plesiomonas shigelloides (Bader 1954) Habs and Schubert 1962
- N.L. fem. dim. n. Shigella, a generic name; L. adj. suff. -oides, ressembling, similar; from Gr. neut. adj. suff. -eides, resembling, similar; from Gr. neut. n. eîdos, that which is seen, form, shape, figure; N.L. adj. shigelloides, Shigella-like
- The description is the same as reported by Janda (2015) with the following modifications:
- The genome size of the type strain is 3.4 Mb and genomic G+C content is 52.0%. Type strain: ATCC 14029; CCUG 410; CIP 63.5; DSM 8224; LMG 4242; NCCB 80007; NCTC 10360.
- Description of Plesiomonas shigelloides subsp. shigelloides subsp. nov.
- N.L. fem. dim. n. Shigella, a generic name; L. adj. suff. -oides, ressembling, similar; from Gr. neut. adj. suff. -eides, resembling, similar; from Gr. neut. n. eîdos, that which is seen, form, shape, figure; N.L. adj. shigelloides, Shigella-like
- The description is as given by Janda (2015) with the following modifications: The genome size of the type strain is 3.4 Mb and genomic G+C content is 52.0%. Genome size of strains in the subspecies ranges from 3.0 to 4.08. Genomic GC content ranges from 50.9 to 52.4%. Type strain: ATCC 14029; CCUG 410; CIP 63.5; DSM 8224; LMG 4242; NCCB 80007; NCTC 10360.
- Description of Plesiomonas shigelloides subsp. oncorhynchi subsp. nov.
- on.co.rhyn’chi. N.L. gen. masc. n. oncorhynchi, of Oncorhynchus, named after the rainbow trout, Oncorhynchus mykiss, from which the type strain was isolated
- Gram-negative, short bacilli, motile, oxidase and catalase positive, glucose fermentative, non-hemolytic on sheep blood. Able to tolerate up to 1.5% NaCl and grow at a temperature range of 4-45°C. Negative for gelatin hydrolysis.
- The genome size is 4.4 Mb and genomic G+C content is 51.1%. The GenBank accession number for the whole genome is JAQFHV000000000.
References
- Steinberg, J.P.; Lutgring, J.D.; Burd, E.M. Other Gram-Negative and Gram-Variable Bacilli - ClinicalKey. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Bennett, J.E., Dolin, R., Blaser, M.J., Eds.; Elsevier, 2020; Vol. 9, pp. 2847–2864.
- Willems, A.; Mergaert, J.; Swings, J. Variovorax. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., DeVos, P., Dedysh, S., Hedlund, B., Kämpfer, P., Rainey, F., Trujillo, M.E., Bowman, J.P., Brown, D.R., Glöckner, F.O., et al., Eds.; American Cancer Society: Hoboken, NJ, USA, 2015; Vol. 324, pp. 1–11; ISBN 978-1-118-96060-8.
- Huber, I.; Spanggaard, B.; Appel, K.F.; Rossen, L.; Nielsen, T.; Gram, L. Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhynchus mykiss, Walbaum). J. Appl. Microbiol. 2004, 96, 117–132. [CrossRef]
- Salgado-Miranda, C.; Palomares, E.; Jurado, M.; Marín, A.; Vega, F.; Soriano-Vargas, E. Isolation and Distribution of Bacterial Flora in Farmed Rainbow Trout from Mexico. J. Aquat. Anim. Health 2010, 22, 244–247. [CrossRef]
- Hu, Q.; Lin, Q.; Shi, C.; Fu, X.; Li, N.; Liu, L.; Wu, S. Isolation and identification of a pathogenic Plesiomonas shigelloides from diseased grass carp. Wei sheng wu xue bao= Acta Microbiol. Sin. 2014, 54, 229–35.
- Pakingking, R.; Palma, P.; Usero, R. Quantitative and qualitative analyses of the bacterial microbiota of tilapia (Oreochromis niloticus) cultured in earthen ponds in the Philippines. World J. Microbiol. Biotechnol. 2015, 31, 265–275. [CrossRef]
- Nadirah, M.; Ruhil, H.H.; Jalal, K.C.A.; Najiah, M. Occurrence of Plesiomonas shigelloides in Cultured Red Hybrid Tilapia (Oreochromis niloticus) from Tropical Rivers, East Coast Malaysia. Pak. J. Biol. Sci. 2012, 15, 600–603. [CrossRef]
- Liu, Z.; Ke, X.; Lu, M.; Gao, F.; Cao, J.; Zhu, H.; Wang, M. Identification and pathological observation of a pathogenic Plesiomonas shigelloides strain isolated from cultured tilapia (Oreochromis niloticus). Wei sheng wu xue bao= Acta Microbiol. Sin. 2015, 55, 96–106.
