1. Introduction
Gaussian Mixture Models (GMMs) are powerful, parametric, and flexible models in the probabilistic realm. GMMs tackle two important problems. The first is known as the “inverse problem,” in which
and
x are not unique one-to-one mappings [
1]. The second issue is the uncertainty problem of both the data (aleatoric) and the model (epistemic) [
2,
3,
4]. When we consider the two problems together, it is easy to see that if we deal with multimodal problems, classic model assumptions start falling apart. Traditionally, we attempt to discover the relationship between input
x and output
y, which is denoted as
with erro
.In a probabilistic sense, the above model assumption is equivalent to
. In terms of inverse and uncertainty problems, the model assumption is
. Under this condition, when
y is no longer following any distribution in a known form, GMM becomes a viable option for model data. If
could be satisfied, then we could use GMM as a general model regardless of whatever distribution
y is following. The question remaining for us to answer is whether GMMs can approximate arbitrary densities. Some studies show that all normal mixture densities are dense in the set of all density functions under the
metric [
5,
6,
7]. A model that can fit an arbitrary distribution density is relatively similar, as it requires the model to imitate all possible shapes of distribution density. In this work, taking ideas from Fourier expansion, we assume that any distribution density can be approximately seen as a combination with a set of finite Gaussian distributions. To put it another way, any density can be approximated by a set of Gaussian distribution bases with fixed means and variances. GMM approximates an unknown density by using data to learn the amplitudes (coefficients) of the Gaussian components.In summary, we have
X as input data,
as output data, and
here
is the function that requires learning. To learn
, which maps input x into GMM parameters, we can use any modeling technique.
While modeling using GMM, we need to perform two stages of learning. To begin, we must find appropriate GMM parameters to approximate the target distribution . Secondly, once we have the values of GMM parameters , we can learn model map to . In this paper, we mainly discuss the first part of learning. How to learn a GMM to approximate the target distribution. Fitting a GMM from the observed data related to two problems: 1) how many mixture components are needed; 2) how to estimate the parameters of the mixture components.
For the first problem, some techniques are introduced [
5]. For the second problem, as early as 1977, the Expectation Maximization (EM) algorithm [
8] was well developed and capable of fitting GMMs based on minimizing the negative log-likelihood (NLL). While the EM algorithm remains one of the most popular choices for learning GMMs, other methods have also been developed for learning GMMs [
9,
10,
11]. The EM algorithm has a few drawbacks. Srebro [
12] asked an open question that questioned the existence of local maxima between an arbitrary distribution and a GMM. That paper also addressed the possibility that the KL-divergence between an arbitrary distribution and mixture models may have non-global local minima. Especially in cases where the target distribution has more than
k components but we fit it with a GMM with fewer than
k components. Améndola at al. [
13] has shown that the likelihood function for GMMs is transcendental. Jin et al. [
14] resolved the problem that Srebro discovered and proved that with random initialization, the EM algorithm will converge to a bad critical point with high probability. The maximum likelihood function is transcendental and will cause arbitrarily many critical points. Intuitively speaking, when handling any distribution with multimodal analysis, if the size of the GMM components is less than the peaks that the target distribution contains, it will lead to an undesirable estimation.
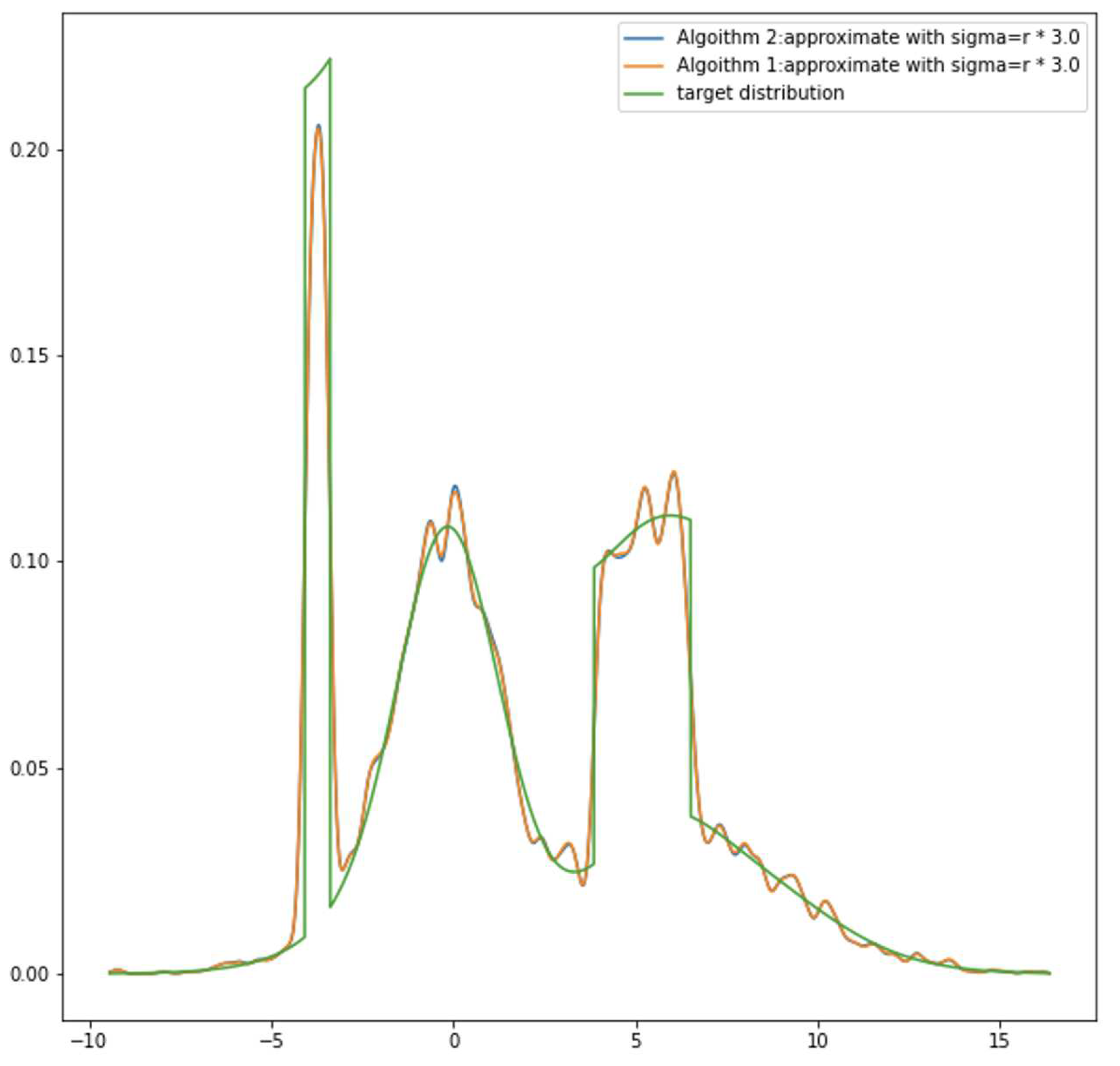
Furthermore, instead of a bell shape, if a peak of the target distribution is present as a flat top like a roof, a single Gaussian component is limited in capturing these kinds of features and is not capable of making a good estimation. It is not a surprise that the likelihood function could have bad local maxima and be arbitrarily worse than any global optimum. Other studies turn their heads to the Wasserstein distance. Instead of minimizing the NLL function or Kullback-Leibler divergence, Wasserstein distances are introduced to replace the NLL function [
11,
15,
16]. These methods avoid the downside of using NLL, but the calculation and formulation of the learning process are relatively complex.
