Submitted:
03 February 2023
Posted:
07 February 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
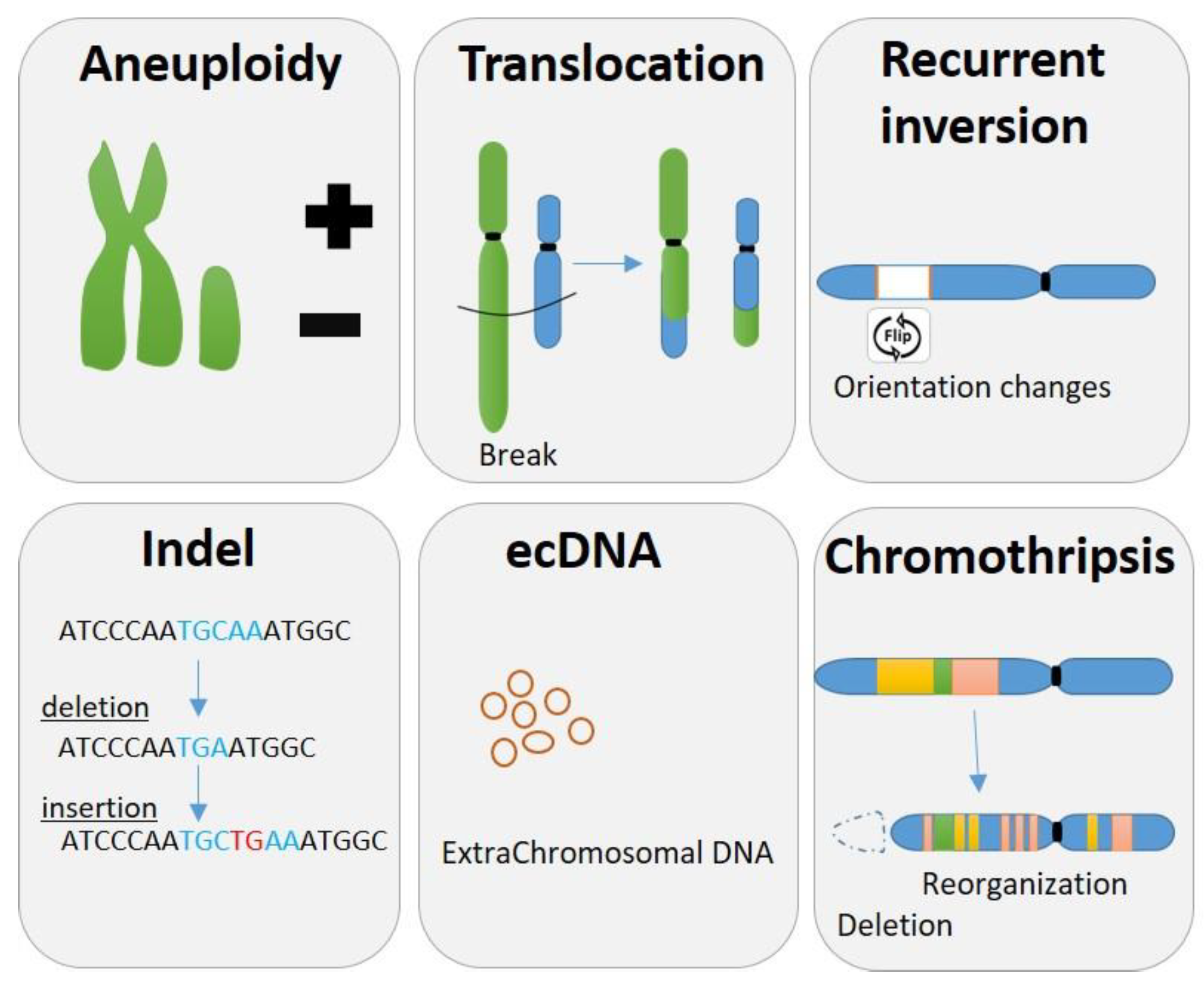
2. Different Pattern of Structure Variants and Chromosomal Instability
2.1. Aneuploidy
2.2. Common Chromosomal Changes
2.3. Indels
2.4. Complex Genomic Rearrangements
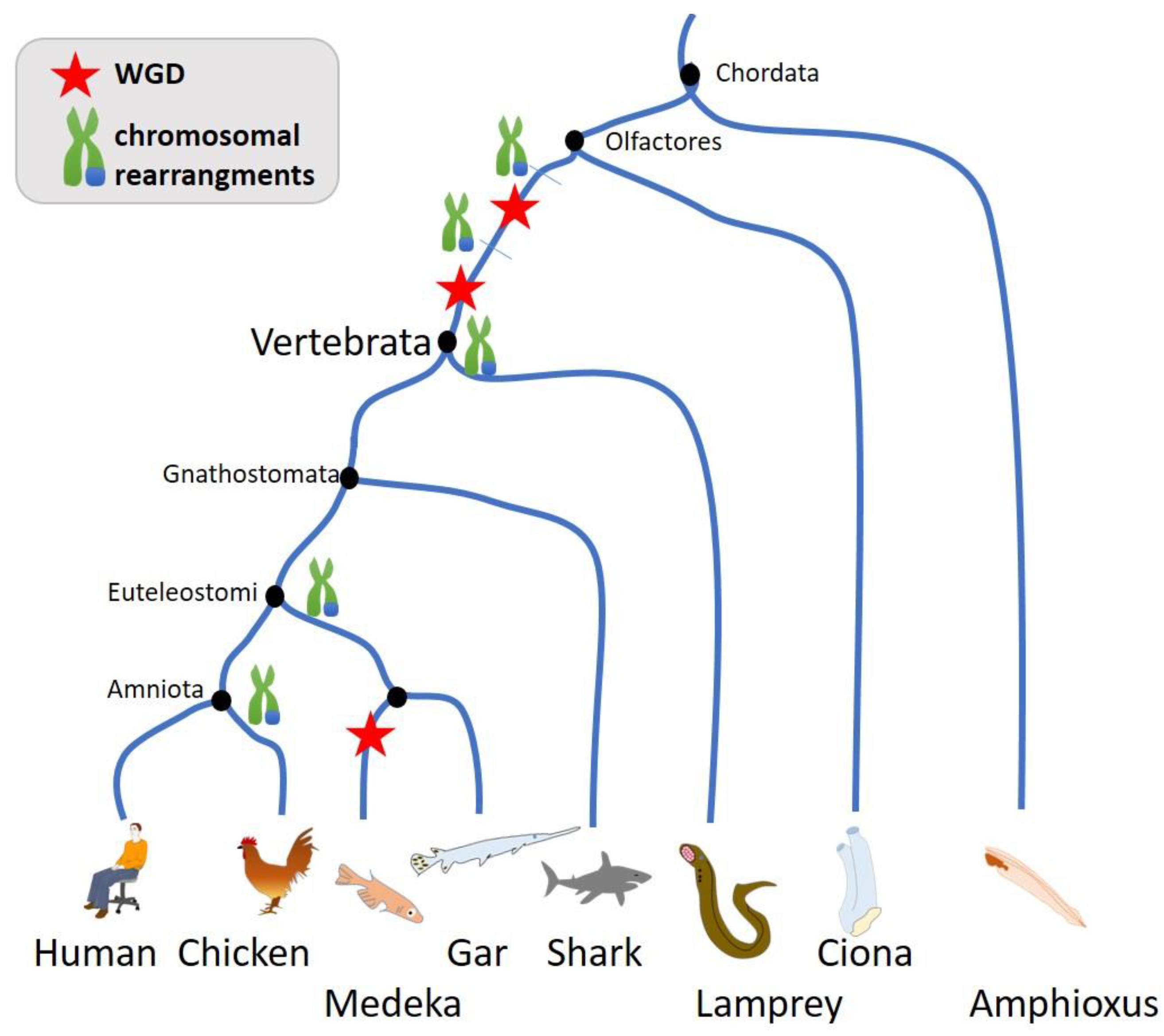
3. Chromosomal Instability during Evolution

3.1. SNPs
3.2. Genome Size Variability and Chromosomal Instability
3.3. Chromosomal Variance in Human
4. Chromosomal Instability in Cancer
5. Possible Origins of Chromosomal Instability
5.1. Mitotic-Cell Cycle Errors
5.2. Transient Nuclear Envelope Rupture (NER)
5.3. Double-Stranded Breaks
5.4. Expression of Meiotic-Specific Proteins
5.5. Tumorigenesis Drive CIN
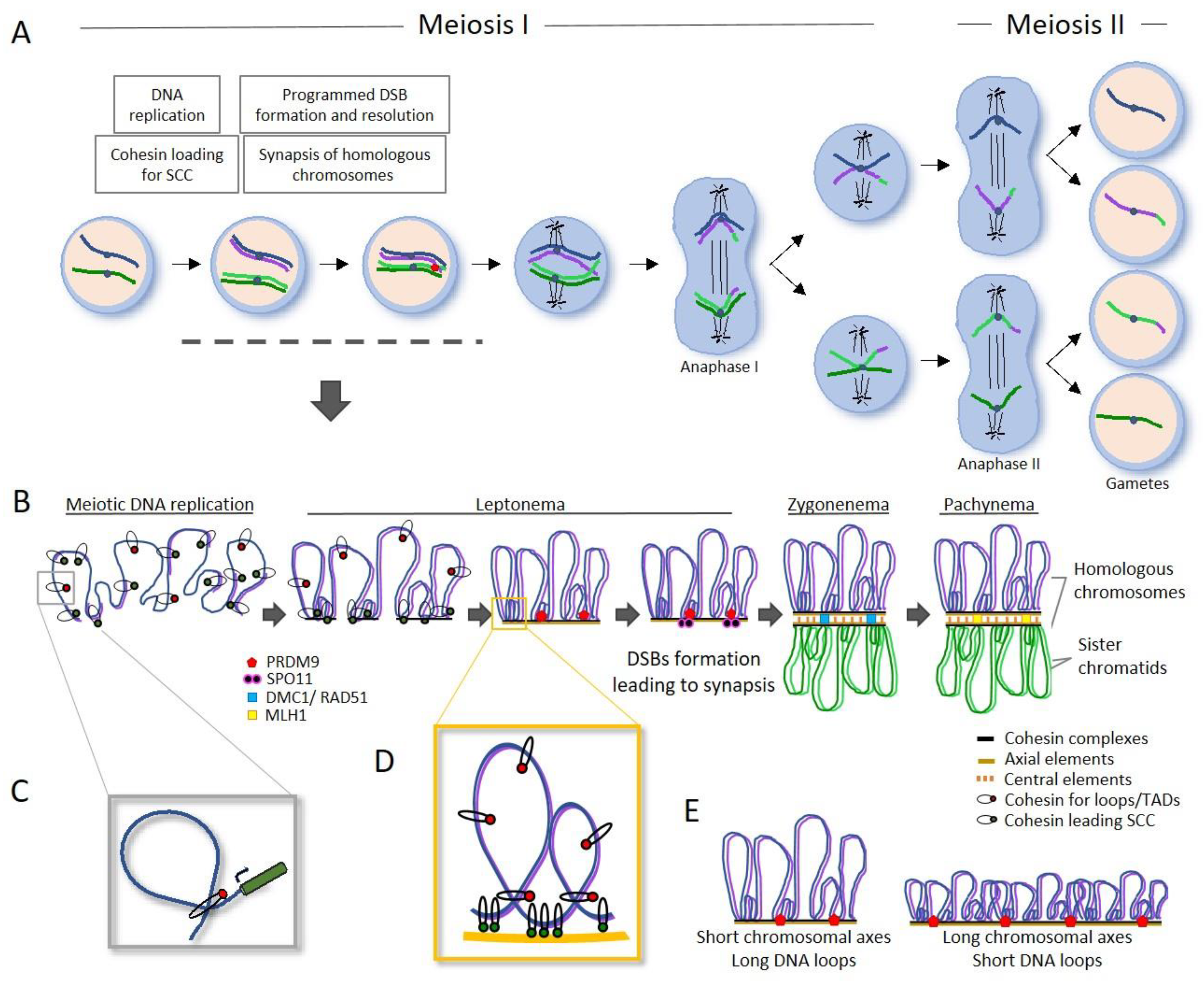
6. Germ Cells and Meiotic Program as Drivers for Genome Evolution
6.1. Chromosomal Axis and Synapsis in Genome Stability and Evolution
6.2. Impact of Three-Dimensional Chromatin Structure on the Control of Gene Expression
6.3. DNA Recombination and Repair as a Mayor Source for Evolution
6.3.1. Recombinational Hotspots as a Factor for Genome Stability and Evolution
6.3.2. Factors and Complexes Defines Recombinational Hotspots
- Cohesin complexes and Synaptonemal Complex axial elements
- PRDM9
- Chromosome organization as a modulator of recombination landscape
6.3.3. Pathways of Repair of Meiotic DSB to Chiasma Formation
6.3.4. Defects in Meiotic Recombination Leads to Chromosome Rearrangements
- Large chromosomal rearrangements as drivers for speciation
- Microchromosomes
- Whole genome duplications and gain in chromosomes
- Chromothripsis
6.3.5. Repetitive DNA Elements as Contributors for Chromosome Evolution and Speciation
6.3.6. Telomeres
7. CIN during Fecundation and First Mitosis
8. Disease Associated to Chromosomal Instability
8.1. Fertility
8.2. Rare Disease
8.2.1. Impairment of DNA Damage Repair Pathways
8.2.2. Germile SV and CCR
9. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Heng, H.H. The genome-centric concept: resynthesis of evolutionary theory. Bioessays 2009, 31, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Liu, G.; Bremer, S.; Ye, K.J.; Stevens, J.; Ye, C.J. Clonal and non-clonal chromosome aberrations and genome variation and aberration. Genome 2006, 49, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Larkin, D.M.; der Wind, A.E.-V.; Bourque, G.; Tesler, G.; Auvil, L.; Beever, J.E.; Chowdhary, B.P.; Galibert, F.; Gatzke, L. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 2005, 309, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Barton, N.H. Chromosomal speciation and molecular divergence--accelerated evolution in rearranged chromosomes. Science 2003, 300, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Pellestor, F.; Gatinois, V.; Puechberty, J.; Geneviève, D.; Lefort, G. Chromothripsis: potential origin in gametogenesis and preimplantation cell divisions. A review. Fertility and sterility 2014, 102, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Capilla, L.; Garcia Caldés, M.; Ruiz-Herrera, A. Mammalian Meiotic Recombination: A Toolbox for Genome Evolution. Cytogenet Genome Res 2016, 150, 1–16. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Guryev, V.; van Roosmalen, M.; Duran, K.J.; de Bruijn, E.; Bakker, S.C.; Letteboer, T.; van Nesselrooij, B.; Hochstenbach, R.; Poot, M. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Human molecular genetics 2011, 20, 1916–1924. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, K.; Oliveros, W.; Gerhardinger, C.; Andergassen, D.; Maass, P.G.; Rinn, J.L.; Melé, M. Cis and trans effects differentially contribute to the evolution of promoters and enhancers. Genome biology 2020, 21, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, Y.; Koshiba-Takeuchi, K. Significance of whole-genome duplications on the emergence of evolutionary novelties. Brief Funct Genomics 2018, 17, 329–338. [Google Scholar] [CrossRef]
- Antonarakis, S.E.; Lyle, R.; Dermitzakis, E.T.; Reymond, A.; Deutsch, S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet 2004, 5, 725–738. [Google Scholar] [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics 2013, 45, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.P.; Petersen, B.L.; Johansen, I.E.; Niu, Y.; Yang, Z.; Chamberlain, C.A.; Met, O.; Wandall, H.H.; Frodin, M. INDEL detection, the ‘Achilles heel’ of precise genome editing: a survey of methods for accurate profiling of gene editing induced indels. Nucleic Acids Res 2020, 48, 11958–11981. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, M.L.; Zhang, C.Z.; Pellman, D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu Rev Genet 2015, 49, 183–211. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M.; et al. Punctuated evolution of prostate cancer genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Bafna, V.; Mischel, P.S. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer 2019, 19, 283–288. [Google Scholar] [CrossRef]
- Nathanson, D.A.; Gini, B.; Mottahedeh, J.; Visnyei, K.; Koga, T.; Gomez, G.; Eskin, A.; Hwang, K.; Wang, J.; Masui, K.; et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 2014, 343, 72–76. [Google Scholar] [CrossRef]
