Submitted:
17 January 2023
Posted:
18 January 2023
You are already at the latest version
Abstract
Keywords:
Introduction
Methods
Data Sources
Empirical Formulas and Biosynthesis Reactions
Thermodynamic Properties of Live Matter and Biosynthesis
½ nN N2 + ¼ nP P4O10 + nS SO3
Thermodynamic Properties of Antigen-Receptor Binding
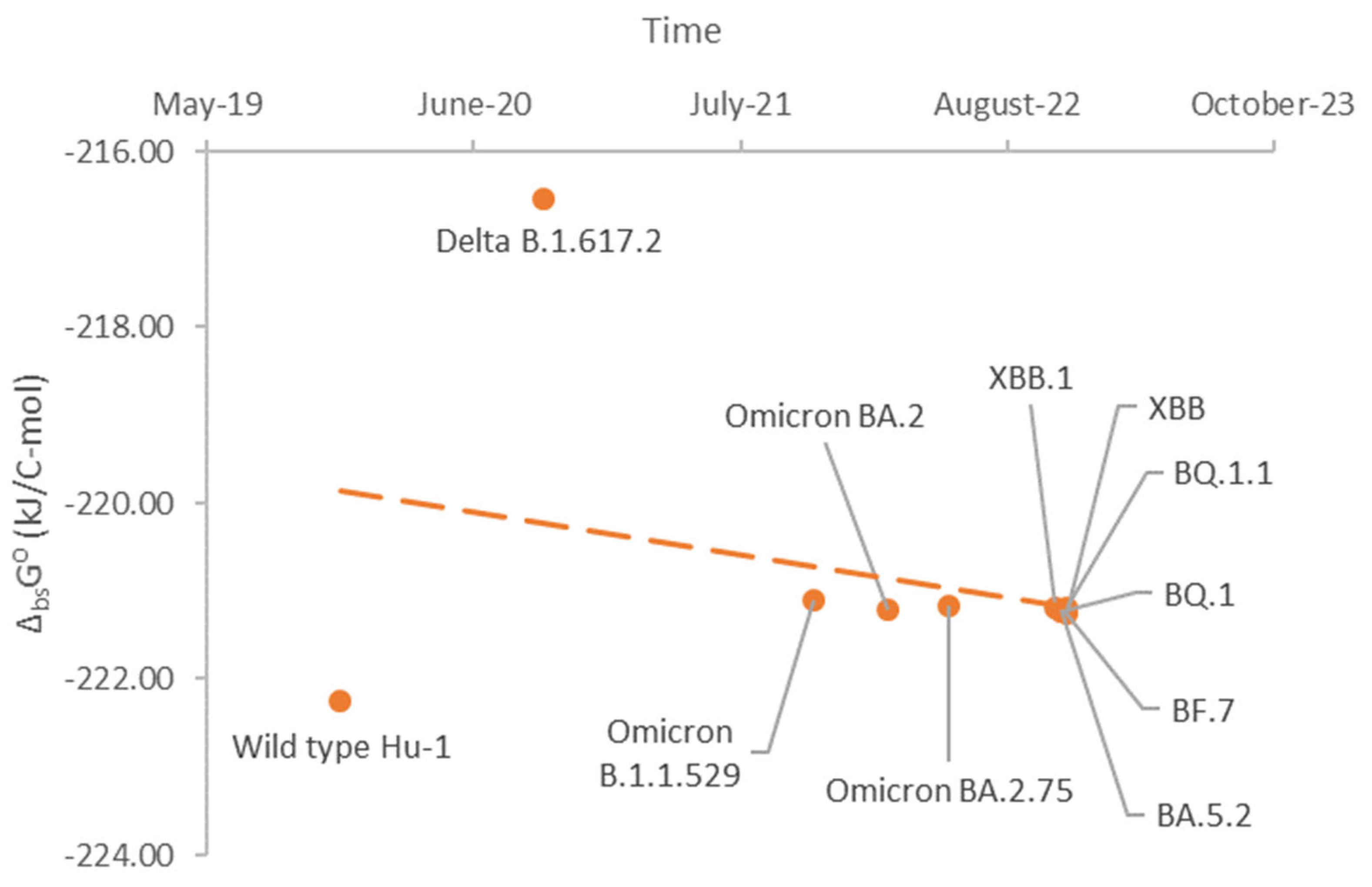
Results
Discussion
Conclusions
Acknowledgements
References
- Aladag, A., Hoffmann, S., Stoldt, M., Bösing, C., Willbold, D., & Schwarten, M. (2014). Hepatitis C virus NS5A is able to competitively displace c-Myc from the Bin1 SH3 domain in vitro. Journal of peptide science : an official publication of the European Peptide Society, 20(5), 334–340. [CrossRef]
- Anasir, M. I., Caria, S., Skinner, M. A., & Kvansakul, M. (2017). Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039. The Journal of biological chemistry, 292(22), 9010–9021. [CrossRef]
- Atkins, P. W., & de Paula, J. (2011). Physical Chemistry for the Life Sciences (2nd edition), W. H. Freeman and Company. ISBN-13: 978-1429231145.
- Atkins, P.W. & de Paula, J. (2014). Physical Chemistry: Thermodynamics, Structure, and Change, 10th Edition. New York: W. H. Freeman and Company. ISBN-13: 978-1429290197.