Instead of following the EM algorithm, we take a different path. The main concept of our work is to take the idea of Fourier expansion and apply it to density decomposition. Similarly, like Fourier expansion, our model needs a set of base components, and their location is predetermined. Instead of using a couple of components, we use a relatively large number of components to construct this base. The benefit of this approach is that these bases represent how much detail our models can capture without going into a calculation process to decide how many GMM components are needed. Another benefit of this idea is that, on a predetermined basis, the learning parameters become relatively simple. We only need to learn an appropriate set of which are the probability coefficients of normal components, to reach an optimum approximation. A simple algorithm without heavy formulation is designed to achieve this task. More detailed explanations, formulations, and proofs are provided in the next section. It is worth mentioning that, like cousin expansion, if base frequencies are not enough, there will be information loss for the target sequence. This method suffers similar information losses from not enough Gaussian components. We provide a measurement of the information loss. In our experiments, our method provided a good estimation result.
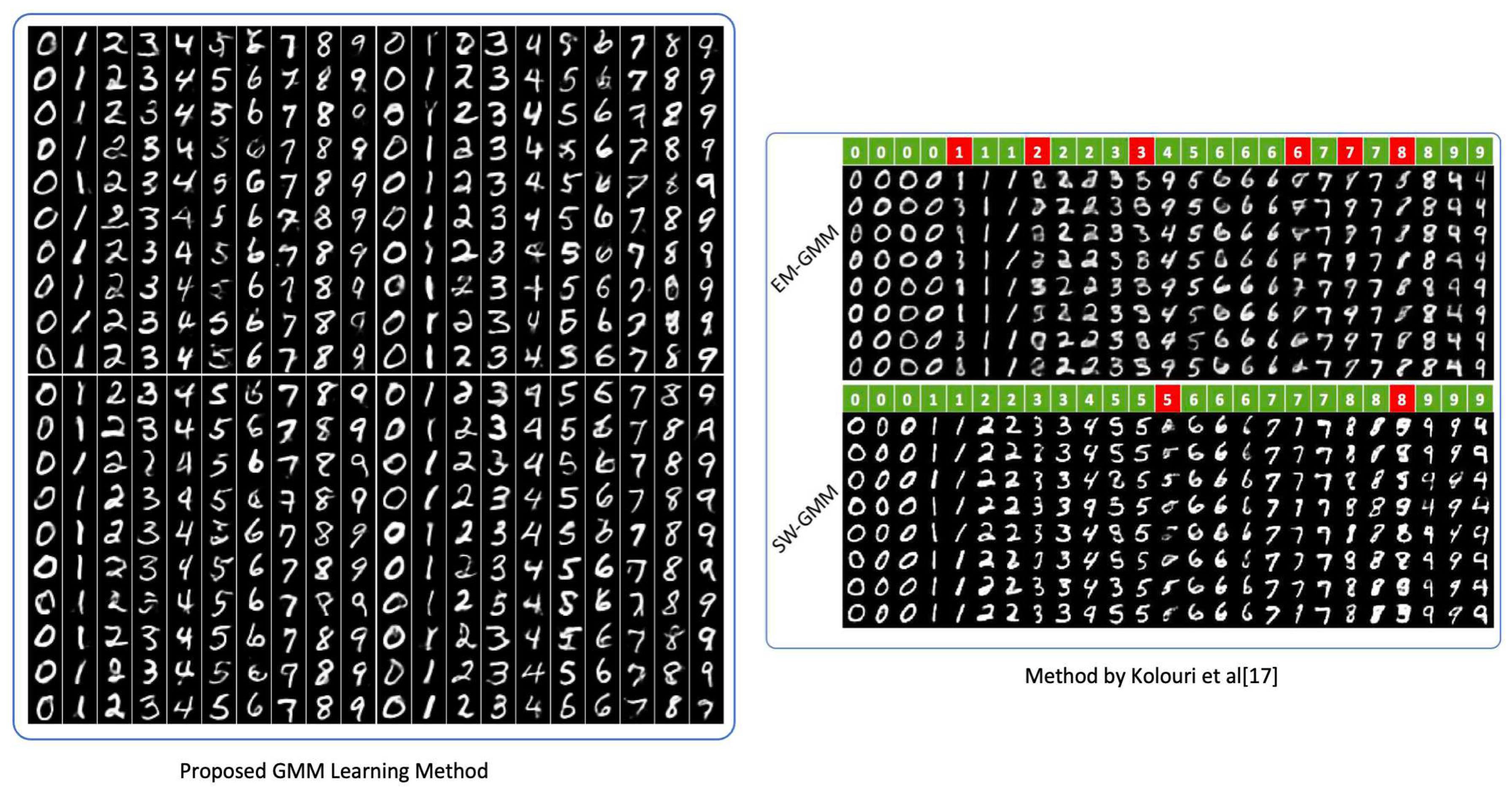
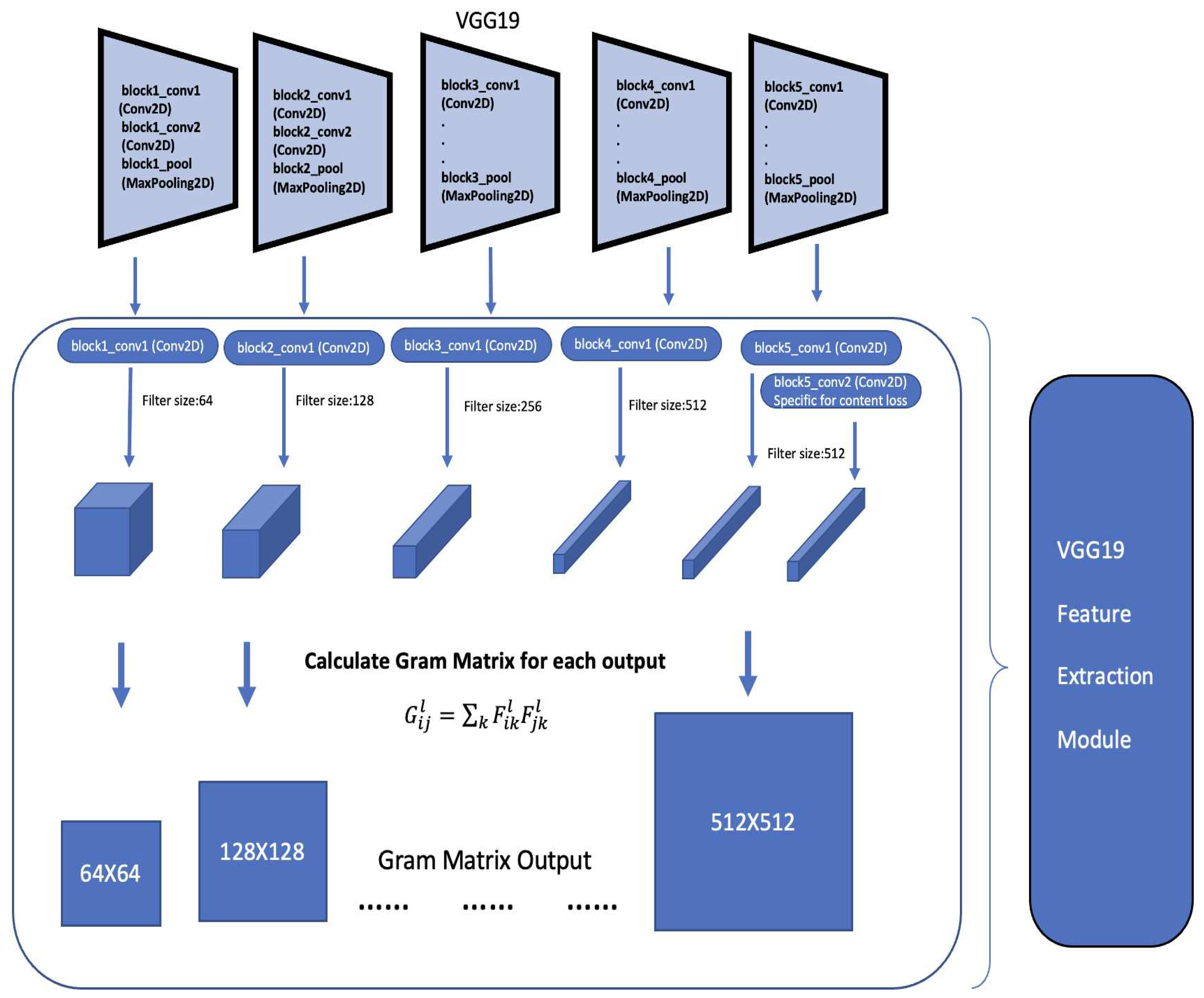
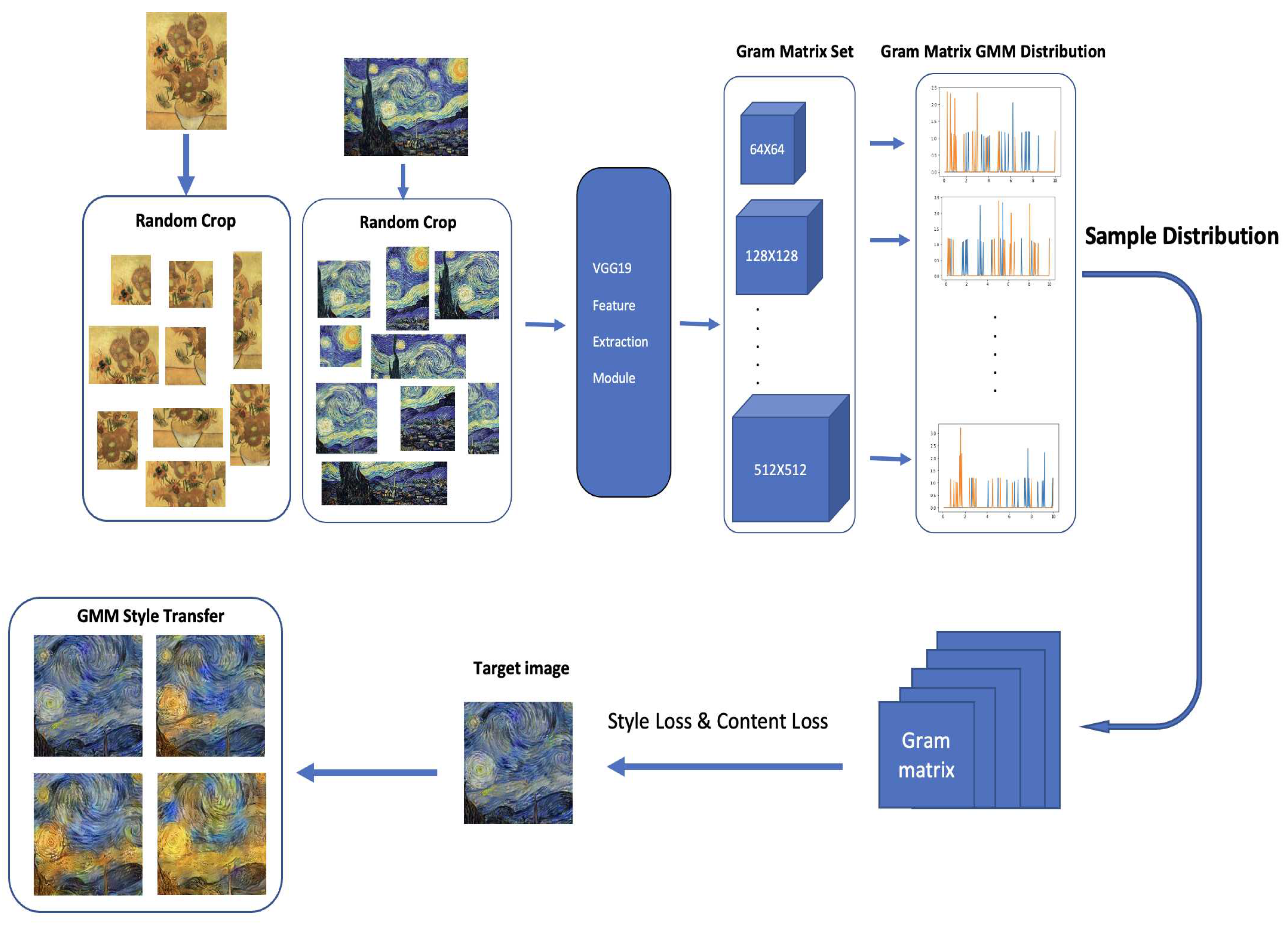
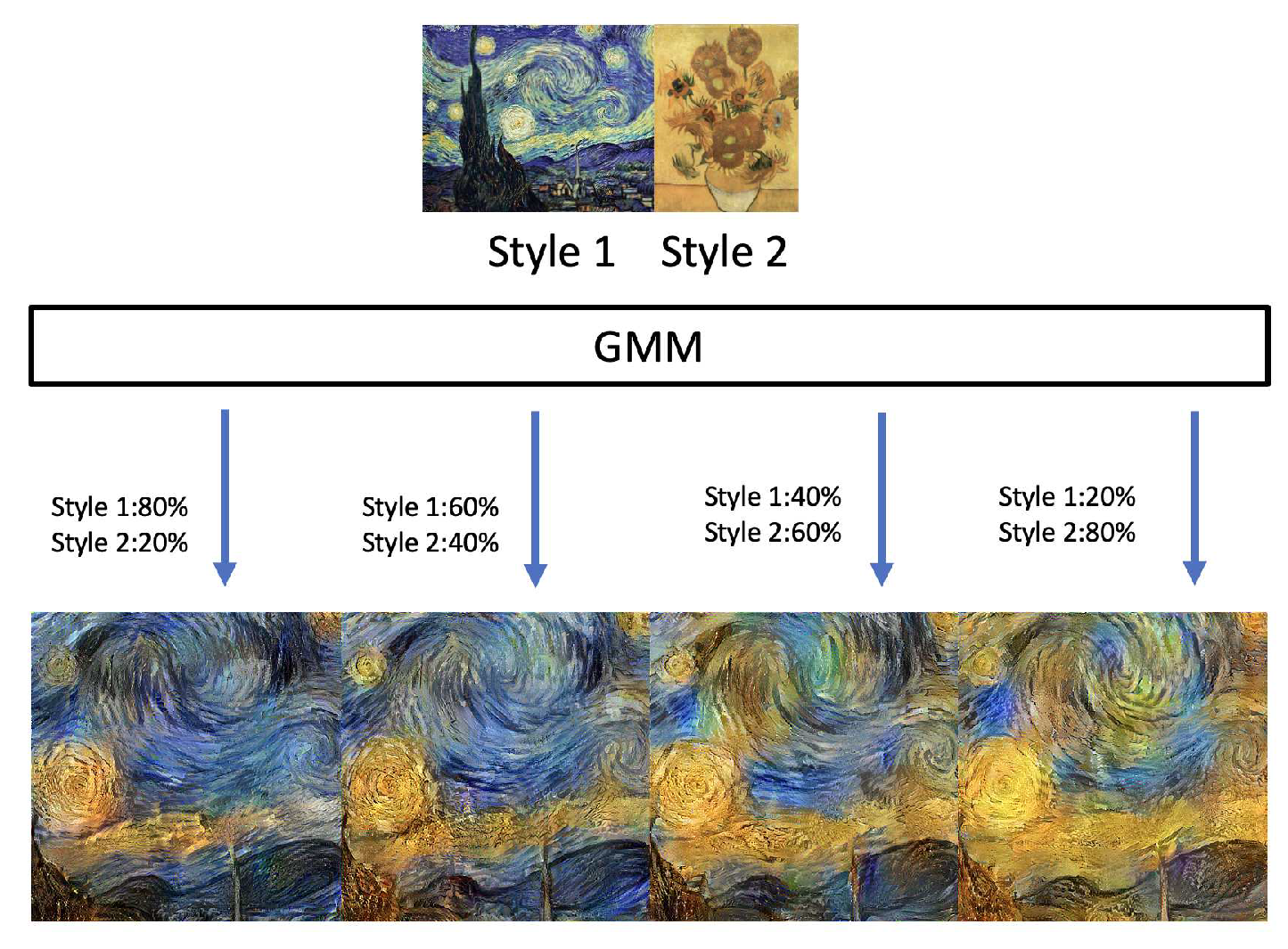
GMMs have been used across computer vision and pattern classification among many others [
17,
18,
19,
20]. Additionally, GMMs can also be used to learn embedded space in neural networks [
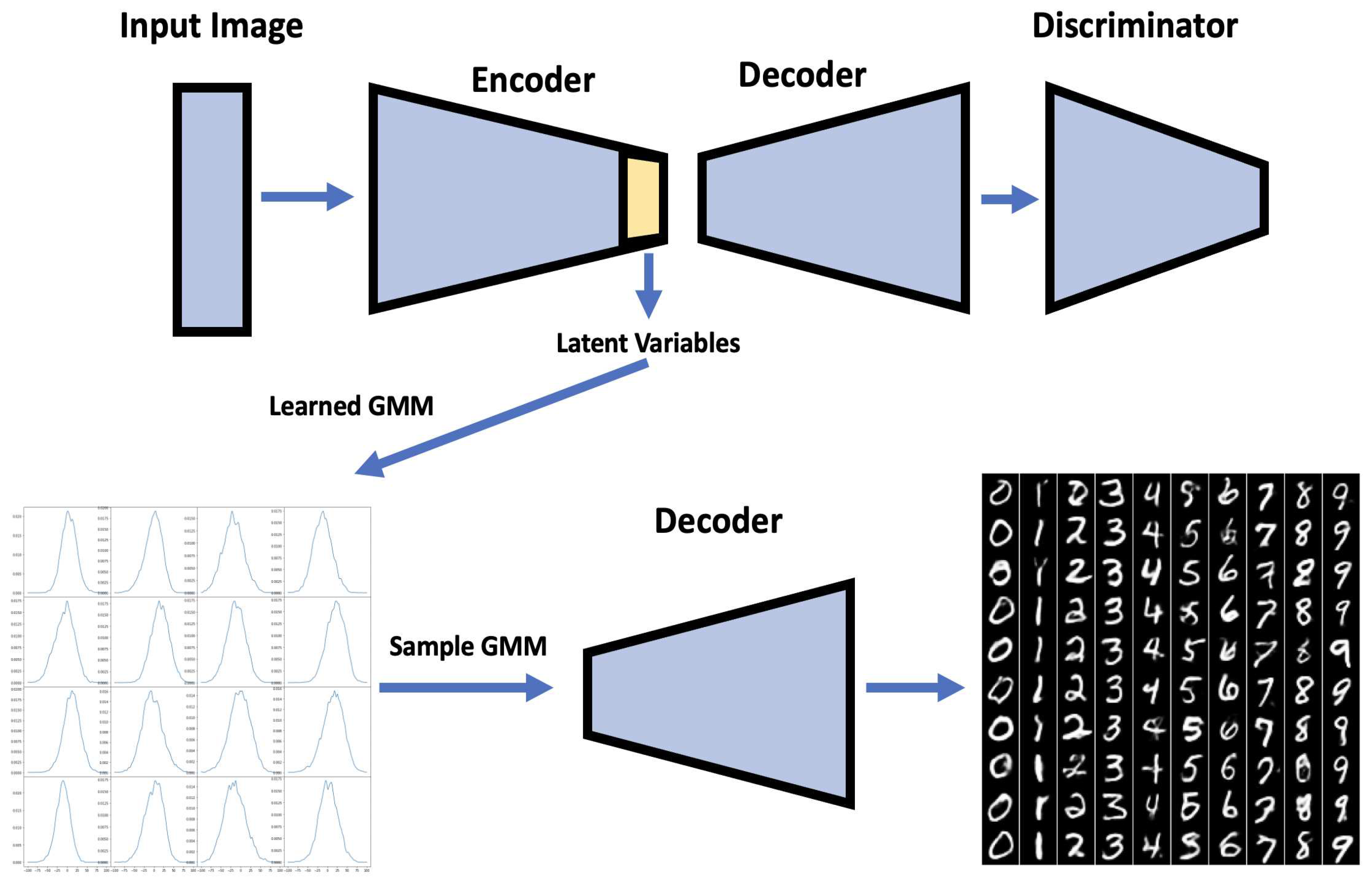
11]. Similarly, as Kolouri et al., we apply our algorithm to learn embedded space in neural networks in two classic neural network applications of handwritten digits generation using autoencoders [