- Storlazzi, C.T.; Fioretos, T.; Surace, C.; Lonoce, A.; Mastrorilli, A.; Strombeck, B.; D’Addabbo, P.; Iacovelli, F.; Minervini, C.; Aventin, A.; et al. MYC-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of MYC as the target gene. Hum Mol Genet 2006, 15, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Gao, Y.; Jiang, G.; Yang, W.; Jin, W.; Gong, J.; Xu, X.; Niu, X. Animal-SNPAtlas: a comprehensive SNP database for multiple animals. Nucleic Acids Res 2022. [Google Scholar] [CrossRef]
- Burraco, P.; Orizaola, G. Ionizing radiation and melanism in Chornobyl tree frogs. Evol Appl 2022, 15, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Klunk, J.; Vilgalys, T.P.; Demeure, C.E.; Cheng, X.; Shiratori, M.; Madej, J.; Beau, R.; Elli, D.; Patino, M.I.; Redfern, R.; et al. Evolution of immune genes is associated with the Black Death. Nature 2022, 611, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Enard, D. Ancient DNA reveals rapid natural selection during the Black Death. Nature 2022, 611, 237–238. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H. Bubonic plague left lingering scars on the human genome. Nature 2022. [Google Scholar] [CrossRef] [PubMed]
- Holland, L.Z.; Ocampo Daza, D. A new look at an old question: when did the second whole genome duplication occur in vertebrate evolution? Genome Biol 2018, 19, 209. [Google Scholar] [CrossRef] [PubMed]
- Sacerdot, C.; Louis, A.; Bon, C.; Berthelot, C.; Roest Crollius, H. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol 2018, 19, 166. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Suh, A.; Feschotte, C. Dynamics of genome size evolution in birds and mammals. Proc Natl Acad Sci USA 2017, 114, E1460–E1469. [Google Scholar] [CrossRef]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics 2014, 289, 1045–1060. [Google Scholar] [CrossRef]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun 2014, 5, 3657. [Google Scholar] [CrossRef]
- Clark, J.W.; Donoghue, P.C.J. Whole-Genome Duplication and Plant Macroevolution. Trends Plant Sci 2018, 23, 933–945. [Google Scholar] [CrossRef]
- Jonsson, H.; Schubert, M.; Seguin-Orlando, A.; Ginolhac, A.; Petersen, L.; Fumagalli, M.; Albrechtsen, A.; Petersen, B.; Korneliussen, T.S.; Vilstrup, J.T.; et al. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc Natl Acad Sci USA 2014, 111, 18655–18660. [Google Scholar] [CrossRef]
- Du, K.; Stock, M.; Kneitz, S.; Klopp, C.; Woltering, J.M.; Adolfi, M.C.; Feron, R.; Prokopov, D.; Makunin, A.; Kichigin, I.; et al. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat Ecol Evol 2020, 4, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Carbonell-Bejerano, P.; Royo, C.; Torres-Perez, R.; Grimplet, J.; Fernandez, L.; Franco-Zorrilla, J.M.; Lijavetzky, D.; Baroja, E.; Martinez, J.; Garcia-Escudero, E.; et al. Catastrophic Unbalanced Genome Rearrangements Cause Somatic Loss of Berry Color in Grapevine. Plant Physiol 2017, 175, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Abel, H.J.; Larson, D.E.; Regier, A.A.; Chiang, C.; Das, I.; Kanchi, K.L.; Layer, R.M.; Neale, B.M.; Salerno, W.J.; Reeves, C.; et al. Mapping and characterization of structural variation in 17,795 human genomes. Nature 2020, 583, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.L.; Brand, H.; Karczewski, K.J.; Zhao, X.; Alfoldi, J.; Francioli, L.C.; Khera, A.V.; Lowther, C.; Gauthier, L.D.; Wang, H.; et al. A structural variation reference for medical and population genetics. Nature 2020, 581, 444–451. [Google Scholar] [CrossRef]
- Porubsky, D.; Hops, W.; Ashraf, H.; Hsieh, P.; Rodriguez-Martin, B.; Yilmaz, F.; Ebler, J.; Hallast, P.; Maria Maggiolini, F.A.; Harvey, W.T.; et al. Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders. Cell 2022, 185, 1986–2005. [Google Scholar] [CrossRef]
- Notta, F.; Chan-Seng-Yue, M.; Lemire, M.; Li, Y.; Wilson, G.W.; Connor, A.A.; Denroche, R.E.; Liang, S.B.; Brown, A.M.; Kim, J.C.; et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 2016, 538, 378–382. [Google Scholar] [CrossRef]
- Yates, L.R.; Knappskog, S.; Wedge, D.; Farmery, J.H.R.; Gonzalez, S.; Martincorena, I.; Alexandrov, L.B.; Van Loo, P.; Haugland, H.K.; Lilleng, P.K.; et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017, 32, 169–184. [Google Scholar] [CrossRef]
- Stachler, M.D.; Taylor-Weiner, A.; Peng, S.; McKenna, A.; Agoston, A.T.; Odze, R.D.; Davison, J.M.; Nason, K.S.; Loda, M.; Leshchiner, I.; et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nature genetics 2015, 47, 1047–1055. [Google Scholar] [CrossRef]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep 2018, 23, 3392–3406. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.A.; Rodriguez, D.A.; Kurtenbach, S.; Kuznetsov, J.N.; Sanchez, M.I.; Decatur, C.L.; Snyder, H.; Feun, L.G.; Livingstone, A.S.; Harbour, J.W. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun 2020, 11, 496. [Google Scholar] [CrossRef]
- Steele, C.D.; Abbasi, A.; Islam, S.M.A.; Bowes, A.L.; Khandekar, A.; Haase, K.; Hames-Fathi, S.; Ajayi, D.; Verfaillie, A.; Dhami, P.; et al. Signatures of copy number alterations in human cancer. Nature 2022, 606, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Drews, R.M.; Hernando, B.; Tarabichi, M.; Haase, K.; Lesluyes, T.; Smith, P.S.; Morrill Gavarro, L.; Couturier, D.L.; Liu, L.; Schneider, M.; et al. A pan-cancer compendium of chromosomal instability. Nature 2022, 606, 976–983. [Google Scholar] [CrossRef]