- Balmer, R.T. (2010). Modern Engineering Thermodynamics, Cambridge, MA: Academic Press. [CrossRef]
- Banerjee, M., Speir, J. A., Kwan, M. H., Huang, R., Aryanpur, P. P., Bothner, B., & Johnson, J. E. (2010). Structure and function of a genetically engineered mimic of a nonenveloped virus entry intermediate. Journal of virology, 84(9), 4737–4746. [CrossRef]
- Battley, E. H., & Stone, J. R. (2000). A comparison of values for the entropy and the entropy of formation of selected organic substances of biological importance in the solid state, as determined experimentally or calculated empirically. Thermochimica acta, 349(1-2), 153-161. [CrossRef]
- Battley, E.H. (1999). An empirical method for estimating the entropy of formation and the absolute entropy of dried microbial biomass for use in studies on the thermodynamics of microbial growth. Thermochimica Acta, 326(1-2), 7-15. [CrossRef]
- Battley, E. H. (1998). The development of direct and indirect methods for the study of the thermodynamics of microbial growth. Thermochimica Acta, 309(1-2), 17-37. [CrossRef]
- Battley, E.H. (1992). On the enthalpy of formation of Escherichia coli K-12 cells, Biotechnology and Bioengineering, 39, 5-12. [CrossRef]
- Bauer, D. W., Li, D., Huffman, J., Homa, F. L., Wilson, K., Leavitt, J. C., Casjens, S. R., Baines, J., & Evilevitch, A. (2015). Exploring the Balance between DNA Pressure and Capsid Stability in Herpesviruses and Phages. Journal of virology, 89(18), 9288–9298. [CrossRef]
- Bauer, D. W., Huffman, J. B., Homa, F. L., & Evilevitch, A. (2013). Herpes virus genome, the pressure is on. Journal of the American Chemical Society, 135(30), 11216–11221. [CrossRef]
- Berg, J.M., Tymoczko, J.L. and Stryer, L. (2002). Biochemistry, 5th ed. New York: W H Freeman. ISBN-13: 978-0716746843.
- Boltzmann, L. (1974). The second law of thermodynamics. In: Theoretical physics and philosophical problems, McGuinnes, B., ed., Boston, MA: D. Riedel Publishing Company, LLC. ISBN 978-90-277-0250-0 (translation of the original version published in 1886).
- Brouillette, C. G., Compans, R. W., Brandts, J. F., & Segrest, J. P. (1982). Structural domains of vesicular stomatitis virus. A study by differential scanning calorimetry, thermal gel analysis, and thermal electron microscopy. The Journal of biological chemistry, 257(1), 12–15. PMID: 6273421. [CrossRef]
- Byrn, R. A., Jones, S. M., Bennett, H. B., Bral, C., Clark, M. P., Jacobs, M. D., Kwong, A. D., Ledeboer, M. W., Leeman, J. R., McNeil, C. F., Murcko, M. A., Nezami, A., Perola, E., Rijnbrand, R., Saxena, K., Tsai, A. W., Zhou, Y., & Charifson, P. S. (2015). Preclinical activity of VX-787, a first-in-class, orally bioavailable inhibitor of the influenza virus polymerase PB2 subunit. Antimicrobial agents and chemotherapy, 59(3), 1569–1582. [CrossRef]
- Degueldre, C. (2021). Single virus inductively coupled plasma mass spectroscopy analysis: A comprehensive study. Talanta, 228, 122211. [CrossRef]
- Demirel, Y. (2014). Nonequilibrium Thermodynamics: Transport and Rate Processes in Physical, Chemical and Biological Systems, 3rd ed. Amsterdam: Elsevier. ISBN: 9780444595812.
- Deschuyteneer, M., Elouahabi, A., Plainchamp, D., Plisnier, M., Soete, D., Corazza, Y., Lockman, L., Giannini, S., & Deschamps, M. (2010). Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix™, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Human vaccines, 6(5), 407–419. [CrossRef]
- Djamali, E., Nulton, J. D., Turner, P. J., Rohwer, F., & Salamon, P. (2012). Heat output by marine microbial and viral communities. Journal of Non-Equilibrium Thermodynamics, 37(3), 291-313. [CrossRef]
- Dodd, T., Botto, M., Paul, F. et al. (2020). Polymerization and editing modes of a high-fidelity DNA polymerase are linked by a well-defined path. Nat Commun 11, 5379. [CrossRef]
- Du, X., Li, Y., Xia, Y. L., Ai, S. M., Liang, J., Sang, P., ... & Liu, S. Q. (2016). Insights into protein–ligand interactions: mechanisms, models, and methods. International journal of molecular sciences, 17(2), 144. [CrossRef]
- Duan, L., Zheng, Q., Zhang, H., Niu, Y., Lou, Y., & Wang, H. (2020). The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens. Frontiers in immunology, 11, 576622. [CrossRef]
- Elbe, S. and Buckland-Merrett, G. (2017) Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenges, 1:33-46. [CrossRef] [PubMed Central]
- Fanaei Pirlar, R., Wagemans, J., Ponce Benavente, L., Lavigne, R., Trampuz, A., & Gonzalez Moreno, M. (2022). Novel Bacteriophage Specific against Staphylococcus epidermidis and with Antibiofilm Activity. Viruses, 14(6), 1340. MDPI AG. [CrossRef]
- Gale P. (2022). Using thermodynamic equilibrium models to predict the effect of antiviral agents on infectivity: Theoretical application to SARS-CoV-2 and other viruses. Microbial risk analysis, 21, 100198. [CrossRef]
- Gao, X., Zhu, K., Qin, B., Olieric, V., Wang, M., & Cui, S. (2021). Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions. Nature communications, 12(1), 2843. [CrossRef]
- Gelman, D., Yerushalmy, O., Alkalay-Oren, S., Rakov, C., Ben-Porat, S., Khalifa, L., Adler, K., Abdalrhman, M., Coppenhagen-Glazer, S., Aslam, S., Schooley, R. T., Nir-Paz, R., & Hazan, R. (2021). Clinical Phage Microbiology: a suggested framework and recommendations for the in-vitro matching steps of phage therapy. The Lancet. Microbe, 2(10), e555–e563. [CrossRef]
- Glansdorff, P. and Prigogine, I. (1971). Thermodynamic Theory of Structure, Stability and Fluctuations. Hoboken, NJ: Wiley. ISBN-13: 978-0471302803.