21,
22] and style transfer [
23]. This helps us turn latent variables into a complex distribution, which allows us to perform sampling to create variation. More detail is provided in
Section 4.
Section 2 introduces Fourier expansion-inspired Gaussian mixture models, shows this method is capable of approximating arbitrary densities, discusses the convergency, defines a suitable metric, and details some of its properties.
Section 3 provides our methodology for fitting a GMM through data, introduces two learning algorithms, and shows learning accuracy through experiments.
Section 4 presents two neural network applications. The final section provides a summary and conclusions.
2. Decompose Arbitrary Density by GMM
In this section, we show our method from scratch, including learning algorithms, as well as prove that the idea of density decomposition by GMM is feasible with bounded approximation error. A metric called density information loss is introduced to measure the approximation error between target density and GMM. Let’s begin with the basic assumption and notation:
Observed a dataset X, which is following a unknown distribution with density ;
Assuming there is a GMM that matches the distribution of X :;
The density of GMM: ;
As general definition of GMM, and .
In this work, we mainly discuss one-dimensional cases. The GMM is mixed with Gaussian distributions rather than multivariate Gaussian distributions.
2.1. General Concept of GMM Decomposition
The idea behind GMM decomposition is a Fourier series in probability that, assuming arbitrary density, can be accurately decomposed by infinite mixtures of Gaussian distributions. As mentioned earlier, a lot of studies prove that the likelihood of GMM has many critical points. There are far too many sets of and that could produce the same likelihood result on a given dataset. Learning three sets of parameters, could increase the complexity of the learning