- Voronina, N.; Wong, J.K.L.; Hubschmann, D.; Hlevnjak, M.; Uhrig, S.; Heilig, C.E.; Horak, P.; Kreutzfeldt, S.; Mock, A.; Stenzinger, A.; et al. The landscape of chromothripsis across adult cancer types. Nat Commun 2020, 11, 2320. [Google Scholar] [CrossRef] [PubMed]
- Shoshani, O.; Brunner, S.F.; Yaeger, R.; Ly, P.; Nechemia-Arbely, Y.; Kim, D.H.; Fang, R.; Castillon, G.A.; Yu, M.; Li, J.S.Z.; et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature 2021, 591, 137–141. [Google Scholar] [CrossRef]
- Ly, P.; Brunner, S.F.; Shoshani, O.; Kim, D.H.; Lan, W.; Pyntikova, T.; Flanagan, A.M.; Behjati, S.; Page, D.C.; Campbell, P.J.; et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nature genetics 2019, 51, 705–715. [Google Scholar] [CrossRef]
- Consortium, I.T.P.-C.A.o.W.G. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Maura, F.; Bolli, N.; Angelopoulos, N.; Dawson, K.J.; Leongamornlert, D.; Martincorena, I.; Mitchell, T.J.; Fullam, A.; Gonzalez, S.; Szalat, R.; et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun 2019, 10, 3835. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef]
- Watkins, T.B.K.; Lim, E.L.; Petkovic, M.; Elizalde, S.; Birkbak, N.J.; Wilson, G.A.; Moore, D.A.; Gronroos, E.; Rowan, A.; Dewhurst, S.M.; et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 2020, 587, 126–132. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Lukow, D.A.; Sausville, E.L.; Suri, P.; Chunduri, N.K.; Wieland, A.; Leu, J.; Smith, J.C.; Girish, V.; Kumar, A.A.; Kendall, J. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev Cell 2021, 56, 2427–2439 e2424. [Google Scholar] [CrossRef]
- Spurr, L.F.; Weichselbaum, R.R.; Pitroda, S.P. Tumor aneuploidy predicts survival following immunotherapy across multiple cancers. Nature genetics 2022, 54, 1782–1785. [Google Scholar] [CrossRef] [PubMed]
- Roosen, M.; Ode, Z.; Bunt, J.; Kool, M. The oncogenic fusion landscape in pediatric CNS neoplasms. Acta Neuropathol 2022, 143, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Sweet-Cordero, E.A.; Biegel, J.A. The genomic landscape of pediatric cancers: Implications for diagnosis and treatment. Science 2019, 363, 1170–1175. [Google Scholar] [CrossRef]
- Levine, M.S.; Holland, A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev 2018, 32, 620–638. [Google Scholar] [CrossRef]
- Gauthier, B.R.; Comaills, V. Nuclear Envelope Integrity in Health and Disease: Consequences on Genome Instability and Inflammation. Int J Mol Sci 2021, 22, 7281. [Google Scholar] [CrossRef]
- Hatch, E.M.; Fischer, A.H.; Deerinck, T.J.; Hetzer, M.W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 2013, 154, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Umbreit, N.T.; Zhang, C.Z.; Lynch, L.D.; Blaine, L.J.; Cheng, A.M.; Tourdot, R.; Sun, L.; Almubarak, H.F.; Judge, K.; Mitchell, T.J.; et al. Mechanisms generating cancer genome complexity from a single cell division error. Science 2020, 368. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; Chatzipli, A.; Dananberg, A.; Chu, K.; Toufektchan, E.; Klimczak, L.J.; Gordenin, D.A.; Campbell, P.J.; de Lange, T. APOBEC3-dependent kataegis and TREX1-driven chromothripsis during telomere crisis. Nature genetics 2020, 52, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; Hatch, E.M. Nuclear Membrane Rupture and Its Consequences. Annu Rev Cell Dev Biol 2020, 36, 85–114. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Schultz, S.W.; Bellanger, A.; Jones, C.M.; Petersen, L.I.; Raiborg, C.; Skarpen, E.; Pedurupillay, C.R.J.; Kjos, I.; Kip, E.; et al. Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat Cell Biol 2020, 22, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Denais, C.M.; Gilbert, R.M.; Isermann, P.; McGregor, A.L.; te Lindert, M.; Weigelin, B.; Davidson, P.M.; Friedl, P.; Wolf, K.; Lammerding, J. Nuclear envelope rupture and repair during cancer cell migration. Science 2016, 352, 353–358. [Google Scholar] [CrossRef]
- Comaills, V.; Kabeche, L.; Morris, R.; Buisson, R.; Yu, M.; Madden, M.W.; LiCausi, J.A.; Boukhali, M.; Tajima, K.; Pan, S.; et al. Genomic Instability Is Induced by Persistent Proliferation of Cells Undergoing Epithelial-to-Mesenchymal Transition. Cell Rep 2016, 17, 2632–2647. [Google Scholar] [CrossRef]
- Raab, M.; Gentili, M.; de Belly, H.; Thiam, H.R.; Vargas, P.; Jimenez, A.J.; Lautenschlaeger, F.; Voituriez, R.; Lennon-Dumenil, A.M.; Manel, N.; et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016, 352, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, B.R.; Lorenzo, P.I.; Comaills, V. Physical Forces and Transient Nuclear Envelope Rupture during Metastasis: The Key for Success? Cancers (Basel) 2021, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Weigelin, B.; den Boer, A.T.; Wagena, E.; Broen, K.; Dolstra, H.; de Boer, R.J.; Figdor, C.G.; Textor, J.; Friedl, P. Cytotoxic T cells are able to efficiently eliminate cancer cells by additive cytotoxicity. Nat Commun 2021, 12, 5217. [Google Scholar] [CrossRef] [PubMed]
