1. Introduction
New scientific discoveries in the life sciences are increasingly driven by computational methods nowadays. Microscopy plays a key role in the life sciences and so does bio-image analysis, a computational part of the life sciences research field. Traditionally, computer science research groups collaborate with wet-lab experimentalists on the biology, biochemistry and biophysics side. Furthermore, some imaging core-facilities hired individuals with computational expertise. In the last decade, we saw an increasing number of dedicated groups appearing in biological research institutes who serve the demand for incorporating quantitative computational image analysis methods into biology-centered research projects. Some of these appear as classical research groups, developing new algorithms and serving collaborators by applying the new methods to specific biological questions. Others are more on the applied science side and focus on translating algorithms to user-friendly software applicable to answer research questions by scientists less proficient with computational methods. Their naming goes along ”bio-image analysis core-facility”, ”bio-image analysis hub” or ”bio-image analysis technology development group”, which highlights that these groups are not pure computer science research groups. While the idea of establishing computationally-oriented core-facilities in biology research institutes is not new [
1], pure bio-image-analysis-focused core-facilities remain rare. Reasons to found bio-image analysis core-facilities are manifold and decision makers are increasingly reaching similar conclusions. The German Bioimaging society currently
1 lists a number 41 groups from various institutions having image analysis among their services. In this article, we introduce the demands calling for such groups to be set up and potential services these groups can offer. We outline typical scenarios and potentially arising conflicts of interests between provided services and the missions of a core-facility.
2. Missions of bio-image analysis core-facilities
Bio-image analysts are faced with manifold tasks: Often-received requests from wet-lab scientists revolve around programming custom workflows or guiding collaborators towards the right workflow for image analysis. Such requests obviously call for wide-range expertise. Secondary tasks, typically demanded by institutional decision makers, aim towards sustainability of services and long-term operation of the group. In this section, we define these terms. We start with the most common requests and transition towards the more invisible tasks that typically also need to be covered.
2.1. Having image analysis expertise on-site
Likely the most common request we bio-image analysts at the Cluster of Excellence ”Physics of Life” at TU Dresden / Germany see, is the demand for a custom image segmentation workflow. Workflows are assemblies of reusable image analysis components serving a specific task [
2]. For this, the workflow designer needs experience with a large number of relevant components and needs general methodological knowledge. Moreover, the workflow should be developed in a way that it can be executed reproducibly on the collaborator’s computer and potentially later on computers of reviewers, readers and others who aim at using the same strategy in their own research. Depending on if the workflow is provided as script, plugin or container, this requires different levels of software engineering and deployment expertise. Lastly, the goal of the analysis needs to be critically reviewed: Is image segmentation necessary for answering the scientific question? A common mistake by early career scientists in the field is focusing on improving the image segmentation strategy, thus loosing the focus on answering the biological question. Computational experts are necessary to guide life scientists through the entire workflow and afterwards identify potential bottlenecks. Such tasks and requests can be best addressed with computational and bio-image analysis experts on site [
3]. Depending on the diversity of tasks and needed expertise, single individuals may not be able fulfill all needs. Hence, there might be a need for dedicated bio-image analysis teams.
2.2. Knowledge conservation and incubation
A major strategic aim of core-facilities is to serve as knowledge incubator. Experts working in such a group are expected to exchange knowledge with each other, the local community and other international experts. This aims to prevent developed workflows from being forgotten and reinvented afterwards. A successful strategy for this is key towards sustainable operation of the group. If experts manage to establish standardized workflows and components, future research projects can be executed more efficiently, giving the staff time to develop advanced tools for upcoming challenges [
4].
Staying up-to-date in the rapidly developing field of bio-image analysis is challenging. For example, deep learning and cloud computing are expected to have major impact on how computational resources are used in projects in the next 5-10 years [
5]. Dedicated actions need to be undertaken by service-providing staff to determine which of the upcoming techniques are relevant for the institute, fulfill certain quality criteria and fit into the available computational infrastructure on campus.
2.3. Software maintenance
Similar to knowledge conservation, developed software needs to be conserved and maintained mid-/long-term for the benefit of its users and the institute [
6]. This includes efforts to ensure that software remains functional in the context of an evolving software ecosystem. Moreover, maintenance efforts undertaken by individuals, groups and institutes ensure a degree of control over the code base of strategic, regularly used software components. Software projects such as CellProfiler [
7], ilastik [
8], QuPath [
9] and Fiji [
10] serve as examples which play an important role not just at the institutes where they were developed, but also internationally.
2.4. Standardization
As the distinct field of bio-image analysis is quite young, standardization of common procedures such as reporting [
11,
12,
13], benchmarking [
14,
15] and file-formats [
16,
17] are still emerging. Standardized workflows for common tasks such as nuclei segmentation or cell shape measurements are not widely established currently. A possible exception to this is given by medical imaging and clinical data, which is typically acquired in highly standardized procedures, rendering the analysis feasible according to strict standard operating procedures [
18]. The present degree of standardization within an institute or field introduces a trade-off between flexibility and comparability. Depending on research questions and input data type, standardized workflows can deliver comparable results that may, however, not yield the right answers. On the other hand, the results of custom workflows are difficult to compare but they can be tailored to the respective project. A core-facility can choose to establish local standards with limited scope even in the absence of common standards. Such local standards allow avoiding redundant efforts in subsequent projects with similar aims and enables comparison of quantitative analysis results between projects.
2.5. Research data management
Computational science relies on research data management (RDM). Crude data storage and management solutions using custom folder structures on individual or group level need to be left behind to enable cross-group RDM standardization and reap the benefits of well organized research data. Even more so as the publication of biological imaging data is expected to further advance the field as a whole [
19]. Journals and funding agencies increasingly require RDM standards. While technical infrastructure for managing biological microscopy imaging data has been developed in the last decade [
20,
21,
22], human resources are necessary for maintaining this infrastructure and training users. A guide towards state-of-the-art research data management are for example the FAIR principles [
23]. Image data that is findable (F), accessible (A), stored in interoperable (I) file formats in a way it can easily be reused (R). The FAIR principles were also recently re-formulated for research software [
24]. It appears natural for this responsibility to be given into the hands of bio-image analysts as they profit from well-organized imaging data. This can simplify data analysis workflow design and allow answering questions that were out of reach before.