- Michael Janda, J.; Abbott, S.L.; McIver, C.J. Plesiomonas Shigelloides Revisited. Clin. Microbiol. Rev. 2016, 29, 349–374. [CrossRef]
- Cruz, J.M.; Saraiva, A.; Eiras, J.C.; Branco, R.; Sousa, J.C. An Outbreak of Plesiomonas Shigelloides in Farmed Rainbow Trout, Salmo Gairdneri Richardson, in Portugal. Bull. Eur. Assoc. Fish Pathol. 1986.
- Vladík, P.; Vítovec, J. Plesiomonas shigelloides in rainbow troup septicemia. Vet. Med. (Praha).1974, 19, 297–301.
- Wang, X.; Xu, L.; Cao, H.; Wang, J.; Wang, S. Identification and drug sensitivity of a Plesiomonas shigelloides isolated from diseased sturgeons. Wei sheng wu xue bao= Acta Microbiol. Sin. 2013, 53, 723–729.
- Liu, Y.; Rzeszutek, E.; van der Voort, M.; Wu, C.-H.; Thoen, E.; Skaar, I.; Bulone, V.; Dorrestein, P.C.; Raaijmakers, J.M.; de Bruijn, I. Diversity of Aquatic Pseudomonas Species and Their Activity against the Fish Pathogenic Oomycete Saprolegnia. PLOS ONE 2015, 10, e0136241. [CrossRef]
- Nisha, R.G.; Rajathi, V.; Manikandan, R.; Prabhu, N.M. Isolation of Plesiomonas Shigelloides from Infected Cichlid Fishes Using 16S RRNA Characterization and Its Control with Probiotic Pseudomonas Sp. Acta Sci. Vet. 2014, 42, 1–7.
- Ruimy, R.; Breittmayer, V.; Elbase, P.; Lafay, B.; Boussemart, O.; Gauthier, M.; Christen, R. Phylogenetic Analysis and Assessment of the Genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas Deduced from Small-Subunit rRNA Sequences. Int. J. Syst. Evol. Microbiol. 1994, 44, 416–426. [CrossRef]
- Salerno, A.; Delétoile, A.; Lefevre, M.; Ciznar, I.; Krovacek, K.; Grimont, P.; Brisse, S. Recombining Population Structure of Plesiomonas shigelloides ( Enterobacteriaceae ) Revealed by Multilocus Sequence Typing. J. Bacteriol. 2007, 189, 7808–7818. [CrossRef]
- Abdelhamed, H.; Ozdemir, O.; Tekedar, H.C.; Arick, M.A.; Hsu, C.-Y.; Karsi, A.; Lawrence, M.L. Complete Genome Sequence of Multidrug-Resistant Plesiomonas shigelloides Strain MS-17-188. Genome Announc. 2018, 6. [CrossRef]
- Piqué, N.; Aquilini, E.; Alioto, T.; Miñana-Galbis, D.; Tomás, J.M. Genome Sequence of Plesiomonas shigelloides Strain 302-73 (Serotype O1). Genome Announc. 2013, 1. [CrossRef]
- Behera, B.K.; Bera, A.K.; Paria, P.; Das, A.; Parida, P.K.; Kumari, S.; Bhowmick, S.; Das, B.K. Identification and pathogenicity of Plesiomonas shigelloides in Silver Carp. Aquaculture 2018, 493, 314–318. [CrossRef]
- Herrera, F.C.; Santos, J.A.; Otero, A.; García-López, M.-L. Occurrence of Plesiomonas shigelloides in displayed portions of saltwater fish determined by a PCR assay based on the hugA gene. Int. J. Food Microbiol. 2006, 108, 233–238. [CrossRef]
- Martins, A.F.M.; Fontana, H.; Moreira, B.M.; Bonelli, R.R. Draft Genome Sequence of a Tetracycline-Resistant Plesiomonas shigelloides Strain Isolated from Aquaculture-Reared Tilapia. Genome Announc. 2018, 7, e00832-18. [CrossRef]
- Duman, M.; Buján, N.; Altun, S.; Romalde, J.L.; Saticioglu, I.B. Population genetic and evolution analysis of Vibrio isolated from Turkish fish farms. Aquaculture 2023, 562, 738728. [CrossRef]
- PubMLST, P. databases for molecular typing and microbial genome diversity Vibrio Spp. | PubMLST Available online: https://pubmlst.org/organisms/vibrio-spp (accessed on 6 March 2023).