- Guosheng, L., Yi, L., Xiangdong, C., Peng, L., Ping, S., & Songsheng, Q. (2003). Study on interaction between T4 phage and Escherichia coli B by microcalorimetric method. Journal of virological methods, 112(1-2), 137–143. [CrossRef]
- Istifli, E. S., Netz, P. A., Sihoglu Tepe, A., Sarikurkcu, C., & Tepe, B. (2022). Understanding the molecular interaction of SARS-CoV-2 spike mutants with ACE2 (angiotensin converting enzyme 2). Journal of biomolecular structure & dynamics, 40(23), 12760–12771. [CrossRef]
- Javorsky, A., Maddumage, J. C., Mackie, E., Soares da Costa, T. P., Humbert, P. O., & Kvansakul, M. (2022). Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM. The FEBS journal, 10.1111/febs.16607. Advance online publication. [CrossRef]
- Johansson, E., & Dixon, N. (2013). Replicative DNA polymerases. Cold Spring Harbor perspectives in biology, 5(6), a012799. [CrossRef]
- Kaniadakis, G., Baldi, M. M., Deisboeck, T. S., Grisolia, G., Hristopulos, D. T., Scarfone, A. M., Sparavigna, A., Wada, T., & Lucia, U. (2020). The κ-statistics approach to epidemiology. Scientific reports, 10(1), 19949. [CrossRef]
- Katen, S., & Zlotnick, A. (2009). The thermodynamics of virus capsid assembly. Methods in enzymology, 455, 395–417. [CrossRef]
- Kawahara, T., Akiba, I., Sakou, M., Sakaguchi, T., & Taniguchi, H. (2018). Inactivation of human and avian influenza viruses by potassium oleate of natural soap component through exothermic interaction. PloS one, 13(9), e0204908. [CrossRef]
- Khare, S., et al (2021) GISAID’s Role in Pandemic Response. China CDC Weekly, 3(49): 1049-1051. [CrossRef] [PubMed Central]
- Krell, T., Manin, C., Nicolaï, M. C., Pierre-Justin, C., Bérard, Y., Brass, O., Gérentes, L., Leung-Tack, P., & Chevalier, M. (2005). Characterization of different strains of poliovirus and influenza virus by differential scanning calorimetry. Biotechnology and applied biochemistry, 41(Pt 3), 241–246. [CrossRef]
- Lee, J., Schwarz, K. J., Kim, D. S., Moore, J. S., & Jewett, M. C. (2020). Ribosome-mediated polymerization of long chain carbon and cyclic amino acids into peptides in vitro. Nature communications, 11(1), 4304. [CrossRef]
- Liu, T., Sae-Ueng, U., Li, D., Lander, G. C., Zuo, X., Jönsson, B., Rau, D., Shefer, I., & Evilevitch, A. (2014). Solid-to-fluid-like DNA transition in viruses facilitates infection. Proceedings of the National Academy of Sciences of the United States of America, 111(41), 14675–14680. [CrossRef]
- Lucia, U., Grisolia, G., & Deisboeck, T. S. (2021). Thermodynamics and SARS-CoV-2: neurological effects in post-Covid 19 syndrome. Atti della Accademia Peloritana dei Pericolanti, 99(2), A3. [CrossRef]
- Lucia, U., Grisolia, G., & Deisboeck, T. S. (2020a). Seebeck-like effect in SARS-CoV-2 bio-thermodynamics. Atti della Accademia Peloritana dei Pericolanti-Classe di Scienze Fisiche, Matematiche e Naturali, 98(2), 6. [CrossRef]
- Lucia, U., Deisboeck, T. S., & Grisolia, G. (2020b). Entropy-based pandemics forecasting. Frontiers in Physics, 8, 274. [CrossRef]
- Maassen, S. J., Huskens, J., & Cornelissen, J. J. L. M. (2019). Elucidating the Thermodynamic Driving Forces of Polyanion-Templated Virus-like Particle Assembly. The journal of physical chemistry. B, 123(46), 9733–9741. [CrossRef]
- Makarov, V. V., Skurat, E. V., Semenyuk, P. I., Abashkin, D. A., Kalinina, N. O., Arutyunyan, A. M., Solovyev, A. G., & Dobrov, E. N. (2013). Structural lability of Barley stripe mosaic virus virions. PloS one, 8(4), e60942. [CrossRef]
- Maskow, T., Kiesel, B., Schubert, T., Yong, Z., Harms, H., & Yao, J. (2010). Calorimetric real time monitoring of lambda prophage induction. Journal of virological methods, 168(1-2), 126–132. [CrossRef]
- Molla, A., Paul, A. V., & Wimmer, E. (1991). Cell-free, de novo synthesis of poliovirus. Science (New York, N.Y.), 254(5038), 1647–1651. [CrossRef]
- Morais, F.M., Buchholz, F., Hartmann, T. et al. (2014). Chip-calorimetric monitoring of biofilm eradication with bacteriophages reveals an unexpected infection-related heat profile. J Therm Anal Calorim 115, 2203–2210. [CrossRef]
- Morowitz, H. J., Kostelnik, J. D., Yang, J., & Cody, G. D. (2000). The origin of intermediary metabolism. Proceedings of the National Academy of Sciences, 97(14), 7704-7708. [CrossRef]
- Morowitz, H. (1995). The emergence of complexity. Complexity, 1(1), 4-5. [CrossRef]
- Morowitz, H.J. (1992). Beginnings of Cellular Life: Metabolism Recapitulates Biogenesis. New Haven, CT: Yale University Press.