3. Scientific services
The above mentioned missions can be tackled by offering specific scientific services. We outline what bio-image analysis core-facilities can offer to satisfy the introduced needs and explain relationships with other services.
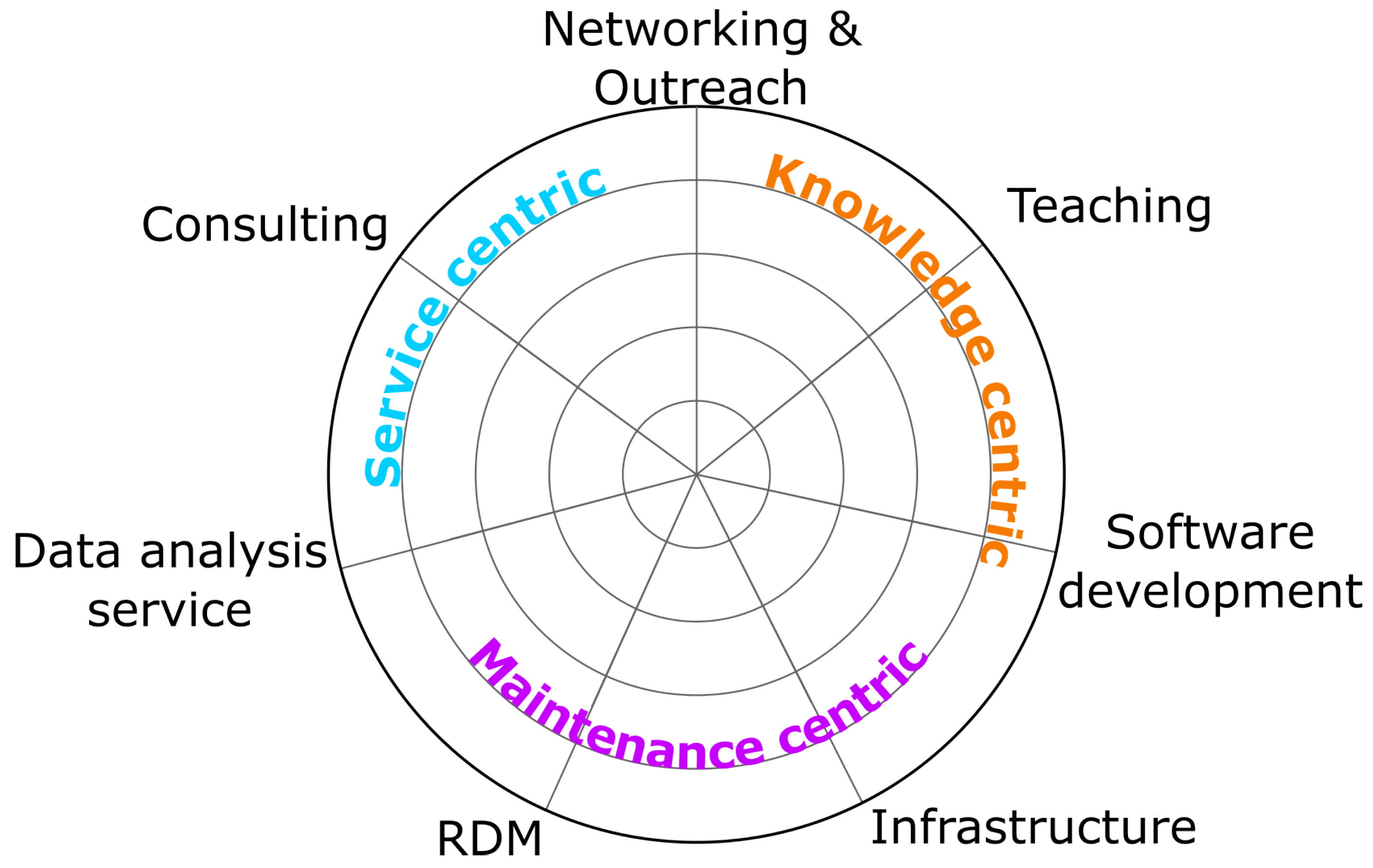
Figure 1 shows a radial enumeration of possible services and categorizes them in accordance with the introduced missions.
3.1. Consulting and co-supervision
Every collaborative project in the bio-image analysis context should start with an open-door consultation. Conducting such a session according to defined guidelines can help to synchronize consulting activities within the facility and outline further activities. Furthermore, regular open-desk hours, email hot-lines and public online forums such as the image science forum
https://image.sc [
25] can be suitable channels for this kind of interaction. It is in general recommended to start discussions about bio-image analysis early during the project. In case image data appears non-suitable for quantifying specific biological properties, modifying the sample preparation and imaging procedures might be required. We also recommend to schedule follow-up appointments to make sure the project develops in the right direction.
3.2. Data analysis as a service
Another potential strategy is pursuing data analysis as a service. A data producer ships entire datasets to a bio-image analyst. The analyst can then independently work with the data. Such a service assumes a shared understanding between data provider and analyst about which tasks are to be conducted. Consequently, it is reasonable to provide such services in case scientific questions can be answered using workflows and techniques that are highly standardized. Bio-image analysts working in such environments are typically also more specialized. A freelancing consultant who works for multiple institutes can focus more on a specific topic than generalists working within a single institute. This can, for example be the case in the pharmaceutical domain, where workflows and used assays are highly standardized.
It may not be possible to provide such a service in exploratory bio-image analysis projects, which typically demand a high level of interaction between data provider and bio-image analyst, and flexibility when designing a bio-image data analysis workflow.