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [CrossRef]
- Mungan, M.D.; Alanjary, M.; Blin, K.; Weber, T.; Medema, M.H.; Ziemert, N. ARTS 2.0: feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining. Nucleic Acids Res. 2020, 48, W546–W552. [CrossRef]
- Bertelli, C.; Gray, K.L.; Woods, N.; Lim, A.C.; Tilley, K.E.; Winsor, G.L.; Hoad, G.R.; Roudgar, A.; Spencer, A.; Peltier, J.; et al. Enabling genomic island prediction and comparison in multiple genomes to investigate bacterial evolution and outbreaks. Microb. Genom. 2022, 8, 000818. [CrossRef]
- A Hitch, T.C.; Riedel, T.; Oren, A.; Overmann, J.; Lawley, T.D.; Clavel, T. Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria. ISME Commun. 2021, 1, 1–16. [CrossRef]
- 2006; 32. CLSI - Clinical and Laboratory Standards Institute VET03-A Methods for Antimicrobial Disk Susceptibility Testing of Bacteria Isolated From Aquatic Animals; Approved Guideline A Guideline for Global Application Developed through the Clinical and Laboratory Standards Institute Consensus Process; 2006;
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [CrossRef]
- Qin, Q.-L.; Xie, B.-B.; Zhang, X.-Y.; Chen, X.-L.; Zhou, B.-C.; Zhou, J.; Oren, A.; Zhang, Y.-Z. A Proposed Genus Boundary for the Prokaryotes Based on Genomic Insights. J. Bacteriol. 2014, 196, 2210–2215. [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132.
- Lagkouvardos, I.; Joseph, D.; Kapfhammer, M.; Giritli, S.; Horn, M.; Haller, D.; Clavel, T. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci. Rep. 2016, 6, srep33721. [CrossRef]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662.e20. [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing—EUCAST. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 15 November 2018).
- Monteil, H.; Harf-Monteil, C. Plesiomonas Shigelloides: Une Bactérie Exotique. La Lett. l’infectiologue 1997, 12.
- Levin, R.E. Plesiomonas shigelloides- An Aquatic Food Borne Pathogen: A Review of its Characteristics, Pathogenicity, Ecology, and Molecular Detection. Food Biotechnol. 2008, 22, 189–202. [CrossRef]
- Miller, M.L.; Koburger, J.A.; MARY L. MILLER and JOHN A. KOBURGER*Food Science and Human Nutrition, University of Florida, Gainesville, Florida 32611; Nedoluha, P.C.; Owens, S.; Russek-Cohen, E.; Westhoff, D.C. Plesiomonas shigelloides: An Opportunistic Food and Waterborne Pathogen. J. Food Prot. 1985, 48, 449–457. [CrossRef]
- Habs, H.; Schubert, R.H. Uber Die Biochemischen Merkmale Und Die Taxonomische Stellung von Pseudomonas Shigelloides (Bader). Zentralblatt Fur Bakteriol. Parasitenkd. Infekt. und Hyg. Abteılung 1 1962, 186, 316.
- Gu, W.; Gonzalez-Rey, C.; Krovacek, K.; Levin, R.E. Genetic Variability among Isolates of Plesiomonas Shigelloides from Fish, Human Clinical Sources and Fresh Water, Determined by RAPD Typing. Food Biotechnol. 2006, 20, 1–12. [CrossRef]
- Holmberg, S.D.; Wachsmuth, I.K.; Hickman-Brenner, F.W.; Blake, P.A.; Farmer, J.J. Plesiomonas Enteric Infections in the United States. Ann. Intern. Med. 1986, 105, 690–694. [CrossRef]
- Kain, K.C.; Kelly, M.T. Clinical features, epidemiology, and treatment of Plesiomonas shigelloides diarrhea. J. Clin. Microbiol. 1989, 27, 998–1001. [CrossRef]
- Jiang, J.; Liu, Y.; Yan, L.; Yan, Q.; Wen, X.; Cao, S.; Huang, Y.; Huang, X.; Ma, X.; Han, X.; et al. Identification and pathogenicity of Plesiomonas shigelloides from Acipenser dabryanus in China. Aquac. Res. 2021, 52, 2286–2293. [CrossRef]