process. Not to mention that these parameters interact with each other. Changing one of them could lead to new sets of optimal solutions for others. If we consider that GMM can decompose any density like the Fourier series into a periodic sequence, we can predetermine the base Gaussian distributions (component distributions).
In practice, can be set to be evenly spread out though the dataset space. The parameter can be treated as a hyperparameter that adjusts overall GMM density smoothness. This means that of the component distribution are non-parametric and do not need to be optimised. The parameter is the only remaining problem for us to solve. Our goal shifted from finding the best set of to finding the best set of on a given dataset. With this idea, our GMM set-up becomes:
is the density of GMM;
, , n is the numbers of normal distributions;
is the interval for locating ;
are means for Gaussian components;
is——;
t is a real number hyper-parameter, usually .
2.2. Approximate Arbitrary Density by GMM
In this subsection, we show that, in certain conditions, if the component size of a mixture Gaussian model goes to infinite, the error of approximating an arbitrary density can go toward zero.
Through the idea of Monte Carlo, we could assume that the accuracy of a fine frequency distribution estimation of the target probability mass through dataset is good enough in general. A frequency distribution could also be seen as a mixture distribution with a uniform distribution components. Elaborate from this, if we swap each uniform distribution into a Gaussian unit and push this toward infinite, we could have a correct probability mass approximation of any target distribution. Because we do not have access to the true density, the dataset is all we have, an accurate probability mass is generally good enough in practice. Let
, we have:
Expand the density of GMM
results
.
It is not hard to notice that, if