- Morowitz, H.J., Heinz, B. & Deamer, D.W. The chemical logic of a minimum protocell. Origins Life Evol Biosphere 18, 281–287 (1988). [CrossRef]
- Morowitz, H.J. (1976). The high cost of being human. The New York Times, February 11, p.45. Available at: https://www.nytimes.com/1976/02/11/archives/the-high-cost-of-being-human.html.
- Morowitz, H.J. (1968). Energy Flow in Biology: Biological Organization as a Problem in Thermal Physics. New York: Academic Press. [CrossRef]
- Morowitz, H.J. (1955). Some order-disorder considerations in living systems. Bulletin of Mathematical Biophysics, 17, 81–86. [CrossRef]
- Nadi, F., & Özilgen, M. (2021). Effects of COVID-19 on energy savings and emission reduction: a case study. International Journal of Global Warming, 25(1), 38-57. [CrossRef]
- National Center for Biotechnology Information (2022). NCBI Database [online]. Available at: https://www.ncbi.nlm.nih.gov/ (Accessed on January 7, 2023).
- Nebel, S., Bartoldus, I., & Stegmann, T. (1995). Calorimetric detection of influenza virus induced membrane fusion. Biochemistry, 34(17), 5705–5711. [CrossRef]
- Noble, C. G., Lim, S. P., Arora, R., Yokokawa, F., Nilar, S., Seh, C. C., Wright, S. K., Benson, T. E., Smith, P. W., & Shi, P. Y. (2016). A Conserved Pocket in the Dengue Virus Polymerase Identified through Fragment-based Screening. The Journal of biological chemistry, 291(16), 8541–8548. [CrossRef]
- Neuman, B.W. and Buchmeier, M.J. (2016). Supramolecular architecture of the coronavirus particle. Advances in Virus Research, 96, 1-27. [CrossRef]
- Neuman, B. W., Kiss, G., Kunding, A. H., Bhella, D., Baksh, M. F., Connelly, S., Droese, B., Klaus, J. P., Makino, S., Sawicki, S. G., Siddell, S. G., Stamou, D. G., Wilson, I. A., Kuhn, P., & Buchmeier, M. J. (2011). A structural analysis of M protein in coronavirus assembly and morphology. Journal of structural biology, 174(1), 11–22. [CrossRef]
- Neuman, B. W., Adair, B. D., Yoshioka, C., Quispe, J. D., Orca, G., Kuhn, P., Milligan, R. A., Yeager, M., & Buchmeier, M. J. (2006). Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. Journal of virology, 80(16), 7918–7928. [CrossRef]
- Ozilgen, M. and Sorgüven, E. (2017). Biothermodynamics: Principles and Applications. Boca Raton: CRC Press. [CrossRef]
- Özilgen, M., & Yilmaz, B. (2021). COVID-19 disease causes an energy supply deficit in a patient. International journal of energy research, 45(2), 1157–1160. [CrossRef]
- Patel, S.A. and Erickson, L.E. (1981). Estimation of heats of combustion of biomass from elemental analysis using available electron concepts. Biotechnology and Bioengineering, 23, 2051-2067. [CrossRef]
- Pinheiro, A. V., Baptista, P., & Lima, J. C. (2008). Light activation of transcription: photocaging of nucleotides for control over RNA polymerization. Nucleic acids research, 36(14), e90. [CrossRef]
- Popovic, M. (2023). Simple but Powerful: Viroids can Hijack their Host Cells’ Metabolism due to Greater Gibbs Energy Dissipation . Preprints, 2023010085. [CrossRef]
- Popovic, M., & Popovic, M. (2022). Strain Wars: Competitive interactions between SARS-CoV-2 strains are explained by Gibbs energy of antigen-receptor binding. Microbial risk analysis, 21, 100202. [CrossRef]
- Popovic M. (2022a). Strain wars 2: Binding constants, enthalpies, entropies, Gibbs energies and rates of binding of SARS-CoV-2 variants. Virology, 570, 35–44. [CrossRef]
- Popovic M. (2022b). Strain wars 3: Differences in infectivity and pathogenicity between Delta and Omicron strains of SARS-CoV-2 can be explained by thermodynamic and kinetic parameters of binding and growth. Microbial risk analysis, 22, 100217. [CrossRef]
- Popovic M. (2022c). Strain wars 4 - Darwinian evolution through Gibbs' glasses: Gibbs energies of binding and growth explain evolution of SARS-CoV-2 from Hu-1 to BA.2. Virology, 575, 36–42. [CrossRef]