- Janda, J.M.; Abbott, S.L. Expression of hemolytic activity by Plesiomonas shigelloides. J. Clin. Microbiol. 1993, 31, 1206–1208. [CrossRef]
- Baratéla, K.C.; Saridakis, H.O.; Gaziri, L.C.J.; Pelayo, J.S. Effects of medium composition, calcium, iron and oxygen on haemolysin production by Plesiomonas shigelloides isolated from water. J. Appl. Microbiol. 2001, 90, 482–487. [CrossRef]
- Falcón, R.; Carbonell, G. V; Figueredo, P.M.S.; Butiao, F.; Saridakis, H.O.; Pelayo, J.S.; Yano, T. Intracellular vacuolation induced by culture filtrates of Plesiomonas shigelloides isolated from environmental sources. J. Appl. Microbiol. 2003, 95, 273–278. [CrossRef]
- Abbott, S.L.; Kokka, R.P.; Janda, J.M. Laboratory investigations on the low pathogenic potential of Plesiomonas shigelloides. J. Clin. Microbiol. 1991, 29, 148–153. [CrossRef]
- Gardner, S.E.; Fowlston, S.E.; George, W.L. In Vitro Production of Cholera Toxin-Like Activity by Plesiomonas shigelloides. J. Infect. Dis. 1987, 156, 720–722. [CrossRef]
- Kaszowska, M.; Stojkovic, K.; Niedziela, T.; Lugowski, C. The O-antigen of Plesiomonas shigelloides serotype O36 containing pseudaminic acid. Carbohydr. Res. 2016, 434, 1–5. [CrossRef]
- Hoel, S.; Vadstein, O.; Jakobsen, A.N. Species Distribution and Prevalence of Putative Virulence Factors in Mesophilic Aeromonas spp. Isolated from Fresh Retail Sushi. Front. Microbiol. 2017, 8, 931. [CrossRef]
- Qian, Q.; Chen, Z.; Xu, J.; Zhu, Y.; Xu, W.; Gao, X.; Jiang, Q.; Zhang, X. Pathogenicity of Plesiomonas shigelloides causing mass mortalities of largemouth bass (Micropterus salmoides) and its induced host immune response. Fish Shellfish. Immunol. 2023, 132, 108487. [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [CrossRef]
| Strain name | Host | Country | Total Genes | Housekeeping Genes | Known Resistance | Virulence Genes |
|---|---|---|---|---|---|---|
| zfcc0051 | Zebra Fish | USA | 3818 | 596 | 31 | 41 |
| V-78 | Rainbow trout | Turkey | 5454 | 297 | 19 | 41 |
| P5462 | Gentoo penguins | Hong Kong | 3287 | 596 | 33 | 40 |
| NCTC10364 | Human | Unknown | 3098 | 584 | 32 | 42 |
| NCTC10363 | Human | Unknown | 3264 | 599 | 30 | 40 |
| NCTC10360 | Dog | Unknown | 2886 | 583 | 28 | 40 |
| MS-17-188 | Catfish | USA | 3426 | 595 | 32 | 42 |
| MD22D9 | Macrobdella decora | USA | 3190 | 595 | 32 | 40 |
| LS1 | Percocypris pingi | China | 3484 | 590 | 33 | 43 |
| GT4 | Oreochromis niloticus | Brazil | 3134 | 600 | 34 | 43 |
| GN7 | Water | Malaysia | 3434 | 596 | 33 | 42 |
| FM82 | Fish | Brazil | 3233 | 593 | 32 | 41 |
| FDAARGOS_725 | Human | USA | 3250 | 597 | 32 | 40 |
| Colony20 | Food | Thailand | 2598 | 576 | 28 | 43 |
| Colony13 | Human | Thailand | 2648 | 580 | 28 | 39 |
| CAPA010 | Oreochromis niloticus | Peru | 3176 | 599 | 33 | 42 |
| 7A | River | Sweden | 3298 | 597 | 32 | 41 |
| 302-73 | Human | Japan | 3398 | 592 | 32 | 40 |
| CAPA003 | Oreochromis niloticus | Peru | 3185 | 596 | 33 | 41 |
| Strain number | Type strain | POCP (%) |
|---|---|---|
| Strain V-78 | Plesiomonas shigelloides | 68.1 |
| Yersinia entomophaga | 43.2 | |
| Obesumbacterium proteus | 42.4 | |
| Hafnia paralvei | 42.1 | |
| Buttiauxella agrestis | 41.8 | |
| Citrobacter werkmanii | 41.8 | |
| Buttiauxella noackiae | 41.8 | |