which means that each Gaussian component is very dense in their region, GMM become a discrete distribution that can be precisely 1:1 clone the estimation probability mass. It becomes:
These simple deductions say that under this set up, a GMM with a large size of component is guaranteed to perform as well as a frequency distribution. Furthermore, changing the mixture component from a regular Gaussian distribution to a truncated normal distribution or uniform distribution could lead to the same conclusion. One good thing about this set up is that it could bring discrete probability mass estimation to continuous density. Allow us to deal with regression problems in a classification manner. We do suffer loss of information at some points in the density, but generally, in practice, the probability mass is what we are looking for. For example, let’s say we are trying to figure out the distribution of people’s height. Instead of looking for a precise density estimation of people’s height, , is what we targeting. Therefore, if we accept a discrete frequency distribution as one of our general approximation models, there is no reason that GMM can not be used to approximate arbitrary density, because it is capable of performing as good as frequency distribution estimation.
2.3. Density Information Loss
Even though GMM can approximate arbitrary density similarly to frequency distribution, density information within remains a problem that needs to be discussed. In another word, even if we learn a model that probability mass is equal to target density , the differentiation is not equal . We could apply KL-divergence or Wasserstain distance to measure the difference between two densities. But in our set up, we introduce a simple metric to measure regional density information loss (DIL). If DIL is small enough under some conditions, then we may be able to conclude that GMM can be a generalised model to decompose arbitrary density.