3.3. Data Stewardship
Managing data and software can be a challenging task, especially in the life sciences which face ever-increasing amounts and sizes of acquired image data. This challenge has been encountered in the bio-image analysis context early [
26,
27,
28]. This led to the development of the concept of data stewardship in order to guarantee research data management standards across institutes and streamlined application of recurrent workflows. Data stewards help groups to comply to common RDM standards. This can be implemented on different levels: On a strategic level, data stewards can help to formulate management guidelines and policies. On an intermediary level, data stewards can be tasked with setting up suitable hardware and software infrastructure. This should be done in close collaboration with the institute’s IT support team. Lastly, data stewards can help in a consultation setting to guide collaborators towards using the existing resources effectively. While bio-image analysts can serve all these roles, their expertise lies close to the image data and the associated biology. Hence, the expertise of bio-image analysts is best used by consulting them on how to manage data.
If the institute is new to having data stewards, a first step could be conducted as consulting. From our perspective, many research groups struggle with defining their own folder structures and RDM plans. Thus, dedicated experts can guide scientists in managing their data. After such basic consulting strategies have been established, a second level of support appears worthwhile from an institutional decision maker’s perspective: A data steward or RDM officer role can be established at the institute. Due to their responsibilities, these individuals may report directly to the board of directors and have the duty to establish common procedures at the institute, which includes rules for implementing standards such as the FAIR principles. Given the agreement of the producers and owners of the data, this officer can have a special right: They may be allowed to browse research data of groups at the institute and take a more active role in guiding the scientists, e.g. contacting them directly in case they see violations of established rules. As this role comes with substantial responsibilities regarding data protection, safety and integrity, it is recommended to hire those persons on permanent positions. Needless to say, proven long-term experience in handling research data in a responsible fashion has to be a key requirement for these positions.
3.4. IT infrastructure maintenance
The maintenance of software and hardware infrastructure is a common task at institutes where computational methods are used frequently.
Workstations: Dedicated hardware infrastructure, such as image processing workstations, are common equipment at research institutes. If the computational hardware is partially under control of bio-image analysts, unique solutions exploiting the available hardware can be developed. For example, if the institute has graphics processing units (GPUs) installed in a number of image processing workstations, software libraries that can harness the computational power of GPUs can be used [
29]. In advanced scenarios, it is reasonable to maintain GPU-accelerated image processing software and hardware infrastructure for the benefit of the institute. Moreover, localizing workstations, potentially with commercial software, in central, open, makerspace-like environments sparks collaboration and induces a culture of cooperation and knowledge exchange among scientists across groups and even interdisciplinarily [
30]. If research groups decide to buy their own computers because booking centrally administered workstations is too expensive, the benefits from shared infrastructure cannot be exploited.
High-performance computing: The maintenance of high-performance computing (HPC) resources is typically beyond the scope of a bio-image analysis core-facility. Nonetheless, its staff should engage in frequent dialogue with the institute’s IT support team. This ensures that HPC infrastructure meets the demand of the life-science community and is being actively used to leverage computation-heavy projects. In case of a narrow research focus, specialized imaging and IT hardware can be purchased for the optimal operation of the core-facility. It may furthermore make sense to have joint groups providing imaging and image analysis services. In such groups, infrastructure can be optimized to have unified workflows ranging from the sample preparation, through standardized image acquisition and established image analysis a in high-performance-computing infrastructure.
3.5. Software development and maintenance
The development and maintenance of software can be considered as the digital counterpart of infrastructure maintenance. Bio-image analysis experts in core-facilities are often the only programming experts at biological research institutes. Thus, they may be responsible for developing software and maintaining it. There may also be dedicated research software engineering positions at the core-facility for this.
Development: In general, we differentiate two kinds of software development projects: 1) Custom workflow development for specific projects and 2) reusable component development for multiple projects. Both can be conducted in a consultation setting or by bio-image analysts taking a more active role in a project, e.g. transforming a custom script developed by a collaborator into a reusable component. The software development process could begin as part of a first exploratory project aimed at programming a custom script for counting cells together with a single collaborator. After receiving requests for similar tasks from a second and third collaborator, the written code can be refactored into a reusable component, for example as a plugin for a common image analysis framework such as Fiji or Napari [
10,
31]. The first strategy, custom script development, allows bio-image analysts to become co-authors on specific research publications and demonstrate their wide-ranging skills in their curriculum vitae. The second strategy, development of reusable components, can lead to independent research projects where the software developers become first-author and the collaborators who provided research data and scientific questions can become co-authors. Either way, the collaboration is of mutual benefit.
Maintenance: Maintaining software is a continuous effort. It needs to be kept ongoing for software upon which scientists rely. In particular, this needs to be ensured for periods of time beyond the duration of the contracts of the core-facility’s members. However, while maintenance is of strategic importance, bio-image analysts are part of the scientific community. They are consequently subject to a continuously high pressure to deliver scientific contributions. Hence, besides diverting effort to the maintenance of projects of strategic importance, employment in a bio-image analysis core-facility needs to offer staff the opportunity to develop their own profiles, projects and careers. Goal is to provide bio-image analysts a path to stay in academia long-term.
Both kinds of efforts, software development and maintenance, are necessary to strengthen the reputation of the core-facility within the local and the international research community. Efforts on maintenance of software relevant to the host institute, but also to the international community, may appear costly to institutional decision makers. However, investing this effort can pay off in the long run: The development and maintenance of tools through a core-facility ensures that the software meets the demand of as many users on campus as possible, avoids re-inventing the wheel and remains up-to-date. Core-facility members maintaining a software-package also typically know it well and can teach it to a local community. As maintaining software and related knowledge can come at high costs, keeping the number of maintained and taught software applications small is key towards sustainable software and knowledge maintenance [
5].
Lastly, the supervision of software and hardware within the scope of a core-facility furthermore allows staff bio-image analysts to oversee the process of projects as well as identify the possible need for further developments. Thus, deciding which software to develop and maintain is a decision that should follow strategic goals set by the core-facility members in consultation with group leaders at the institute.
3.6. Networking and outreach
In order to use resources of core-facilities efficiently, efforts should be routed to internal and external networking.