- Popovic M. (2022d). Strain wars 5: Gibbs energies of binding of BA.1 through BA.4 variants of SARS-CoV-2. Microbial risk analysis, 22, 100231. [CrossRef]
- Popovic M. (2022e). Beyond COVID-19: Do biothermodynamic properties allow predicting the future evolution of SARS-CoV-2 variants?. Microbial risk analysis, 22, 100232. [CrossRef]
- Popovic, M. (2022f). Omicron BA.2.75 Sublineage (Centaurus) Follows the Expectations of the Evolution Theory: Less Negative Gibbs Energy of Biosynthesis Indicates Decreased Pathogenicity. Microbiology Research, 13(4), 937–952. MDPI AG. [CrossRef]
- Popovic, M. (2022g). Omicron BA.2.75 Subvariant of SARS-CoV-2 Is Expected to Have the Greatest Infectivity Compared with the Competing BA.2 and BA.5, Due to Most Negative Gibbs Energy of Binding. BioTech, 11(4), 45. MDPI AG. [CrossRef]
- Popovic, M. (2022h). Never Ending Story? Biothermodynamic Properties of Biosynthesis and Binding of Omicron BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2. Preprints, 2022120122. [CrossRef]
- Popovic M. (2022i). Biothermodynamics of Viruses from Absolute Zero (1950) to Virothermodynamics (2022). Vaccines, 10(12), 2112. [CrossRef]
- Popovic M. (2022j). Why doesn't Ebola virus cause pandemics like SARS-CoV-2?. Microbial risk analysis, 22, 100236. [CrossRef]
- Popovic M. (2022k). Atom counting method for determining elemental composition of viruses and its applications in biothermodynamics and environmental science. Computational biology and chemistry, 96, 107621. [CrossRef]
- Popovic, M. (2022L). Formulas for death and life: Chemical composition and biothermodynamic properties of Monkeypox (MPV, MPXV, HMPXV) and Vaccinia (VACV) viruses. Thermal Science, 26(6A). [CrossRef]
- Popovic, M. (2022m). Everything you Always Wanted to Know about the Biothermodynamic Background of Herpes Simplex Virus Type 1 – Host Interaction. Preprints, 2022120063. [CrossRef]
- Popovic, M. (2022n). Standard Gibbs Energy of Binding of the gp120 Antigen of HIV-1 to the CD4 Receptor. Preprints, 2022110482. [CrossRef]
- Popovic, M. (2022p). Biothermodynamic Key Opens the Door of Life Sciences: Bridging the Gap between Biology and Thermodynamics. Preprints, 2022100326. [CrossRef]
- Popovic, M. (2022q). Thermodynamics of Bacteria-Phage Interactions: T4 and Lambda Bacteriophages, and E. Coli Can Coexist in Natural Ecosystems due to the Ratio of their Gibbs Energies of Biosynthesis. Preprints, 2022110327. [CrossRef]
- Popovic, M., & Minceva, M. (2021a). Coinfection and Interference Phenomena Are the Results of Multiple Thermodynamic Competitive Interactions. Microorganisms, 9(10), 2060. [CrossRef]
- Popovic, M., & Minceva, M. (2020a). A thermodynamic insight into viral infections: do viruses in a lytic cycle hijack cell metabolism due to their low Gibbs energy?. Heliyon, 6(5), e03933. [CrossRef]
- Popovic, M., & Minceva, M. (2020b). Thermodynamic insight into viral infections 2: empirical formulas, molecular compositions and thermodynamic properties of SARS, MERS and SARS-CoV-2 (COVID-19) viruses. Heliyon, 6(9), e04943. [CrossRef]
- Popovic, M. (2018a). Living organisms from Prigogine’s perspective: an opportunity to introduce students to biological entropy balance. Journal of Biological Education, 52(3), 294-300. [CrossRef]
- Popovic, M. (2018b). Research in entropy wonterland: A review of the entropy concept. Thermal Science, 22(2), 1163-1178. [CrossRef]
- Prins, K. C., Binning, J. M., Shabman, R. S., Leung, D. W., Amarasinghe, G. K., & Basler, C. F. (2010). Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. Journal of virology, 84(20), 10581–10591. [CrossRef]
- Prigogine, I. and Wiame, J.M. (1946). Biologie et thermodynamique des phénomènes irréversibles. Experientia, 2, 451–453. [CrossRef]
- Prigogine, I. (1977). Nobel lecture: Time, Structure and Fluctuations. [Online] Available at: https://www.nobelprize.org/prizes/chemistry/1977/prigogine/lecture/ (Accessed on: January 5, 2023).