| Citrobacter pasteurii | 41.7 | |
| Salmonella enterica | 41.7 | |
| Buttiauxella brennerae | 41.6 | |
| Citrobacter freundii | 41.5 | |
| Enterobacter soli | 41.2 | |
| Serratia marcescens | 40.9 | |
| Serratia grimesii | 40.8 | |
| Pectobacterium carotovorum | 40.8 | |
| Serratia proteamaculans | 40.6 | |
| Buttiauxella ferragutiae | 40.6 | |
| Serratia nematodiphila | 40.5 | |
| Citrobacter braakii | 40.5 | |
| Serratia odorifera | 40.3 | |
| Chania multitudinisentens | 40.3 | |
| Serratia quinivorans | 40.2 | |
| Yersinia pestis | 40.2 | |
| Serratia fonticola | 40.2 | |
| Buttiauxella gaviniae | 40.1 | |
| Enterobacter hormaechei | 40.0 | |
| Proteus hauseri | 39.9 | |
| Serratia ficaria | 39.9 | |
| Serratia plymuthica | 39.9 | |
| Cronobacter sakazakii | 39.8 | |
| Citrobacter amalonaticus | 39.7 | |
| Pectobacterium parmentieri | 39.5 | |
| Enterobacter cloacae | 39.4 | |
| Dickeya chrysanthemi | 39.4 | |
| Raoultella planticola | 39.0 | |
| Klebsiella pneumoniae | 38.9 | |
| Raoultella ornithinolytica | 38.8 | |
| Lonsdalea quercina | 38.3 | |
| Pantoea agglomerans | 38.1 | |
| Enterobacter ludwigii | 38.0 | |
| Rouxiella chamberiensis | 38.0 | |
| Pantoea allii | 37.2 | |
| Rouxiella silvae | 37.2 | |
| Rouxiella badensis | 37.0 | |
| Sodalis praecaptivus | 36.3 | |
| Pantoea cypripedii | 35.3 |
| Environment | Detection ratio (%) | Mean relative abundance (%) | Standard deviation (%) |
|---|---|---|---|
| Rhizosphere | 33.9 | 0.71 | 3.47 |
| Freshwater | 28.7 | 3.29 | 12.49 |
| Insect gut | 25.5 | 4.42 | 16.15 |
| Wastewater | 19.5 | 0.20 | 0.55 |
| Plant | 13.7 | 3.26 | 11.07 |
| Soil | 11.1 | 0.20 | 1.37 |
| Activated sludge | 10.0 | 0.02 | 0.05 |
| Coral | 8.5 | 0.38 | 0.66 |
| Chicken gut | 8.2 | 0.23 | 1.17 |
| Human gut | 7.9 | 6.17 | 16.59 |
| Human skin | 7.4 | 0.22 | 0.67 |
| Pig gut | 5.7 | 0.04 | 0.08 |
| Human vaginal | 5.0 | 0.02 | 0.04 |
| Marine sediment | 4.9 | 0.10 | 0.21 |
| Mouse gut | 4.8 | 0.15 | 0.33 |
| Human lung | 4.3 | 0.38 | 1.61 |
| Marine | 4.2 | 0.05 | 0.22 |
| Bovine gut | 2.5 | 0.03 | 0.05 |
| Human oral | 0.9 | 0.20 | 0.36 |
| Family | Antibiotics |
Disk content (µg) |
A. salmonicida subps. salmonicida ATCC 33658 |
E. coli ATCC 25922 |
P. shigelloides | ||
| V63 | V78 | AF160 | |||||
| Sulfonamides | Trimethoprim/Sulfamethoxazole (SXT) | 25 | 23 | 27 | 25 (S) | 25 (S) | 21 (S) |
| Aminopenicillins | Amoxicillin (AML)* | 25 | 32 | 23 | 12 | 13 | 25 |
| Tetracyclines | Doxycycline (DO)* | 30 | 31 | 25 | 22 | 20 | 15 |
| Tetracycline (TE)* | 30 | 35 | 30 | 15 | 13 | 12 | |
| Oxytetracycline (OT)* | 30 | 30 | 29 | 10 | 0 (R) | 0 (R) | |
| Quinolone | Oxolinic Acid (OA)* | 2 | 35 | 28 | 24 | 25 | 10 |
| Fluoroquinolone | Enrofloxacin (ENR)* | 5 | 40 | 37 | 31 | 35 | 25 |
| Ciprofloxacin (CIP) | 5 | 50 | 40 | 33 (S) | 33 (S) | 25 (S) | |
| Flumequine (UB)* | 30 | 40 | 35 | 26 | 30 | 28 | |
| Cephalosporins | Cefalexin (CN)* | 10 | 25 | 25 | 20 | 20 | 21 |
| Penicillins | Ampicillin (AMP)* | 10 | 32 | 20 | 11 | 12 | 10 |
| Macrolides | Erythromycin (E)* | 15 | 22 | 8 | 16 | 11 | 10 |
| Clindamycin | Lincomycin (MY)* | 15 | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) |
| Chloramphenicol | Florfenicol (FFC)* | 30 | 40 | 24 | 11 | 12 | 12 |
| *Cut-off values are not available on EUCAST | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