First of all, assume we have already learned a GMM which achieves that probability mass is correctly mapped to the target distribution. We split the data space into n regions denoted as
the same as
Section 2.3.
The total density information loss is sum of absolute error.
Because the integration is not particularly easy for us to calculate, here we divide
further into m smaller regions to estimate DIL.
Where
is the discrete approximation of density information loss within region
. If
is universally bounded in some conditions for all
, the overall DIL is bounded. For any
from
, if
we have
In fact,
is bound within a comfortable condition. In
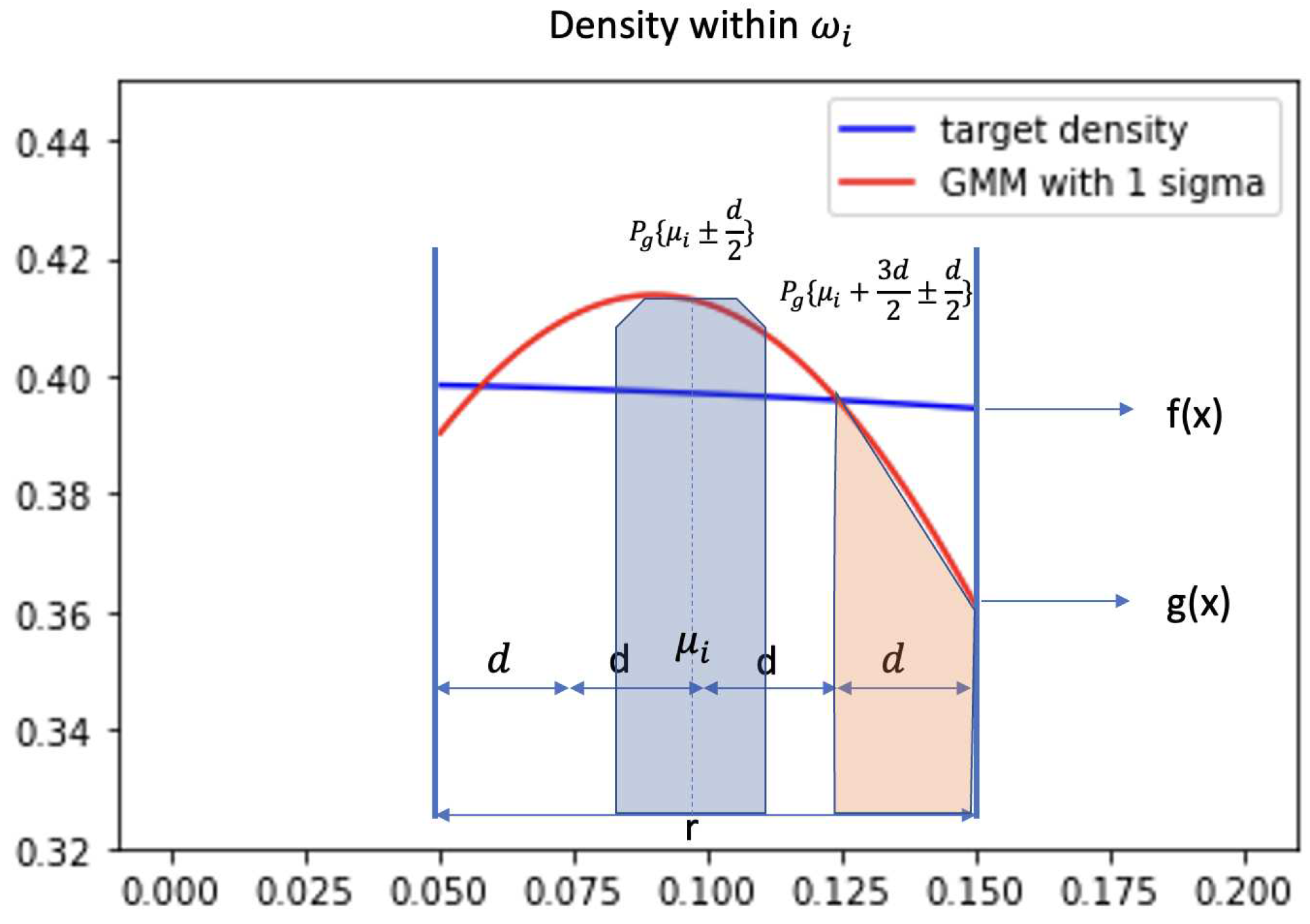
Figure 1, the plot demonstrates the density of target distribution(blue) and GMM(red) in a small region
. The probability
is equal to
. Evenly split
into 4 smaller areas and each area has length
d. When we look at this geometrical relationship between target distribution
and GMM
, we discover that for each small region d,
is bounded. Intuitively, the blue area which is shown in
Figure 1 will always be larger than the orange area. Because we have learned a GMM that
and our GMM model is constructed by evenly spreading out Gaussian components, at each
generally will share similar geometrical structure as
Figure 1.
In brief, giving some conditions, the blue area subtracts the orange area in
Figure 1 is guarantee larger than
. We provide the proof below.
Given condition:
Condition 1:
Condition 2: Because is small, target density is approximately monotone and linear within . Where
Condition 3: GMM density within double cross the target density. Two cross points are named a, b and .
Set the spilt length
. Given condition that GMM density double cross the target density. Since
, for all
, and
we have
and
By condition 2, if target density is monotonically decreasing, i.e.,
, for all
and
Therefore,
With Equation (
2), we have:
Similarly, if target density is monotonically increasing:
The benefit of these two properties of Equations (
3) and (
3) is that even though we do not know what the target density is, this system still guarantees the difference between GMM and target density, which is bounded by Equations (
3) and (
3). As long as we are able to find a good approximation that satisfies condition 1, we will be able to reach an approximation that is not worse than a fully discrete frequency distribution, while keeping the density continuous. This will lead us to a simple algorithm to optimise GMM which shown in the next subsection.
Conditions 1-3 also provide another practical benefit. As long as these 3 conditions are satisfied,
of each Gaussian component becomes a universal smoothness parameter which controls the kurtosis of each normal component. A larger
produces an overall smoother density.