- Prigogine I. (1947). Etude thermodynamique des phénomènes irréversibles. Paris: Dunod. WorldCat ID: 421502786.
- Privalov, P. L. (2012). Microcalorimetry of macromolecules: the physical basis of biological structures. Hoboken, NJ: John Wiley & Sons. ISBN: 978-1-118-10451-4, . [CrossRef]
- Ridgway, H.; Chasapis, C.T.; Kelaidonis, K.; Ligielli, I.; Moore, G.J.; Gadanec, L.K.; Zulli, A.; Apostolopoulos, V.; Mavromoustakos, T.; Matsoukas, J.M. Understanding the Driving Forces That Trigger Mutations in SARS-CoV-2: Mutational Energetics and the Role of Arginine Blockers in COVID-19 Therapy. Viruses 2022, 14, 1029. [CrossRef]
- Sarge, S. M., Höhne, G. W., & Hemminger, W. (2014). Calorimetry: fundamentals, instrumentation and applications. Hoboken, NJ: John Wiley & Sons. ISBN: 978-3-527-32761-4.
- Sayers, E. W., Bolton, E. E., Brister, J. R., Canese, K., Chan, J., Comeau, D. C., Connor, R., Funk, K., Kelly, C., Kim, S., Madej, T., Marchler-Bauer, A., Lanczycki, C., Lathrop, S., Lu, Z., Thibaud-Nissen, F., Murphy, T., Phan, L., Skripchenko, Y., Tse, T., … Sherry, S. T. (2022). Database resources of the national center for biotechnology information. Nucleic acids research, 50(D1), D20–D26. [CrossRef]
- Schrödinger, E. (1944). What is life? The physical aspect of the living cell, Cambridge: Cambridge university press. ISBN: 0-521-42708-8.
- Scialo, F., Daniele, A., Amato, F., Pastore, L., Matera, M. G., Cazzola, M., Castaldo, G., & Bianco, A. (2020). ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung, 198(6), 867–877. [CrossRef]
- Shadrick, W. R., Mukherjee, S., Hanson, A. M., Sweeney, N. L., & Frick, D. N. (2013). Aurintricarboxylic acid modulates the affinity of hepatitis C virus NS3 helicase for both nucleic acid and ATP. Biochemistry, 52(36), 6151–6159. [CrossRef]
- Sigg, A. P., Mariotti, M., Grütter, A. E., Lafranca, T., Leitner, L., Bonkat, G., & Braissant, O. (2022). A Method to Determine the Efficacy of a Commercial Phage Preparation against Uropathogens in Urine and Artificial Urine Determined by Isothermal Microcalorimetry. Microorganisms, 10(5), 845. MDPI AG. [CrossRef]
- Şimşek, B., Özilgen, M., & Utku, F. Ş. (2021). How much energy is stored in SARS-CoV-2 and its structural elements?. Energy Storage, e298. [CrossRef]
- Sharma, R., Fatma, B., Saha, A., Bajpai, S., Sistla, S., Dash, P. K., Parida, M., Kumar, P., & Tomar, S. (2016). Inhibition of chikungunya virus by picolinate that targets viral capsid protein. Virology, 498, 265–276. [CrossRef]
- Shu, Y. and McCauley, J. (2017) GISAID: from vision to reality. EuroSurveillance, 22(13) . [CrossRef] [PubMed Central]
- Stauffer, H., Srinivasan, S., & Lauffer, M. A. (1970). Calorimetric studies on polymerization-depolymerization of tobacco mosaic virus protein. Biochemistry, 9(2), 193–200. [CrossRef]
- Sturtevant, J. M., Velicelebi, G., Jaenicke, R., & Lauffer, M. A. (1981). Scanning calorimetric investigation of the polymerization of the coat protein of tobacco mosaic virus. Biochemistry, 20(13), 3792–3800. [CrossRef]
- Tkhilaishvili, T. (2022). Bacteriophages as an alternative strategy in the treatment and prevention of implant-associated infections (Doctoral dissertation). [CrossRef]
- Tkhilaishvili, T., Wang, L., Tavanti, A., Trampuz, A., & Di Luca, M. (2020a). Antibacterial Efficacy of Two Commercially Available Bacteriophage Formulations, Staphylococcal Bacteriophage and PYO Bacteriophage, Against Methicillin-Resistant Staphylococcus aureus: Prevention and Eradication of Biofilm Formation and Control of a Systemic Infection of Galleria mellonella Larvae. Frontiers in microbiology, 11, 110. [CrossRef]
- Tkhilaishvili, T., Wang, L., Perka, C., Trampuz, A., & Gonzalez Moreno, M. (2020b). Using Bacteriophages as a Trojan Horse to the Killing of Dual-Species Biofilm Formed by Pseudomonas aeruginosa and Methicillin Resistant Staphylococcus aureus. Frontiers in microbiology, 11, 695. [CrossRef]
- Tkhilaishvili, T., Di Luca, M., Abbandonato, G., Maiolo, E. M., Klatt, A. B., Reuter, M., Möncke-Buchner, E., & Trampuz, A. (2018a). Real-time assessment of bacteriophage T3-derived antimicrobial activity against planktonic and biofilm-embedded Escherichia coli by isothermal microcalorimetry. Research in microbiology, 169(9), 515–521. [CrossRef]
- Tkhilaishvili, T., Lombardi, L., Klatt, A. B., Trampuz, A., & Di Luca, M. (2018b). Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. International journal of antimicrobial agents, 52(6), 842–853. [CrossRef]
- Tkhilaishvili, T., Di Luca, M., & Trampuz, A. (2018c). SIMULTANEOUS AND SEQUENTIAL APPLICATIONS OF PHAGES AND CIPROFLOXACIN IN KILLING MIXED-SPECIES BIOFILM OF PSEUDOMONAS AERUGINOSA AND STAPHYLOCOCCUS AUREUS. In Orthopaedic Proceedings (Vol. 100, No. SUPP_17, pp. 65-65). The British Editorial Society of Bone & Joint Surgery.