Local: Most importantly, the core-facility members must be well-connected to all relevant actors on their local campus: Knowing the data analysis needs of the local research community is important for planning and organizing all consulting, development, maintenance and teaching efforts. Core-facility members need to talk to local and internal algorithm developers to know what new tools are in the making and to guide their developments towards scientific questions that are highly relevant for local collaborators from the biology side.
Cross-institutional: It is an obvious course of action to team up with colleagues from the same region and conduct consulting, development and teaching projects together. Nonetheless, core-facility members should strive for international activities that have been shown to be highly efficient in teaching bio-image analysis and in establishing the profile of the bio-image analysts themselves [
32]. If core-facility members are involved in inter-institutional open-source software development projects, they can influence the direction of these projects towards the interests of their institute. This is a social task rather than a technical one: It is reasonable that the core-facility members are involved in relevant societies internationally, such as the Network of European BioImage Analysts (NEUBIAS) and associations of light microscopy core-facilities such a EuroBioimaging. Also, national and regional associations, in our case the German BioImaging society (GerBi) and the Biopolis Dresden Imaging Platform (BioDIP) are invaluable partners in the networking context.
In some research contexts, a higher degree of standardization of image analysis tasks and acquired data is present, e.g. in the clinical research context. Then, such institutions can be established on a national and trans-national level [
33,
34] to synchronize efforts between participating institutions. This allows for analyzing data in a standardized fashion at reduced costs and better comparability. If the same data analysis workflow is applied to data produced in many research projects, the need for consulting collaborators is lower and less custom scripts need to be programmed.
In the time of free online courses, open access shared image data and image analysis software running on every computer, it is obvious that state-of-the-art image data science is not exclusively happening in the rich countries. Networking activities enable us to work closely together with scientists world-wide, including community members from developing countries. Thus, we would like to motivate everyone to get involved in the foundation of the global Society for BioImage Analysis (SoBIAS) as we aim for this society to become a major player that can synchronize efforts world-wide and also represent the community towards funding bodies, research associations and governments.
3.7. Teaching and Education
Teaching and education pertain to all tasks regarding the distribution, incubation and conservation of knowledge. These are of particular importance in environments of high personnel fluctuation such as academic research institutes where scientists only get non-permanent positions [
35]. Activities include lectures, workshops and pair programming sessions. Each of these is suitable for a particular audience size, depth of teaching as well as focus regarding content.
Lectures: For large audiences, lectures on bio-image analysis techniques and programming basics are a suitable way to leverage the overall proficiency on campus in a long-term and thus, sustainable fashion. Naturally, scientists with basic knowledge in image analysis and programming will find themselves having an easier time in analyzing their own data. The content focus and level should be chosen to be of interest to a large target audience and provide entry points of expertise to the attendees. Since lectures may be part of universities’ bachelor and master programs, their content should be standardized also in an international context. Bio-image analysis textbooks can play a key-role in defining these standards [
36,
37].
Workshops: For smaller groups of collaborators with a narrower area of expertise, single or multi-day workshops represent a suitable format of education in more constrained topics. Bio-image analysts can teach a single research group on topics that are relevant to their specific needs. This strategy is considered more time-efficient than lectures at a university which take an entire semester to complete. On the other hand, customizing teaching materials for such workshops also comes at higher costs. Thus, while lectures in universities can be attended for free in many places, workshops could be financed through fees, and contribute to the sustainable long-term operation of a bio-image analysis core-facility. Advanced train-the-trainer workshop formats help to improve the knowledge transfer between attending individuals and experienced, trained-in-teaching staff [
38]. Selecting participants strategically ensures a lasting incubation of knowledge within the participants’ groups and institutes by empowering them to relay principles of learned skills to a broader audience, a task which would be difficult for members of a bio-image analysis core-facility to shoulder alone. The focus of offered workshop should be chosen in alignment with the research interests of the intended target audience.
Pair programming: Pair programming is an established technique in the computational sciences [
39]. An expert in programming and a collaborator meet to work for some time on one screen together. Typically, the non-expert types while the expert speaks out what they would do if they were alone. If the sessions take longer, it is also recommended to switch the roles regularly. This very involved knowledge transfer technique allows for boosting education of collaborators and advancing a particular research project at the same time. It is an effective method for introducing collaborators to specific tools they need to achieve their particular goals and acquire bio-image analysis expertise. Depending on the degree of involvement in the project, the expert should be considered for co-senior authorship similar to the consulting strategy introduced above. Technically, pair-programming is hands-on consulting.
Job shadowing: When substantial knowledge transfer is necessary, e.g. for establishing a routine technique in a research lab, job shadowing is a common technique we also exercise in our institute. Embedding bio-image analysts temporarily in a research group, locally referred to as rent-a-bio-image-analyst, allows interaction on a level far beyond consulting. Also, wet-lab scientists spending a couple of weeks in the offices of bio-image analysts, locally referred to as mini-sabbatical, represents an excellent strategy to make both sub-communities of life scientists to find a common language and collaborate efficiently. In both settings, projects can be pushed forward substantially and bring mutual benefit. Flexible payment schemes might be necessary to establish such a strategy on a larger scale.
Educational activities also include training the core-facility’s own staff. If experts are hard to get on the job-market, they have to be trained within a core-facility. Postdoctoral fellowship programs are one opportunity to train core-facility employees in advanced techniques. This strategy has been exercised in imaging core-facilities and appears transferable to other technologies [
40]. In the bio-image analysis and data science context it appears even more beneficial: Coming back to the above introduced mini-sabbaticals, which core-facility employees can spend in research labs, they may decide their future career path during these close collaborations.
Data scientists are hard to hire in research groups, too. Hiring a core-facility-trained bio-image analyst on a staff scientist position in a research group can transfer expertise into these groups and enable image data science projects that are unthinkable in smaller PhD or postdoctoral projects.
4. Potential sources of conflicts of interest
In this manuscript, we introduced typical needs in the context of bio-image analysis which life science research institutes express and services that dedicated bio-image analysis core-facilities could offer to serve these needs. In the following, we discuss potential caveats arising from design choices to be aware of.