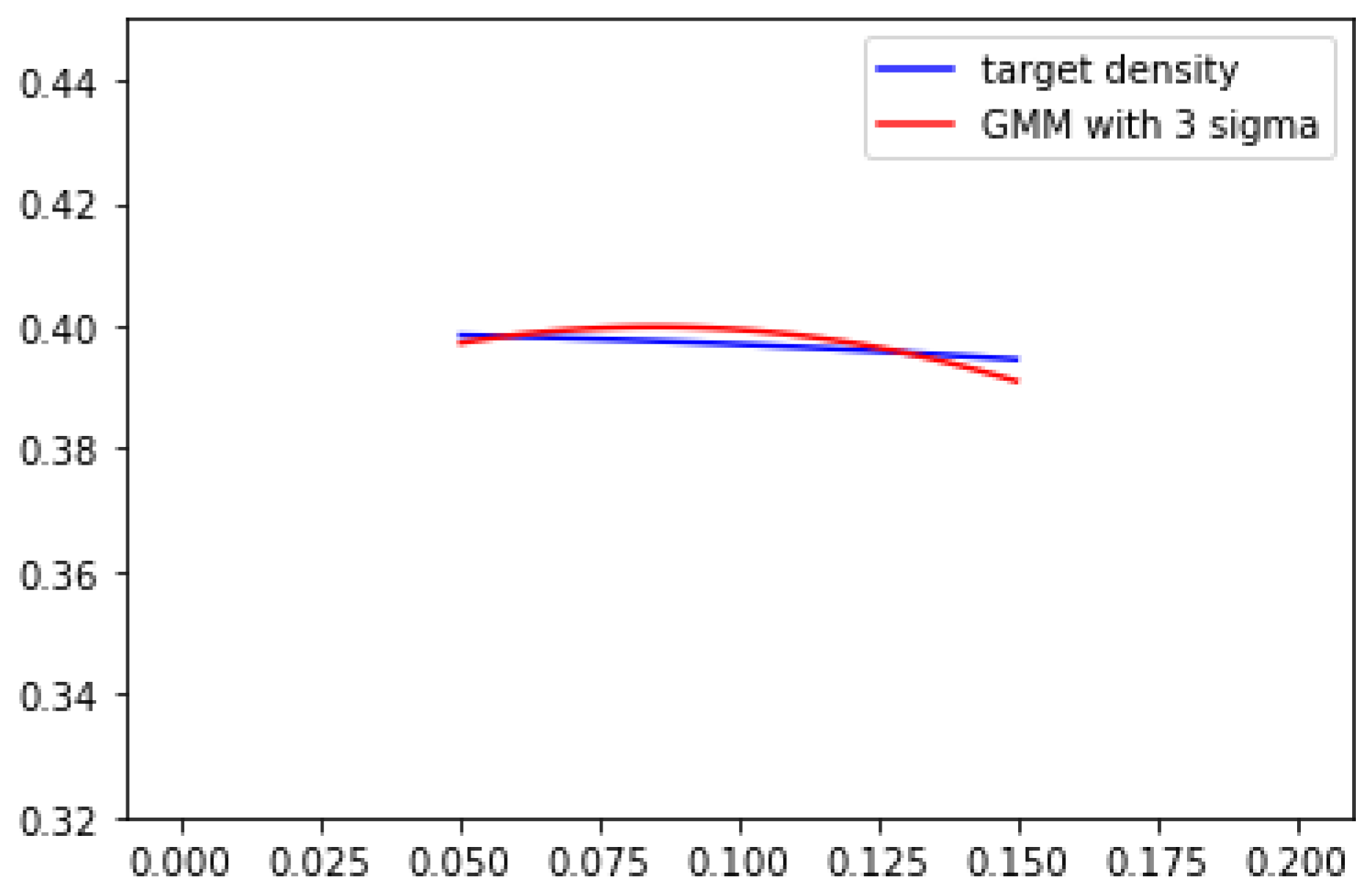
can be treated as a hyper-parameter which does not affect the learning process. GMM in
Figure 1 uses
. If we tune sigma larger, we will have the result shown in
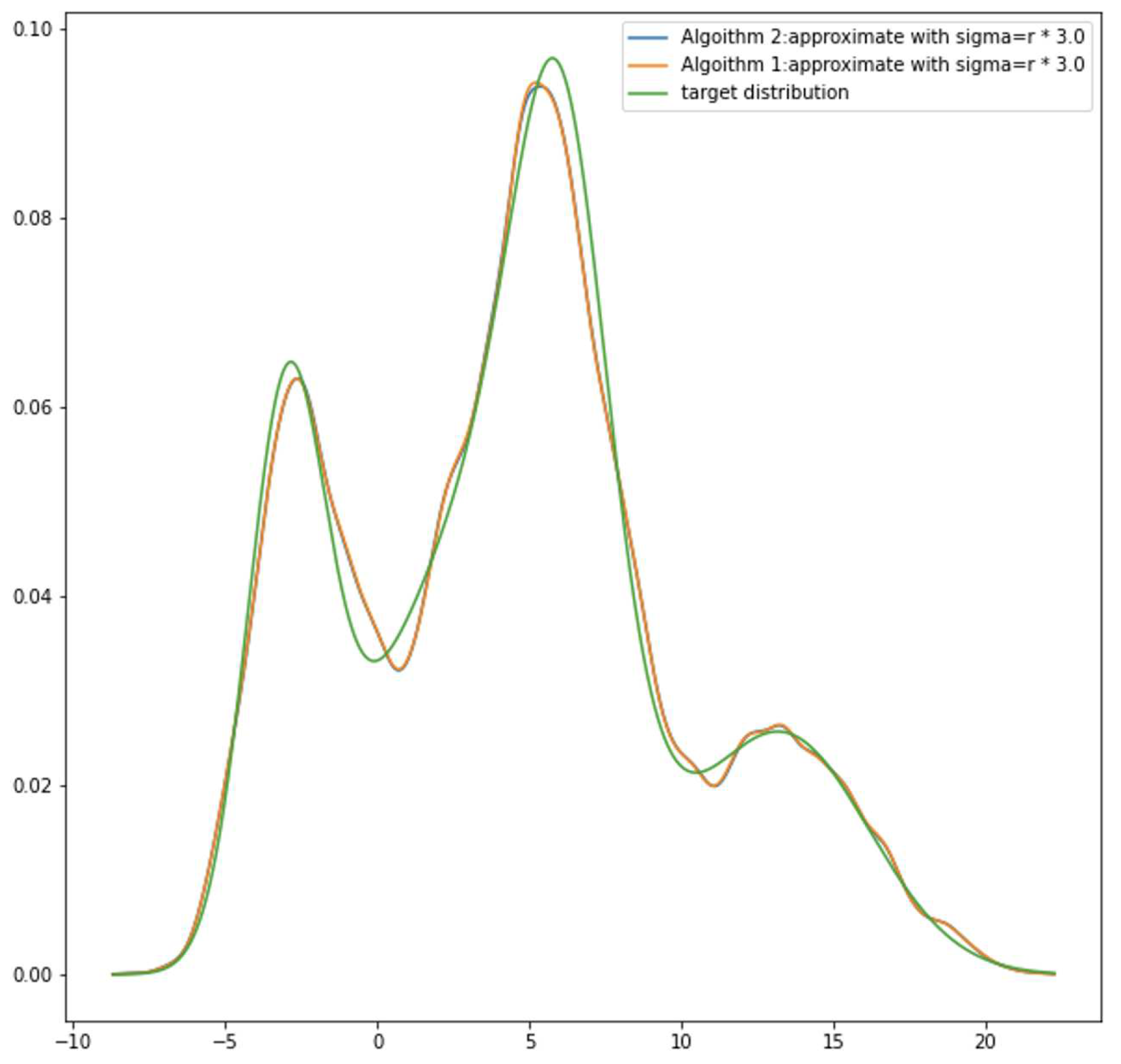
Figure 2, which is
. The differences between target density and GMM are clearly smaller compared to 1 and 2. It also implies that if peaks of target density are not overly sharp (condituion 2), by stretching GMM components, resulting a better estimation of target density.