- Toinon, A., Greco, F., Moreno, N., Claire Nicolai, M., Guinet-Morlot, F., Manin, C., & Ronzon, F. (2015). Study of rabies virus by Differential Scanning Calorimetry. Biochemistry and biophysics reports, 4, 329–336. [CrossRef]
- Virudachalam, R., Harrington, M., & Markley, J. L. (1985a). Thermal stability of cowpea mosaic virus components: differential scanning calorimetry studies. Virology, 146(1), 138–140. [CrossRef]
- Virudachalam, R., Low, P. S., Argos, P., & Markley, J. L. (1985b). Turnip yellow mosaic virus and its capsid have thermal stabilities with opposite pH dependence: studies by differential scanning calorimetry and 31P nuclear magnetic resonance spectroscopy. Virology, 146(2), 213–220. [CrossRef]
- Von Bertalanffy, L. (1950). The theory of open systems in physics and biology. Science, 111(2872), 23-29. [CrossRef]
- Von Bertalanffy, L. (1971). General System Theory: Foundations, Development, Applications. New York, NY: George Braziller Inc. ISBN-13: 978-0807604533.
- Von Stockar, U. (2013a). Live cells as open non-equilibrium systems. In Urs von Stockar, ed., Biothermodynamics: The Role of Thermodynamics in Biochemical Engineering, Lausanne: EPFL Press, 475-534. [CrossRef]
- Von Stockar, U. (2013b). Biothermodynamics of live cells: energy dissipation and heat generation in cellular structures. In: Biothermodynamics: the role of thermodynamics in Biochemical Engineering, von Stockar, U., ed., Lausanne: EPFL Press, pp. 475-534. [CrossRef]
- von Stockar, U., & Liu, J. (1999). Does microbial life always feed on negative entropy? Thermodynamic analysis of microbial growth. Biochimica et biophysica acta, 1412(3), 191–211. [CrossRef]
- Vorobieva, N., Sanina, N., Vorontsov, V., Kostetsky, E., Mazeika, A., Tsybulsky, A., Kim, N., & Shnyrov, V. (2014). On the possibility of lipid-induced regulation of conformation and immunogenicity of influenza a virus H1/N1 hemagglutinin as antigen of TI-complexes. Journal of molecular microbiology and biotechnology, 24(3), 202–209. [CrossRef]
- Wang, Q., Li, Z., Ho, J., Guo, Y., Yeh, A. Y., Mohri, H., Liu, M., Wang, M., Yu, J., Shah, J. G., Chang, J. Y., Herbas, F., Yin, M. T., Sobieszczyk, M. E., Sheng, Z., Liu, L., & Ho, D. D. (2022). Resistance of SARS-CoV-2 omicron subvariant BA.4.6 to antibody neutralisation. The Lancet. Infectious diseases, 22(12), 1666–1668. [CrossRef]
- Wang, L., Tkhilaishvili, T., & Trampuz, A. (2020a). Adjunctive Use of Phage Sb-1 in Antibiotics Enhances Inhibitory Biofilm Growth Activity versus Rifampin-Resistant Staphylococcus aureus Strains. Antibiotics, 9(11), 749. MDPI AG. [CrossRef]
- Wang, L., Tkhilaishvili, T., Trampuz, A., & Gonzalez Moreno, M. (2020b). Evaluation of Staphylococcal Bacteriophage Sb-1 as an Adjunctive Agent to Antibiotics Against Rifampin-Resistant Staphylococcus aureus Biofilms. Frontiers in microbiology, 11, 602057. [CrossRef]
- Wang, L., Tkhilaishvili, T., Bernal Andres, B., Trampuz, A., & Gonzalez Moreno, M. (2020c). Bacteriophage-antibiotic combinations against ciprofloxacin/ceftriaxone-resistant Escherichia coli in vitro and in an experimental Galleria mellonella model. International journal of antimicrobial agents, 56(6), 106200. [CrossRef]
- Wang, G., Wang, H. J., Zhou, H., Nian, Q. G., Song, Z., Deng, Y. Q., Wang, X., Zhu, S. Y., Li, X. F., Qin, C. F., & Tang, R. (2015). Hydrated silica exterior produced by biomimetic silicification confers viral vaccine heat-resistance. ACS nano, 9(1), 799–808. [CrossRef]
- WHO (2022). WHO Coronavirus (COVID-19) Dashboard [Online] World Health Organization. Available at: https://covid19.who.int/ (Accessed on January 5, 2023).