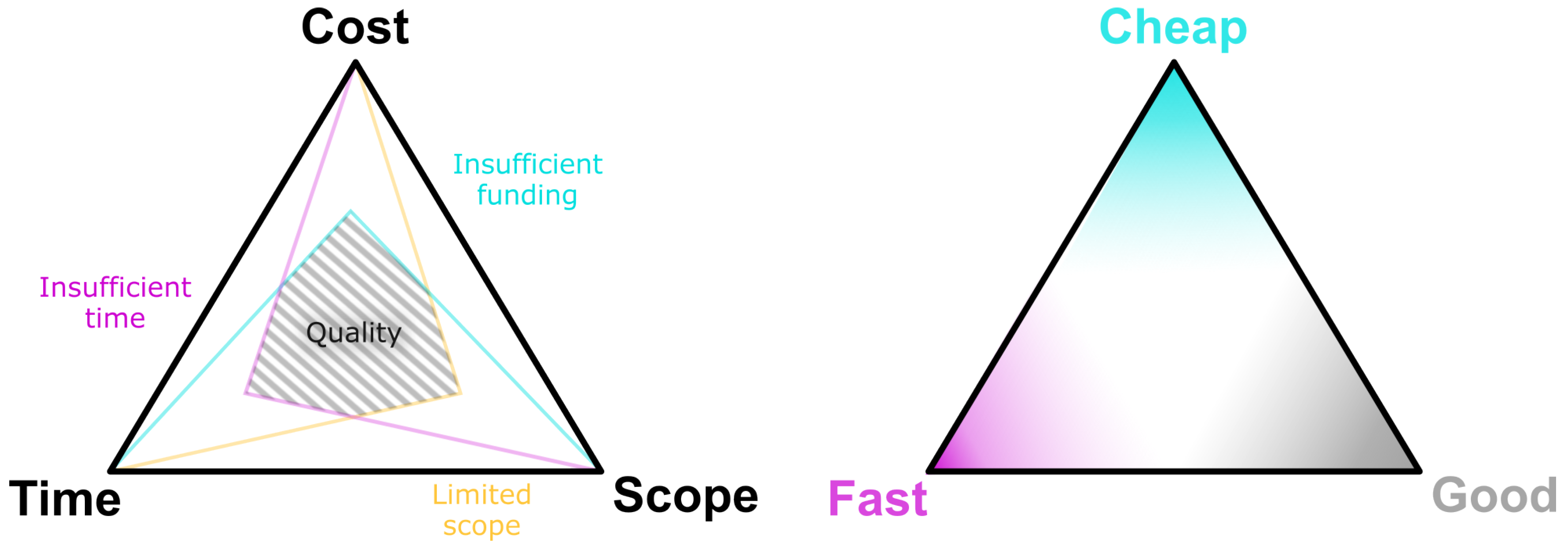
First, both decision-makers considering to fund a core-facility and collaborators requesting services from a core-facility should be aware of the so-called ”unattainable triangle of project management” [
41] as shown in
Figure 2. It states, in short, that the quality of a project is constrained by time, cost and performance. However, not all key parameters of a project can be optimized at the same time: Fast-developed and well-designed solutions are expensive. If the costs are too high, either scope or development time must be sacrificed.
Collaboration between life scientists and software developers
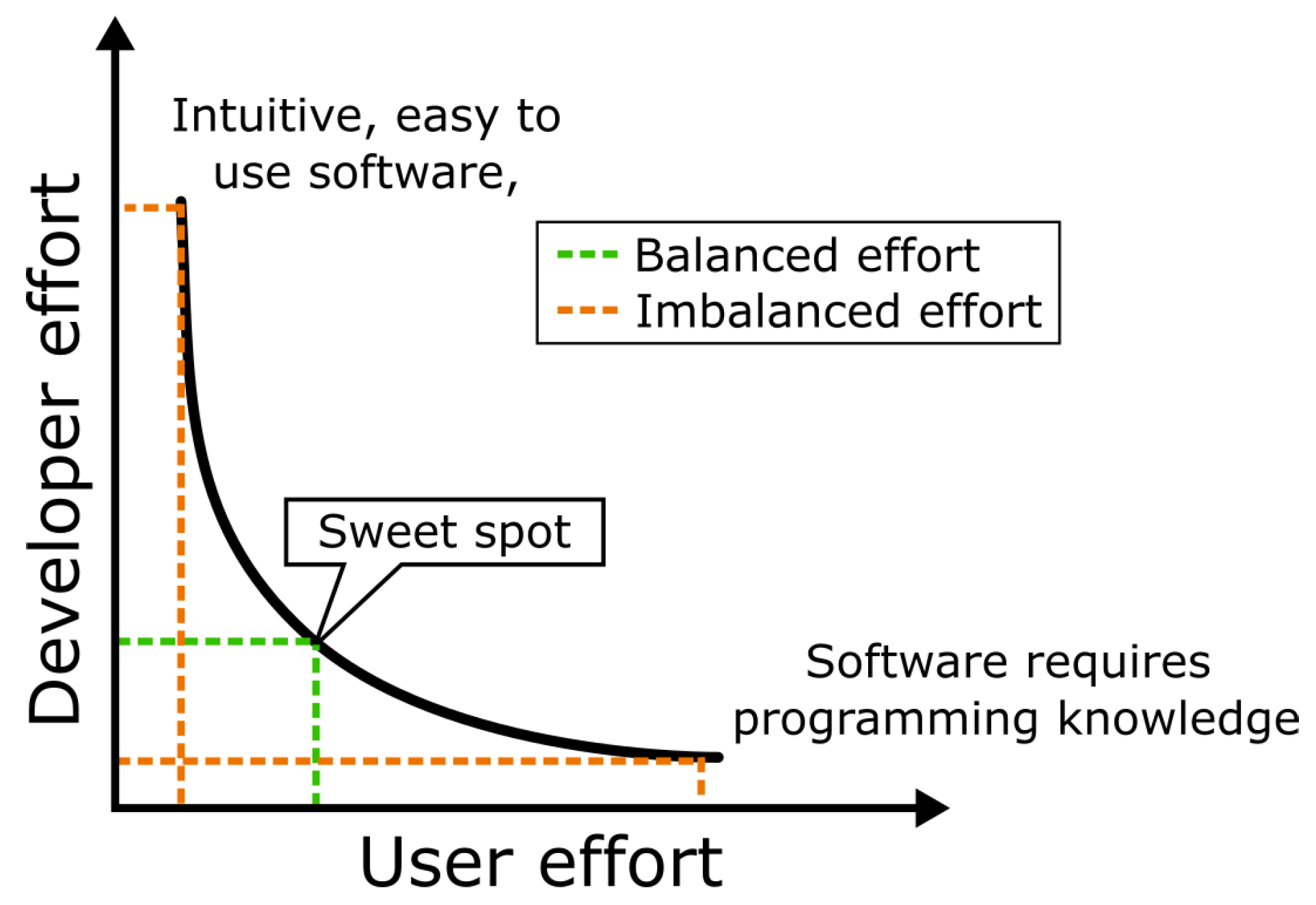
When considering tasks such as software development and user education, it is apparent that putting the focus exclusively on either of these two activities is sub-optimal regarding effort distribution between software users and software developers. A bio-image analysis component that requires an end-user to learn a programming language causes maximum effort on user’s side while minimizing effort on developer’s side. In contrary, crafting a intuitive, user-friendly software with a graphical user interface (GUI), maximizes the effort for the developer while keeping the workload minimal for the user. Algorithm developers may not see it as their responsibility to invest the increased effort for developing GUIs. This may be the case even more if the design elements are specific to particular biological research questions. Within the bio-image analysis community, the term
going the extra mile established for investing this effort, which is often undertaken by core-facility employees. Finding the right balance between these two strategies is key. We claim that the total effort of developers and users is minimal in case the developers spend some time on adding accessible user-interfaces to their software while users acquire some basic programming skills. The challenge is displayed in
Figure 3.
Recharging
When it comes to billing services such as bio-image analysis consulting and software development support, the right model must be negotiated, potentially on a project-by-project basis. Commonly, initial consultation is free. After a first discussion, an evaluation of the benefits for individuals, research groups and the institute could be the next step towards identifying the right model for billing. The more people profit from the core-facility executing a collaborative project, the lower the price could be for the inquiring research group. Such discounts or a general open-door policy can only be awarded if other funding sources are available.
The major aim of a core-facility, per definition, is to serve the entire institute and treating groups equally. If working hours are billed, the focus of the core-facility will naturally shift towards the research groups with larger budget, which are typically senior research groups. To enable young investigator groups to benefit equally, flatrate models can be established: All groups have a certain amount of annual working hours they can ask core-facility staff to work for them.
In the absence of long-term core-funding, a core-facility may need to bill its collaborators while also claiming co-authorships. We want to highlight that we consider this appropriate both in terms of good scientific practice and responsibility [
42] as well as towards securing the above-described missions of the core-facility in a sustainable fashion. This policy should always be communicated clearly within projects at an early stage.