Here we can conclude that as long as the size of component distributions are large enough and satisfy given conditions 1-3, GMM can well approximate arbitrary distributions. The loss of density information between target density and GMM is controlled by Equations (
3) and (
3).
2.4. Learning Algorithm
Most of the learning algorithms apply gradients iteratively to update target parameters until reaching an optimum position. There are no differences from ours. The key of the learning process is to find the right gradient for each parameter. In our model set up, this has become relatively simple. Begin with defining the loss function for the learning process.
We want to find a set of parameters
which:
The Equation (
5) comes from Equation (
1). Because we do not have any access to the overall density information to calculate DIL, we change the region input
to the data points. From inequality in Equations (
3) and (
3), instead of finding the precise gradient at each
, we apply Equations (
3) or (
3) in (
5):
or
where
is the closest
to
. Then,
where
,
is the density of
component distribution,
, and
or
.
Equation (
7), the result is intuitively interpretative. For Gaussian distribution components where means are further away from data point
, gradient
will be smaller and vice versa. For every data point, we perform Equation (
8) to update parameters.
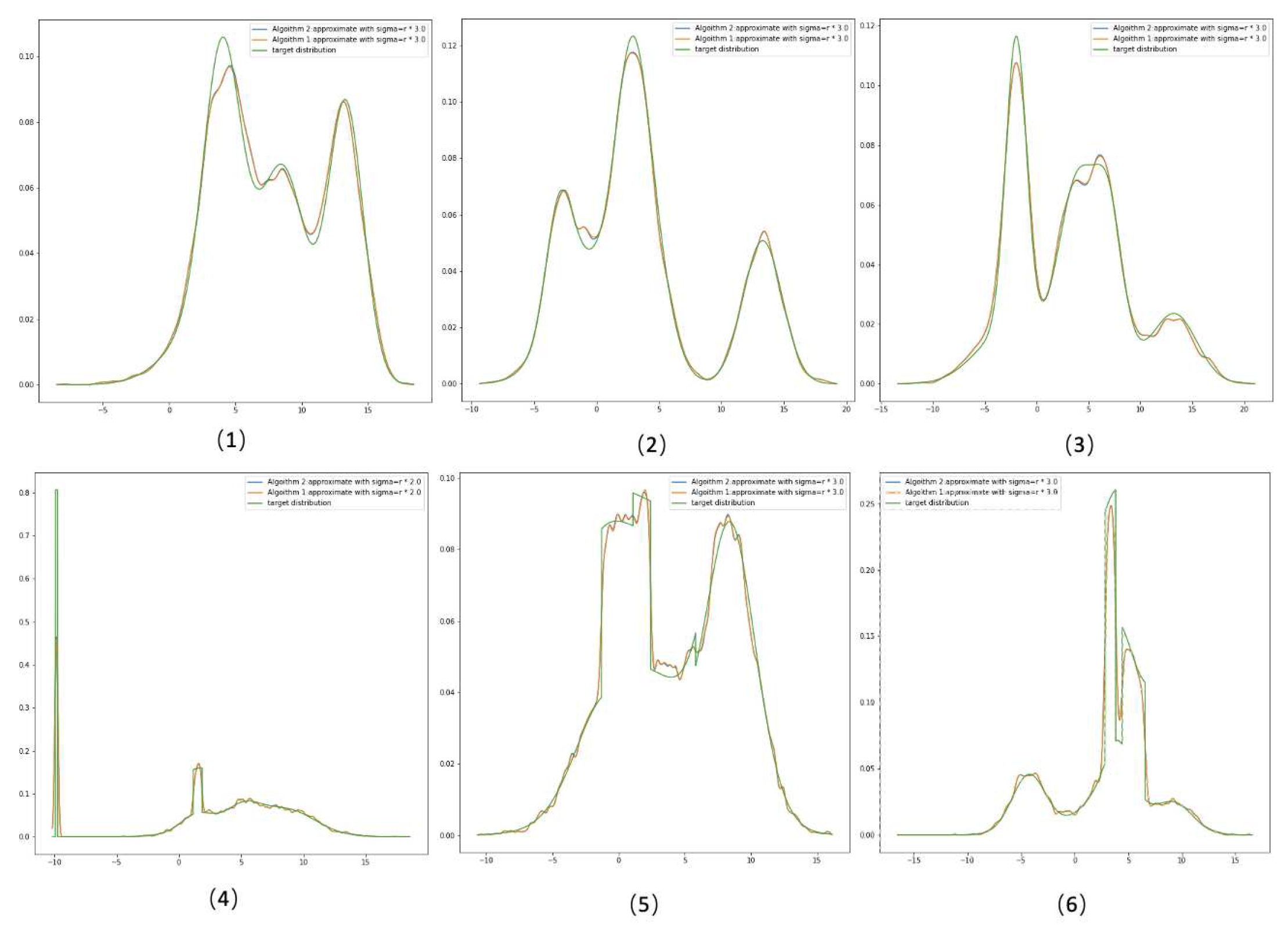
Based on Equation (
7), Here we developed two simple algorithms for learning GMM. Algorithm 1 computes the gradient following Equation (
7). Algorithm 2 is a simplified version where we only update the
which is the coefficient of
that has
closest to
. Algorithms’ performance comparison is shown in the next section. It is worth mentioning that
is initialized uniformly. After learning the whole dataset, the final step is re-normalization to ensure
.
| Algorithm 1: |
 |
| Algorithm 2: |
 |