- Nader, G.P.F.; Aguera-Gonzalez, S.; Routet, F.; Gratia, M.; Maurin, M.; Cancila, V.; Cadart, C.; Palamidessi, A.; Ramos, R.N.; San Roman, M. Compromised nuclear envelope integrity drives TREX1-dependent DNA damage and tumor cell invasion. Cell 2021, 184, 5230–5246 e5222. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Stokasimov, E.; Cui, Y.; Pellman, D. Breakage of cytoplasmic chromosomes by pathological DNA base excision repair. Nature 2022, 606, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.J.; Sun, L.; Yao, Y.; Zhang, C.Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nature genetics 2021. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, J.W.; Koster, J.; Lodder, P.; Repping, S.; Hamer, G. Massive expression of germ cell-specific genes is a hallmark of cancer and a potential target for novel treatment development. Oncogene 2018, 37, 5694–5700. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Cragg, M.S.; Salmina, K.; Hausmann, M.; Scherthan, H. The role of meiotic cohesin REC8 in chromosome segregation in gamma irradiation-induced endopolyploid tumour cells. Exp Cell Res 2009, 315, 2593–2603. [Google Scholar] [CrossRef]
- Feichtinger, J.; Larcombe, L.; McFarlane, R.J. Meta-analysis of expression of l(3)mbt tumor-associated germline genes supports the model that a soma-to-germline transition is a hallmark of human cancers. Int J Cancer 2014, 134, 2359–2365. [Google Scholar] [CrossRef]
- Kalejs, M.; Ivanov, A.; Plakhins, G.; Cragg, M.S.; Emzinsh, D.; Illidge, T.M.; Erenpreisa, J. Upregulation of meiosis-specific genes in lymphoma cell lines following genotoxic insult and induction of mitotic catastrophe. BMC Cancer 2006, 6, 6. [Google Scholar] [CrossRef]
- Lindsey, S.F.; Byrnes, D.M.; Eller, M.S.; Rosa, A.M.; Dabas, N.; Escandon, J.; Grichnik, J.M. Potential role of meiosis proteins in melanoma chromosomal instability. Journal of skin cancer 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Folco, H.; Chalamcharla, V.R.; Sugiyama, T.; Thillainadesan, G.; Zofall, M.; Balachandran, V.; Dhakshnamoorthy, J.; Mizuguchi, T.; Grewal, S.I. Untimely expression of gametogenic genes in vegetative cells causes uniparental disomy. Nature 2017, 543, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Houle, A.A.; Gibling, H.; Lamaze, F.C.; Edgington, H.A.; Soave, D.; Fave, M.J.; Agbessi, M.; Bruat, V.; Stein, L.D.; Awadalla, P. Aberrant PRDM9 expression impacts the pan-cancer genomic landscape. Genome Res 2018, 28, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Baslan, T.; Morris, J.P.t.; Zhao, Z.; Reyes, J.; Ho, Y.J.; Tsanov, K.M.; Bermeo, J.; Tian, S.; Zhang, S.; Askan, G.; et al. Ordered and deterministic cancer genome evolution after p53 loss. Nature 2022, 608, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Kleckner, N.; Storlazzi, A.; Zickler, D. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends in Genetics 2003, 19, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, A.; Vozdova, M.; Fernández, J.; Sebestova, H.; Capilla, L.; Frohlich, J.; Vara, C.; Hernández-Marsal, A.; Sipek, J.; Robinson, T.J.; et al. Recombination correlates with synaptonemal complex length and chromatin loop size in bovids-insights into mammalian meiotic chromosomal organization. Chromosoma 2017, 126, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, H. Meiosis: an overview of key differences from mitosis. Cold Spring Harb Perspect Biol 2015, 7. [Google Scholar] [CrossRef]
- Bannister, L.A.; Reinholdt, L.G.; Munroe, R.J.; Schimenti, J.C. Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis 2004, 40, 184–194. [Google Scholar] [CrossRef]
- Revenkova, E.; Eijpe, M.; Heyting, C.; Hodges, C.A.; Hunt, P.A.; Liebe, B.; Scherthan, H.; Jessberger, R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol 2004, 6, 555–562. [Google Scholar] [CrossRef]
- Winkel, K.; Alsheimer, M.; Öllinger, R.; Benavente, R. Protein SYCP2 provides a link between transverse filaments and lateral elements of mammalian synaptonemal complexes. Chromosoma 2009, 118, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; De La Fuente, R.; Leu, N.A.; Baumann, C.; McLaughlin, K.J.; Wang, P.J. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol 2006, 173, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Henderson, K.A.; Keeney, S. Synaptonemal complex formation: where does it start? BioEssays 2005, 27, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Page, S.L.; Hawley, R.S. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 2004, 20, 525–558. [Google Scholar] [CrossRef]
- Romanienko, P.J.; Camerini-Otero, R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000, 6, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Hassold, T.; Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001, 2, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, A.C.; Berman, J. Shift and adapt: the costs and benefits of karyotype variations. Curr Opin Microbiol 2015, 26, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, M.P.; Martinez, D.A.; Sakthikumar, S.; Anderson, M.Z.; Berlin, A.; Gujja, S.; Zeng, Q.; Zisson, E.; Wang, J.M.; Greenberg, J.M.; et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res 2015, 25, 413–425. [Google Scholar] [CrossRef]
- Kaya, A.; Mariotti, M.; Tyshkovskiy, A.; Zhou, X.; Hulke, M.L.; Ma, S.; Gerashchenko, M.V.; Koren, A.; Gladyshev, V.N. Molecular signatures of aneuploidy-driven adaptive evolution. Nature Communications 2020, 11, 588. [Google Scholar] [CrossRef]
- Morgan, C.; Knight, E.; Bomblies, K. The meiotic cohesin subunit REC8 contributes to multigenic adaptive evolution of autopolyploid meiosis in Arabidopsis arenosa. PLoS Genet 2022, 18, e1010304. [Google Scholar] [CrossRef]