Challenging sources for conflicts of interest appear in the context of billing collaborators for custom data analysis. If workflow development and consulting are billed per hour, collaborators might tend to exchange workflows among each other without consulting the developers in order to spare money. However, the limitations of custom image analysis scripts might not be easy to judge for non-experts. If custom scripts don’t work on other data, this poses the risk for exposing a bad picture of the work done in the core-facility. This scenario can be prevented by two major actions: 1) Asking questions in a basic consulting setting should always be free. Nobody should fear to receive a bill because they asked a question. 2) Documentation for custom image analysis scripts must explicitly state the intended use. It is also recommended to add a disclaimer to the documentation hinting the user of the script that the developer of the script is not liable or responsible for usage of the script. Common open-source software licenses such as BSD or MIT contain these disclaimers and additionally give the collaborators, who potentially paid for the software development, the legal right to publish the code along with their scientific publication without consulting the author [
6].
Joining efforts between core-facilities and research groups is also a suitable approach to secure core-facility operations sustainably: Grant proposals written by collaborating groups can feature additional staff and infrastructure funding for data management and data science services, which could then be diverted to the core-facility. Since research data management, scientific data analysis support and consulting are increasingly recommended or even mandatory to include in funding proposals, adding resources to sustain a responsible core-facility is of mutual benefit. As funding agencies are aware of the needs, mentioning such items in a grant proposal may increase the chance of securing the funding.
Project prioritization
Another important point of consideration lies in the nature of projects a core-facility is tasked with. If the core-facility’s core-budget is limited and employees are paid from grants, members of the core-facility are typically expected to contribute to the field in the form of publications. In this scenario, core-facility members have to prioritize support requests. A criterion for deciding between research projects to work on is the exploratory or exploitative nature of the project. In case a suitable bio-image data analysis workflow is roughly known and just needs to be further developed to be exploited, e.g. by being executable on HPC infrastructure, the efforts for the project are fairly predictable and allow an informed decision about the project costs. In case no image data analysis workflow is known yet and the outcome hard to predict, the effort for this kind of exploratory project is harder to estimate and thus, potentially not the right situation for a core-facility to invest a lot of time. We recommend consulting and pair-programming as methods of choice for exploratory scenarios and software development efforts for exploitative scenarios.
An important aim of a bio-image analysis core-facility is to enable software solutions to be available long-term. If a PhD project resulted in a new algorithm that appears beneficial beyond the group it was developed in, the core-facility should support maintaining this software. Software developers working hours invested in making software of a research group sustainable, should not entirely be billed to the group as the institute benefits from this work. Also in this case, larger groups who can put more substantial effort in developing new methods have a higher chance to benefit from the core-facilities support in this context. Thus, the decision making process for which software projects to support should be made in close collaboration with group leaders,– potentially democratic – for example in a core-facility committee.