- Wimmer E. (2006). The test-tube synthesis of a chemical called poliovirus. The simple synthesis of a virus has far-reaching societal implications. EMBO reports, 7 Spec No(Spec No), S3–S9. [CrossRef]
- Yang, Y., Song, Y., Lin, X., Li, S., Li, Z., Zhao, Q., Ma, G., Zhang, S., & Su, Z. (2020). Mechanism of bio-macromolecule denaturation on solid-liquid surface of ion-exchange chromatographic media - A case study for inactivated foot-and-mouth disease virus. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences, 1142, 122051. [CrossRef]
- Yang, Y., Zhao, Q., Li, Z., Sun, L., Ma, G., Zhang, S., & Su, Z. (2017). Stabilization study of inactivated foot and mouth disease virus vaccine by size-exclusion HPLC and differential scanning calorimetry. Vaccine, 35(18), 2413–2419. [CrossRef]
- Yilmaz, B., Ercan, S., Akduman, S., & Özilgen, M. (2020). Energetic and exergetic costs of COVID-19 infection on the body of a patient. International Journal of Exergy, 32(3), 314-327. [CrossRef]
- Yu, M., Zhang, S., Zhang, Y., Yang, Y., Ma, G., & Su, Z. (2015). Microcalorimetric study of adsorption and disassembling of virus-like particles on anion exchange chromatography media. Journal of chromatography. A, 1388, 195–206. [CrossRef]
- Zhou, J., Rong, X. L., Cao, X., Tang, Q., Liu, D., Jin, Y. H., Shi, X. X., Zhong, M., Zhao, Y., & Yang, Y. (2022). Assembly of Poly(ethylene glycol)ylated Oleanolic Acid on a Linear Polymer as a Pseudomucin for Influenza Virus Inhibition and Adsorption. Biomacromolecules, 23(8), 3213–3221. [CrossRef]

| Variant | C | H | O | N | P | S | Mr (g/mol) | Mr(nc) (MDa) |
|---|---|---|---|---|---|---|---|---|
| BA.5.2 nucleocapsid - Australia | 1 | 1.573784 | 0.342390 | 0.312321 | 0.005956 | 0.003361 | 23.7419 | 117.0958 |
| BA.5.2 nucleocapsid - Japan | 1 | 1.573557 | 0.342676 | 0.312372 | 0.006022 | 0.003359 | 23.7489 | 117.2088 |
| BA.5.2 nucleocapsid - USA | 1 | 1.573566 | 0.342668 | 0.312369 | 0.00602 | 0.003359 | 23.7487 | 117.2050 |
| BF.7 nucleocapsid - China | 1 | 1.573531 | 0.342715 | 0.312374 | 0.006031 | 0.003358 | 23.7498 | 117.2227 |
| BF.7 nucleocapsid - Greece | 1 | 1.573511 | 0.342740 | 0.312379 | 0.006036 | 0.003358 | 23.7504 | 117.2326 |
| BF.7 nucleocapsid - Serbia | 1 | 1.573528 | 0.342718 | 0.312375 | 0.006031 | 0.003358 | 23.7499 | 117.2241 |
| Variant | Reactants | → | Products | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Amino acid | O2 | HPO42- | HCO3- | Bio | SO42- | H2O | H2CO3 | ||
| BA.5.2 nucleocapsid - Australia | 1.3898 | 0.4908 | 0.0060 | 0.0438 | → | 1 | 0.0279 | 0.0536 | 0.4337 |
| BA.5.2 nucleocapsid - Japan | 1.3901 | 0.4912 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4338 |
| BA.5.2 nucleocapsid - USA | 1.3900 | 0.4912 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4338 |
| BF.7 nucleocapsid - China | 1.3901 | 0.4913 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4338 |
| BF.7 nucleocapsid - Greece | 1.3901 | 0.4913 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4338 |
| BF.7 nucleocapsid - Serbia | 1.3901 | 0.4913 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4338 |
| Variant | ΔfH⁰ (kJ/C-mol) | S⁰m (J/C-mol K) | ΔfG⁰ (kJ/C-mol) |
|---|---|---|---|
| BA.5.2 nucleocapsid - Australia | -75.32 | 32.49 | -33.21 |
| BA.5.2 nucleocapsid - Japan | -75.39 | 32.49 | -33.27 |
| BA.5.2 nucleocapsid - USA | -75.39 | 32.49 | -33.27 |
| BF.7 nucleocapsid - China | -75.40 | 32.49 | -33.28 |
| BF.7 nucleocapsid - Greece | -75.41 | 32.49 | -33.29 |
| BF.7 nucleocapsid - Serbia | -75.40 | 32.49 | -33.28 |
| Variant | ΔbsH⁰ (kJ/C-mol) | ΔbsS⁰ (J/C-mol K) | ΔbsG⁰ (kJ/C-mol) |
|---|---|---|---|
| BA.5.2 nucleocapsid - Australia | -232.14 | -37.29 | -221.06 |
| BA.5.2 nucleocapsid - Japan | -232.32 | -37.34 | -221.23 |
| BA.5.2 nucleocapsid - USA | -232.31 | -37.34 | -221.22 |
| BF.7 nucleocapsid - China | -232.33 | -37.34 | -221.24 |
| BF.7 nucleocapsid - Greece | -232.35 | -37.35 | -221.25 |
| BF.7 nucleocapsid - Serbia | -232.33 | -37.34 | -221.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