- Matveevsky, S.; Bakloushinskaya, I.; Tambovtseva, V.; Atsaeva, M.; Grishaeva, T.; Bogdanov, A.; Kolomiets, O. Nonhomologous Chromosome Interactions in Prophase I: Dynamics of Bizarre Meiotic Contacts in the Alay Mole Vole Ellobius alaicus (Mammalia, Rodentia). Genes 2022, 13, 2196. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Vara, C.; Paytuví-Gallart, A.; Cuartero, Y.; Le Dily, F.; Garcia, F.; Salvà-Castro, J.; Gómez, H.L.; Julià, E.; Moutinho, C.; Aiese Cigliano, R.; et al. Three-Dimensional Genomic Structure and Cohesin Occupancy Correlate with Transcriptional Activity during Spermatogenesis. Cell Rep 2019, 28, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Alavattam, K.G.; Maezawa, S.; Sakashita, A.; Khoury, H.; Barski, A.; Kaplan, N.; Namekawa, S.H. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat Struct Mol Biol 2019, 26, 175–184. [Google Scholar] [CrossRef] [PubMed]
- de Massy, B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet 2013, 47, 563–599. [Google Scholar] [CrossRef] [PubMed]
- Lam, I.; Keeney, S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 2014, 7, a016634. [Google Scholar] [CrossRef] [PubMed]
- Bompadre, O.; Andrey, G. Chromatin topology in development and disease. Curr Opin Genet Dev 2019, 55, 32–38. [Google Scholar] [CrossRef]
- Farré, M.; Robinson, T.J.; Ruiz-Herrera, A. An Integrative Breakage Model of genome architecture, reshuffling and evolution: The Integrative Breakage Model of genome evolution, a novel multidisciplinary hypothesis for the study of genome plasticity. Bioessays 2015, 37, 479–488. [Google Scholar] [CrossRef]
- Franke, M.; Gómez-Skarmeta, J.L. An evolutionary perspective of regulatory landscape dynamics in development and disease. Curr Opin Cell Biol 2018, 55, 24–29. [Google Scholar] [CrossRef]
- Veller, C.; Kleckner, N.; Nowak, M.A. A rigorous measure of genome-wide genetic shuffling that takes into account crossover positions and Mendel’s second law. Proc Natl Acad Sci USA 2019, 116, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bourbon, H.-M.; de Massy, B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes & development 2010, 24, 1266–1280. [Google Scholar]
- Lange, J.; Yamada, S.; Tischfield, S.E.; Pan, J.; Kim, S.; Zhu, X.; Socci, N.D.; Jasin, M.; Keeney, S. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell 2016, 167, 695–708 e616. [Google Scholar] [CrossRef] [PubMed]
- Rockman, M.V.; Kruglyak, L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet 2009, 5, e1000419. [Google Scholar] [CrossRef]
- Barlow, A.L.; Benson, F.E.; West, S.C.; Hulten, M.A. Distribution of the Rad51 recombinase in human and mouse spermatocytes. The EMBO journal 1997, 16, 5207–5215. [Google Scholar] [CrossRef] [PubMed]
- Cole, F.; Kauppi, L.; Lange, J.; Roig, I.; Wang, R.; Keeney, S.; Jasin, M. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 2012, 14, 424–430. [Google Scholar] [CrossRef]
- Myers, S.; Bowden, R.; Tumian, A.; Bontrop, R.E.; Freeman, C.; MacFie, T.S.; McVean, G.; Donnelly, P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 2010, 327, 876–879. [Google Scholar] [CrossRef] [PubMed]
- Smagulova, F.; Gregoretti, I.V.; Brick, K.; Khil, P.; Camerini-Otero, R.D.; Petukhova, G.V. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 2011, 472, 375–378. [Google Scholar] [CrossRef]
- Cappelletti, E.; Piras, F.M.; Badiale, C.; Bambi, M.; Santagostino, M.; Vara, C.; Masterson, T.A.; Sullivan, K.F.; Nergadze, S.G.; Ruiz-Herrera, A.; et al. CENP-A binding domains and recombination patterns in horse spermatocytes. Sci Rep 2019, 9, 15800. [Google Scholar] [CrossRef]
- Brick, K.; Smagulova, F.; Khil, P.; Camerini-Otero, R.D.; Petukhova, G.V. Genetic recombination is directed away from functional genomic elements in mice. Nature 2012, 485, 642–645. [Google Scholar] [CrossRef]
- Parvanov, E.D.; Petkov, P.M.; Paigen, K. Prdm9 controls activation of mammalian recombination hotspots. Science 2010, 327, 835. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zickler, D.; Kleckner, N.; Zhang, L. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell cycle 2015, 14, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. Meiotic chromosomes: integrating structure and function. Annual review of genetics 1999, 33, 603. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Caballero, C.; Herrán, Y.; Sánchez-Martín, M.; Suja, J.Á.; Barbero, J.L.; Llano, E.; Pendás, A.M. Identification and molecular characterization of the mammalian α-kleisin RAD21L. Cell cycle 2011, 10, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Baudat, F.; Buard, J.; Grey, C.; Fledel-Alon, A.; Ober, C.; Przeworski, M.; Coop, G.; De Massy, B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 2010, 327, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Mihola, O.; Trachtulec, Z.; Vlcek, C.; Schimenti, J.C.; Forejt, J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 2009, 323, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Pratto, F.; Brick, K.; Khil, P.; Smagulova, F.; Petukhova, G.V.; Camerini-Otero, R.D. Recombination initiation maps of individual human genomes. Science 2014, 346, 1256442. [Google Scholar] [CrossRef] [PubMed]
- Smagulova, F.; Brick, K.; Pu, Y.; Camerini-Otero, R.D.; Petukhova, G.V. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes & development 2016, 30, 266–280. [Google Scholar]
- Oliver, P.L.; Goodstadt, L.; Bayes, J.J.; Birtle, Z.; Roach, K.C.; Phadnis, N.; Beatson, S.A.; Lunter, G.; Malik, H.S.; Ponting, C.P. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS genetics 2009, 5, e1000753. [Google Scholar] [CrossRef]