Avoiding bottlenecks in scientific workflows
As knowledge incubation and transfer is a major mission of core-facilities, bio-image analysis as a service introduces some conflict of interest as well: If the employees of a core-facility are the only experts that can conduct a certain bio-image data analysis workflow, their service can become a bottleneck. On the one hand, exploiting this bottleneck by billing for the data analysis service is a potential source for funding. It is on the other hand a risk for the institute: Core-facility employees who are constantly overwhelmed by repetitive requests bore out and may seek for new opportunities outside the institute. The experience may be lost and the core-facility then failed in conserving the knowledge. On the other hand, a similar argument can be brought forth from the point of view of good scientific practice: If the expertise to execute a developed workflow resides exclusively with core-facility experts, how reproducible are the results in practice? To circumvent these issues, we recommend researchers to acquire bio-image analysis skills via learning-by-doing under guidance from core-facility employees. The core-facility members can also take user feedback from these sessions and further develop their tools. All involved parties, the software users, its developers and the core-facility profit from related publications as they ensure the continuous influx of third-party funding.
Teaching and education
The importance of teaching shall also be highlighted. Unfortunately, general computational skills, data management and data science are not part of undergraduate curricula in the life sciences and thus, core-facility staff often explains computational basics to researchers. Depending on the size of a core-facility with respect to the number of potential collaborators on campus, more or less consulting and custom script development services can be offered. In case of an overwhelming number of basic questions and support requests, increasing teaching activities can be considered to answer common bio-image analysis questions in a broadcasting fashion. Establishing a bio-image analysis lecture open to students but also PhD candidates and postdocs is worth the effort. Good computational practice will spread in the research groups on campus and the general situation improves. We are exercising this for the fourth year on our campus and the feedback from local researchers is very positive. Our impression goes in line with a recently executed, international survey: More and more scientists seek to learn bio-image analysis skills themselves and thus, there is a large need for tutorials and workshops [
43].
Long-term strategy development
Considering the local needs and demands on a research campus, it may appear to be a feasible course of action to obtain bio-image analysis expertise on a per-project basis: Paying a freelance consultant or image analysis consulting company for a particular project might be more cost effective for many clients compared to creating expensive internal facilities or hiring staff if image analysis is not constantly highly demanded. However, this comes at the disadvantage that an external expert may not always be available if needed and maintenance can not be ensured.
If a core-facility is established as shown above in the various contexts, the staff of a core-facility has far-reaching responsibilities. Together with the value a core-facility provides to its collaborators, balancing the different services, both in terms of scope and its recipients, is a critical task that should be evaluated regularly. These considerations could be addressed, for example by an external advisory board (EAB) of core-facility leaders from other institutes. Using their external perspective, they could on the one hand help the core-facility to adjust their goals and missions. Furthermore, an EAB report could give the core-facility and the responsible steering bodies a powerful instrument when arguing about further development of the core-facility and internal funding with institutional decision makers.
5. Conclusions
Founders and managers of bio-image analysis core-facilities face challenging tasks in shaping their work environment: primarily the institute communities’ needs have to be fulfilled using a well-adjusted selection of services. At the same time, the career perspectives of the core-facility employees are of high importance as the core-facility cannot operate without motivated personnel who strives to improve their skills and develop along with the bio-image analysis field. The core-facility is not just responsible for maintaining existing tools but also for developing workflows for future research projects. From our perspective, core-facilities who develop technologies work best if they are partially funded through third-party research funding, which may turn the core-facility partially into a research group. This group structure compromise deserves an own name, which is why we refer to our group as a ”Bio-image Analysis Technology Development Group”.
Funding
We acknowledge support by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy - EXC2068 - Cluster of Excellence Physics of Life of TU Dresden.
Acknowledgments
We would like to thank Florian Jug (Human Technopole, Milano, Italy), Nicolas Chiaruttini (EPFL, Lausanne, Switzerland), Tatiana Woller (VIB Bio Imaging Core, Zwijnaarde, Belgium), Sebastian Munck (VIB Bio Imaging Core, Zwijnaarde, Belgium), Stéphane Rigaud (Image Analysis Hub, Institut Pasteur, Paris, France) and Jan Brocher (BioVoxxel, Ludwigshafen, Germany) for their constructive feedback during the mansucript preparation.
Abbreviations
BIA, Bio-image Analysis; RDM, Research Data Management; RSE, Research Software Engineer
References
- Lewitter F, Rebhan M. Establishing a successful bioinformatics core facility team. PLOS computational biology 2009, 5, e1000368.
- Miura K, Paul-Gilloteaux P, Tosi S, Colombelli J. Workflows and components of bioimage analysis. In: Bioimage Data Analysis Workflows Springer, Cham; 2020.p. 1–7.
- Lippens S, Audenaert D, Botzki A, Derveaux S, Ghesquière B, Goeminne G, et al. How tech-savvy employees make the difference in core facilities: Recognizing core facility expertise with dedicated career tracks. EMBO reports 2022, 23, e55094.
- Lippens S, D’Enfert C, Farkas L, Kehres A, Korn B, Morales M, et al. One step ahead: innovation in core facilities. EMBO reports 2019, 20, e48017.
- Haase R, Fazeli E, Legland D, Doube M, Culley SA, Belevich I, et al. A Hitchhiker’s guide through the bio-image analysis software universe. FEBS Letters 2022 7; https://onlinelibrary.wiley.com/doi/full/10.1002/1873-3468.14451 https://onlinelibrary.wiley.com/doi/abs/10.1002/1873-3468.14451 https://febs.onlinelibrary.wiley.com/doi/10.1002/1873-3468.14451.
- Levet F, Carpenter AE, Eliceiri KW, Kreshuk A, Bankhead P, Haase R. Developing open-source software for bioimage analysis: opportunities and challenges. F1000Research 2021, 10.
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology 2006, 7, 1–11.
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, et al. Ilastik: interactive machine learning for (bio) image analysis. Nature methods 2019, 16, 1226–1232.
- Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Scientific reports 2017, 7, 1–7.
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nature methods 2012, 9, 676–682.
- Marqués G, Pengo T, Sanders MA. Imaging methods are vastly underreported in biomedical research. Elife 2020, 9.
- Miura K, Nørrelykke SF. Reproducible image handling and analysis. The EMBO journal 2021, 40, e105889.
- Sarkans U, Chiu W, Collinson L, Darrow MC, Ellenberg J, Grunwald D, et al. REMBI: Recommended Metadata for Biological Images—enabling reuse of microscopy data in biology. Nature methods 2021, 18, 1418–1422.
- Rubens U, Mormont R, Paavolainen L, Bäcker V, Pavie B, Scholz LA, et al. BIAFLOWS: A collaborative framework to reproducibly deploy and benchmark bioimage analysis workflows. Patterns 2020, 1, 100040.
- Reinke, A. Metrics Reloaded-A new recommendation framework for biomedical image analysis validation. In: Medical Imaging with Deep Learning; 2022.
- Moore J, Linkert M, Blackburn C, Carroll M, Ferguson RK, Flynn H, et al. OMERO and Bio-Formats 5: flexible access to large bioimaging datasets at scale. In: Medical Imaging 2015: Image Processing, vol. 9413 SPIE; 2015. p. 37–42.
- Moore J, Allan C, Besson S, Burel JM, Diel E, Gault D, et al. OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies. Nature methods 2021, 18, 1496–1498.
- Boyce, B. An update on the validation of whole slide imaging systems following FDA approval of a system for a routine pathology diagnostic service in the United States. Biotechnic & Histochemistry 2017, 92, 381–389. [Google Scholar]
- Ellenberg J, Swedlow JR, Barlow M, Cook CE, Sarkans U, Patwardhan A, et al. A call for public archives for biological image data. Nature Methods 2018, 15, 849–854.
- Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S, et al. OMERO: flexible, model-driven data management for experimental biology. Nature methods 2012, 9, 245–253.
- Williams E, Moore J, Li SW, Rustici G, Tarkowska A, Chessel A, et al. Image Data Resource: a bioimage data integration and publication platform. Nature methods 2017, 14, 775–781.
- Hartley M, Kleywegt GJ, Patwardhan A. The BioImage Archive – Building a Home for Life-Sciences Microscopy Data. Journal of Molecular Biology 2022, 43, 167505 https://wwwsciencedirectcom/science/article/pii/S0022283622000791, computation Resources for Molecular Biology. [Google Scholar] [CrossRef]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific data 2016, 3, 1–9.
- Barker M, Chue Hong NP, Katz DS, Lamprecht AL, Martinez-Ortiz C, Psomopoulos F, et al. Introducing the FAIR Principles for research software. Scientific Data 2022, 9, 1–6.
- Rueden CT, Ackerman J, Arena ET, Eglinger J, Cimini BA, Goodman A, et al. Scientific Community Image Forum: A discussion forum for scientific image software. PLoS biology 2019, 17, e3000340.
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging 2013, 26, 1045–1057.
- Patwardhan A, Carazo JM, Carragher B, Henderson R, Heymann JB, Hill E, et al. Data management challenges in three-dimensional EM. Nature structural & molecular biology 2012, 19, 1203–1207.
- Müller A, Schmidt D, Xu CS, Pang S, D’Costa JV, Kretschmar S, et al. 3D FIB-SEM reconstruction of microtubule–organelle interaction in whole primary mouse β cells. Journal of Cell Biology 2021, 220.
- Haase R, Royer LA, Steinbach P, Schmidt D, Dibrov A, Schmidt U, et al. CLIJ: GPU-accelerated image processing for everyone. Nature methods 2020, 17, 5–6.
- Mersand, S. The state of makerspace research: A review of the literature. TechTrends 2021, 65, 174–186. [Google Scholar] [CrossRef]
- Contributors, N. napari: a multi-dimensional image viewer for Python. Zenodo, 2019; 3555620. [Google Scholar]
- Martins GG, Cordelières FP, Colombelli J, D’antuono R, Golani O, Guiet R, et al. Highlights from the 2016-2020 NEUBIAS training schools for Bioimage Analysts: a success story and key asset for analysts and life scientists. F1000Research 2021, 10.
- Eggermont AM, Apolone G, Baumann M, Caldas C, Celis JE, de Lorenzo F, et al. Cancer Core Europe: a translational research infrastructure for a European mission on cancer. Molecular oncology 2019, 13, 521–527.
- Joos S, Nettelbeck DM, Reil-Held A, Engelmann K, Moosmann A, Eggert A, et al. German Cancer Consortium (DKTK)–A national consortium for translational cancer research. Molecular oncology 2019, 13, 535–542.
- Bahr A, Blume C, Eichhorn K, Kubon S. With# IchBinHanna, German academia protests against a law that forces researchers out. Nature human behaviour 2021, 5, 1114–1115.
- Wu Q, Merchant F, Castleman K. Microscope image processing. Elsevier; 2010.
- Miura K, Sladoje N. Bioimage data analysis workflows. Springer Nature; 2020.
- Via A, Attwood TK, Fernandes PL, Morgan SL, Schneider MV, Palagi PM, et al. A new pan-European Train-the-Trainer programme for bioinformatics: pilot results on feasibility, utility and sustainability of learning. Briefings in bioinformatics 2019, 20, 405–415.
- Hannay JE, Dybå T, Arisholm E, Sjøberg DI. The effectiveness of pair programming: A meta-analysis. Information and software technology 2009, 51, 1110–1122.
- Waters, JC. A novel paradigm for expert core facility staff training. Trends in Cell Biology 2020, 30, 669–672. [Google Scholar] [CrossRef]
- Van Wyngaard CJ, Pretorius JHC, Pretorius L. Theory of the triple constraint—A conceptual review. In: 2012 IEEE International Conference on Industrial Engineering and Engineering Management IEEE; 2012. p. 1991–1997.
- McNutt MK, Bradford M, Drazen JM, Hanson B, Howard B, Jamieson KH, et al. Transparency in authors’ contributions and responsibilities to promote integrity in scientific publication. Proceedings of the National Academy of Sciences 2018, 115, 2557–2560.
- Jamali N, Dobson ET, Eliceiri KW, Carpenter AE, Cimini BA. 2020 BioImage Analysis Survey: Community experiences and needs for the future. Biological imaging 2021, 1, e4.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).