- Davies, B.; Hatton, E.; Altemose, N.; Hussin, J.G.; Pratto, F.; Zhang, G.; Hinch, A.G.; Moralli, D.; Biggs, D.; Diaz, R. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 2016, 530, 171–176. [Google Scholar] [CrossRef]
- Myers, S.; Freeman, C.; Auton, A.; Donnelly, P.; McVean, G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nature genetics 2008, 40, 1124–1129. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Galtier, N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annual review of genomics and human genetics 2009, 10, 285–311. [Google Scholar] [CrossRef] [PubMed]
- Janoušek, V.; Munclinger, P.; Wang, L.; Teeter, K.C.; Tucker, P.K. Functional organization of the genome may shape the species boundary in the house mouse. Molecular biology and evolution 2015, 32, 1208–1220. [Google Scholar] [CrossRef] [PubMed]
- Baker, Z.; Schumer, M.; Haba, Y.; Bashkirova, L.; Holland, C.; Rosenthal, G.G.; Przeworski, M. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. Elife 2017, 6, e24133. [Google Scholar] [CrossRef] [PubMed]
- Capilla, L.; Medarde, N.; Alemany-Schmidt, A.; Oliver-Bonet, M.; Ventura, J.; Ruiz-Herrera, A. Genetic recombination variation in wild Robertsonian mice: on the role of chromosomal fusions and Prdm9 allelic background. Proceedings of the Royal Society B: Biological Sciences 2014, 281, 20140297. [Google Scholar]
- Faria, R.; Navarro, A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends in ecology & evolution 2010, 25, 660–669. [Google Scholar]
- Feulner, P.G.; Chain, F.J.; Panchal, M.; Huang, Y.; Eizaguirre, C.; Kalbe, M.; Lenz, T.L.; Samonte, I.E.; Stoll, M.; Bornberg-Bauer, E. Genomics of divergence along a continuum of parapatric population differentiation. PLoS genetics 2015, 11, e1004966. [Google Scholar] [CrossRef]
- Sodeland, M.; Jorde, P.E.; Lien, S.; Jentoft, S.; Berg, P.R.; Grove, H.; Kent, M.P.; Arnyasi, M.; Olsen, E.M.; Knutsen, H. “Islands of Divergence” in the Atlantic cod genome represent polymorphic chromosomal rearrangements. Genome biology and evolution 2016, 8, 1012–1022. [Google Scholar] [CrossRef]
- Ullastres, A.; Farré, M.; Capilla, L.; Ruiz-Herrera, A. Unraveling the effect of genomic structural changes in the rhesus macaque-implications for the adaptive role of inversions. BMC genomics 2014, 15, 1–13. [Google Scholar] [CrossRef]
- Dumas, D.; Britton-Davidian, J. Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 2002, 162, 1355–1366. [Google Scholar] [CrossRef]
- Kuo, L.J.; Yang, L.-X. γ-H2AX-a novel biomarker for DNA double-strand breaks. In vivo 2008, 22, 305–309. [Google Scholar]
- Murakami, H.; Keeney, S. Regulating the formation of DNA double-strand breaks in meiosis. Genes & development 2008, 22, 286–292. [Google Scholar]
- Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Human aneuploidy: mechanisms and new insights into an age-old problem. Nature Reviews Genetics 2012, 13, 493–504. [Google Scholar] [CrossRef]
- Sasaki, M.; Lange, J.; Keeney, S. Genome destabilization by homologous recombination in the germ line. Nature reviews Molecular cell biology 2010, 11, 182–195. [Google Scholar] [CrossRef]
- Vara, C.; Paytuví-Gallart, A.; Cuartero, Y.; Álvarez-González, L.; Marín-Gual, L.; Garcia, F.; Florit-Sabater, B.; Capilla, L.; Sanchéz-Guillén, R.A.; Sarrate, Z. The impact of chromosomal fusions on 3D genome folding and recombination in the germ line. Nature communications 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Britton-Davidian, J.; Catalan, J.; da Graça Ramalhinho, M.; Ganem, G.; Auffray, J.-C.; Capela, R.; Biscoito, M.; Searle, J.B.; da Luz Mathias, M. Rapid chromosomal evolution in island mice. Nature 2000, 403, 158–158. [Google Scholar] [CrossRef] [PubMed]
- Dutrillaux, B. Chromosomal evolution in primates: tentative phylogeny from Microcebus murinus (Prosimian) to man. Human genetics 1979, 48, 251–314. [Google Scholar] [CrossRef]
- Yunis, J.J.; Sawyer, J.R.; Dunham, K. The striking resemblance of high-resolution G-banded chromosomes of man and chimpanzee. Science 1980, 208, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Newman, T.L.; Tuzun, E.; Morrison, V.A.; Hayden, K.E.; Ventura, M.; McGrath, S.D.; Rocchi, M.; Eichler, E.E. A genome-wide survey of structural variation between human and chimpanzee. Genome research 2005, 15, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Pevzner, P.; Tesler, G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proceedings of the National Academy of Sciences 2003, 100, 7672–7677. [Google Scholar] [CrossRef] [PubMed]
- Dennis, M.Y.; Harshman, L.; Nelson, B.J.; Penn, O.; Cantsilieris, S.; Huddleston, J.; Antonacci, F.; Penewit, K.; Denman, L.; Raja, A. The evolution and population diversity of human-specific segmental duplications. Nature ecology & evolution 2017, 1, 1–10. [Google Scholar]
- Marín-Gual, L.; González-Rodelas, L.; Pujol, G.; Vara, C.; Martín-Ruiz, M.; Berríos, S.; Fernández-Donoso, R.; Pask, A.; Renfree, M.B.; Page, J.; et al. Strategies for meiotic sex chromosome dynamics and telomeric elongation in Marsupials. PLoS Genet 2022, 18, e1010040. [Google Scholar] [CrossRef]
- Waters, P.D.; Patel, H.R.; Ruiz-Herrera, A.; Álvarez-González, L.; Lister, N.C.; Simakov, O.; Ezaz, T.; Kaur, P.; Frere, C.; Grützner, F.; et al. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc Natl Acad Sci USA 2021, 118. [Google Scholar] [CrossRef]
- Comai, L. The advantages and disadvantages of being polyploid. Nat Rev Genet 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat Rev Genet 2009, 10, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat Rev Genet 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Crow, K.D.; Wagner, G.P. What is the role of genome duplication in the evolution of complexity and diversity? Molecular biology and evolution 2005, 23, 887–892. [Google Scholar] [CrossRef]
- Kellis, M.; Birren, B.W.; Lander, E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 2004, 428, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Shields, D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 1997, 387, 708–713. [Google Scholar] [CrossRef]
- Mottes, F.; Villa, C.; Osella, M.; Caselle, M. The impact of whole genome duplications on the human gene regulatory networks. PLoS Comput Biol 2021, 17, e1009638. [Google Scholar] [CrossRef]
- Ohno, S. The enormous diversity in genome sizes of fish as a reflection of natureˈs extensive experiments with gene duplication. Transactions of the American Fisheries Society 1970, 99, 120–130. [Google Scholar] [CrossRef]
- D’Antonio, M.; Ciccarelli, F.D. Integrated analysis of recurrent properties of cancer genes to identify novel drivers. Genome biology 2013, 14, 1–17. [Google Scholar] [CrossRef]
- Singh, P.P.; Arora, J.; Isambert, H. Identification of ohnolog genes originating from whole genome duplication in early vertebrates, based on synteny comparison across multiple genomes. PLoS computational biology 2015, 11, e1004394. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; McLysaght, A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proceedings of the National Academy of Sciences 2010, 107, 9270–9274. [Google Scholar] [CrossRef] [PubMed]
- Acharya, D.; Ghosh, T.C. Global analysis of human duplicated genes reveals the relative importance of whole-genome duplicates originated in the early vertebrate evolution. BMC genomics 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Huminiecki, L.; Heldin, C.H. 2R and remodeling of vertebrate signal transduction engine. BMC biology 2010, 8, 1–21. [Google Scholar] [CrossRef]
- Cai, H.; Kumar, N.; Bagheri, H.C.; von Mering, C.; Robinson, M.D.; Baudis, M. Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens. BMC genomics 2014, 15, 1–13. [Google Scholar] [CrossRef]
- Weckselblatt, B.; Rudd, M.K. Human structural variation: mechanisms of chromosome rearrangements. Trends in Genetics 2015, 31, 587–599. [Google Scholar] [CrossRef] [PubMed]
- De Pagter, M.S.; Van Roosmalen, M.J.; Baas, A.F.; Renkens, I.; Duran, K.J.; Van Binsbergen, E.; Tavakoli-Yaraki, M.; Hochstenbach, R.; Van Der Veken, L.T.; Cuppen, E. Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. The American Journal of Human Genetics 2015, 96, 651–656. [Google Scholar] [CrossRef]
- Pellestor, F.; Gatinois, V. Chromoanagenesis: a piece of the macroevolution scenario. Molecular Cytogenetics 2020, 13, 1–9. [Google Scholar] [CrossRef]
- Zhang, C.-Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Kehrer-Sawatzki, H.; Cooper, D.N. Structural divergence between the human and chimpanzee genomes. Human genetics 2007, 120, 759–778. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Bourque, G. Recovering genome rearrangements in the mammalian phylogeny. Genome research 2009, 19, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Farré, M.; Bosch, M.; López-Giráldez, F.; Ponsà, M.; Ruiz-Herrera, A. Assessing the role of tandem repeats in shaping the genomic architecture of great apes. PLoS One 2011, 6, e27239. [Google Scholar] [CrossRef] [PubMed]
- Kehrer-Sawatzki, H.; Sandig, C.; Goidts, V.; Hameister, H. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenetic and Genome Research 2005, 108, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Carbone, L.; Harris, R.A.; Mootnick, A.R.; Milosavljevic, A.; Martin, D.I.; Rocchi, M.; Capozzi, O.; Archidiacono, N.; Konkel, M.K.; Walker, J.A. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome biology and evolution 2012, 4, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: old ideas, new approaches. Current opinion in genetics & development 2018, 49, 70–78. [Google Scholar]
- Ferreira, D.; Meles, S.; Escudeiro, A.; Mendes-da-Silva, A.; Adega, F.; Chaves, R. Satellite non-coding RNAs: the emerging players in cells, cellular pathways and cancer. Chromosome Research 2015, 23, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Larracuente, A.M. The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC evolutionary biology 2014, 14, 1–12. [Google Scholar] [CrossRef]
- Nazaryan-Petersen, L.; Bertelsen, B.; Bak, M.; Jønson, L.; Tommerup, N.; Hancks, D.C.; Tümer, Z. Germline chromothripsis driven by L1-mediated retrotransposition and Alu/Alu homologous recombination. Human mutation 2016, 37, 385–395. [Google Scholar] [CrossRef]
- Klein, S.J.; O’Neill, R.J. Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosome Research 2018, 26, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Research 2018, 26, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Carbone, L.; Alan Harris, R.; Gnerre, S.; Veeramah, K.R.; Lorente-Galdos, B.; Huddleston, J.; Meyer, T.J.; Herrero, J.; Roos, C.; Aken, B. Gibbon genome and the fast karyotype evolution of small apes. Nature 2014, 513, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.J.; Held, U.; Nevonen, K.A.; Klawitter, S.; Pirzer, T.; Carbone, L.; Schumann, G.G. The flow of the gibbon LAVA element is facilitated by the LINE-1 retrotransposition machinery. Genome biology and evolution 2016, 8, 3209–3225. [Google Scholar] [CrossRef] [PubMed]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-Carvalho, M.; Chaves, R. Decoding the role of satellite DNA in genome architecture and plasticity—An evolutionary and clinical affair. Genes 2020, 11, 72. [Google Scholar] [CrossRef]
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: models, mechanisms and implications. Nature reviews genetics 2010, 11, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bailey, S.M.; Okuka, M.; Muñoz, P.; Li, C.; Zhou, L.; Wu, C.; Czerwiec, E.; Sandler, L.; Seyfang, A.; et al. Telomere lengthening early in development. Nat Cell Biol 2007, 9, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Reig-Viader, R.; Brieño-Enríquez, M.A.; Khoriauli, L.; Toran, N.; Cabero, L.; Giulotto, E.; Garcia-Caldés, M.; Ruiz-Herrera, A. Telomeric repeat-containing RNA and telomerase in human fetal oocytes. Hum Reprod 2013, 28, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Reig-Viader, R.; Garcia-Caldés, M.; Ruiz-Herrera, A. Telomere homeostasis in mammalian germ cells: a review. Chromosoma 2016, 125, 337–351. [Google Scholar] [CrossRef]
- Reig-Viader, R.; Vila-Cejudo, M.; Vitelli, V.; Buscà, R.; Sabaté, M.; Giulotto, E.; Caldés, M.G.; Ruiz-Herrera, A. Telomeric repeat-containing RNA (TERRA) and telomerase are components of telomeres during mammalian gametogenesis. Biol Reprod 2014, 90, 103. [Google Scholar] [CrossRef]
- Cho, N.W.; Dilley, R.L.; Lampson, M.A.; Greenberg, R.A. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 2014, 159, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Ingles, E.D.; Deakin, J.E. Telomeres, species differences, and unusual telomeres in vertebrates: presenting challenges and opportunities to understanding telomere dynamics. Aims Genetics 2016, 3, 001–024. [Google Scholar] [CrossRef]
- Hoang, S.M.; O’Sullivan, R.J. Alternative Lengthening of Telomeres: Building Bridges To Connect Chromosome Ends. Trends Cancer 2020, 6, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Liu, L. Linking Telomere Regulation to Stem Cell Pluripotency. Trends Genet 2017, 33, 16–33. [Google Scholar] [CrossRef] [PubMed]
- Canfield, P.J.; Hartley, W.J.; Reddacliff, G.L. Spontaneous proliferations in Australian marsupials--a survey and review. 2. Dasyurids and bandicoots. J Comp Pathol 1990, 103, 147–158. [Google Scholar] [CrossRef]
- Cavazza, T.; Takeda, Y.; Politi, A.Z.; Aushev, M.; Aldag, P.; Baker, C.; Choudhary, M.; Bucevicius, J.; Lukinavicius, G.; Elder, K.; et al. Parental genome unification is highly error-prone in mammalian embryos. Cell 2021, 184, 2860–2877 e2822. [Google Scholar] [CrossRef]
- Nonaka, T.; Takahashi, M.; Nonaka, C.; Enomoto, T.; Takakuwa, K. The analysis of chromosomal abnormalities in patients with recurrent pregnancy loss, focusing on the prognosis of patients with inversion of chromosome (9). Reprod Med Biol 2019, 18, 296–301. [Google Scholar] [CrossRef]
- Webster, A.L.H.; Sanders, M.A.; Patel, K.; Dietrich, R.; Noonan, R.J.; Lach, F.P.; White, R.R.; Goldfarb, A.; Hadi, K.; Edwards, M.M.; et al. Genomic signature of Fanconi anaemia DNA repair pathway deficiency in cancer. Nature 2022, 612, 495–502. [Google Scholar] [CrossRef]
- Webster, A.L.H.; Sanders, M.A.; Patel, K.; Dietrich, R.; Noonan, R.J.; Lach, F.P.; White, R.R.; Goldfarb, A.; Hadi, K.; Edwards, M.M.; et al. Genomic signature of Fanconi anaemia DNA repair pathway deficiency in cancer. Nature 2022. [Google Scholar] [CrossRef]
- Gregory, J.J., Jr.; Wagner, J.E.; Verlander, P.C.; Levran, O.; Batish, S.D.; Eide, C.R.; Steffenhagen, A.; Hirsch, B.; Auerbach, A.D. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci USA 2001, 98, 2532–2537. [Google Scholar] [CrossRef]
- Gross, M.; Hanenberg, H.; Lobitz, S.; Friedl, R.; Herterich, S.; Dietrich, R.; Gruhn, B.; Schindler, D.; Hoehn, H. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res 2002, 98, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Werling, D.M.; Brand, H.; An, J.Y.; Stone, M.R.; Zhu, L.; Glessner, J.T.; Collins, R.L.; Dong, S.; Layer, R.M.; Markenscoff-Papadimitriou, E.; et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nature genetics 2018, 50, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Brandler, W.M.; Antaki, D.; Gujral, M.; Kleiber, M.L.; Whitney, J.; Maile, M.S.; Hong, O.; Chapman, T.R.; Tan, S.; Tandon, P.; et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science 2018, 360, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nature genetics 2017, 49, 27–35. [Google Scholar] [CrossRef]
- van Belzen, I.A.E.M.; Cai, C.; van Tuil, M.; Badloe, S.; Strengman, E.; Janse, A.; Verwiel, E.T.; van der Leest, D.F.M.; Kester, L.; Molenaar, J.J.; et al. Systematic discovery of gene fusions in pediatric cancer by integrating RNA-seq and WGS. bioRxiv, 2008; 2021.2008.2031.458342. [Google Scholar] [CrossRef]
- Schuy, J.; Grochowski, C.M.; Carvalho, C.M.B.; Lindstrand, A. Complex genomic rearrangements: an underestimated cause of rare diseases. Trends Genet 2022, 38, 1134–1146. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




